Abstract
The Cancer Genomics Hub (CGHub) is the online repository of the sequencing programs of the National Cancer Institute (NCI), including The Cancer Genomics Atlas (TCGA), the Cancer Cell Line Encyclopedia (CCLE) and the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) projects, with data from 25 different types of cancer. The CGHub currently contains >1.4 PB of data, has grown at an average rate of 50 TB a month and serves >100 TB per week. The architecture of CGHub is designed to support bulk searching and downloading through a Web-accessible application programming interface, enforce patient genome confidentiality in data storage and transmission and optimize for efficiency in access and transfer. In this article, we describe the design of these three components, present performance results for our transfer protocol, GeneTorrent, and finally report on the growth of the system in terms of data stored and transferred, including estimated limits on the current architecture. Our experienced-based estimates suggest that centralizing storage and computational resources is more efficient than wide distribution across many satellite labs.
Database URL: https://cghub.ucsc.edu
Introduction and background
When the first human genome draft sequence was built in the year 2000, its in silico representation was made publicly available as a File Transfer Protocol (FTP) download from the University of California, Santa Cruz (1). Its data size was ∼2.5 GB, and its initial download rate reached an aggregate spike of ∼60 megabits per second (Mbps). In 2012, the Cancer Genomics Hub (CGHub) opened officially for downloads (2), with sequence and alignment data from the genomes of >3300 cancer patients. This time, the data size was 170 TB and the aggregate download rate spiked to >4 gigabits per second (Gbps) in the first year. In the 2+ years since CGHub came online, >400 cancer researchers have initiated downloads of ∼20 PB of cancer sequence data derived from >10 000 cancer patients, with peaks of >14 Gbps aggregate download traffic.
Sequencing technology has made significant advances in the past 14 years, allowing an explosion in the size and number of genomics projects. The rate of decrease in sequencing cost has outstripped Moore’s law, which states that the cost of transistors in digital electronics will decrease by a factor of 2 every 2 years (3). However, these advances in sequencing come with significant costs in computational and data management resources. A key example is the National Center for Biotechnology Information (NCBI), the central repository of genomics data in the USA. The NCBI was created both as a public archive and to ease the strain on the research community from accessing and transferring genomics ‘Big Data’. It grows at ∼4 TB a day or ∼1.4 PB a year as reported in 2013 by David Lipman, director of NCBI.
The CGHub stores data for a more focused set of information made up of cancer genomes. These data specifically come from sequencing projects that are managed by the National Cancer Institute (NCI), including The Cancer Genome Atlas (TCGA), the Cancer Cell Line Encyclopedia (CCLE) and the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) project, among others. The TARGET project in particular focuses on pediatric cancer cases. The amount of cancer data these projects have generated and are still generating makes CGHub the largest repository of broadly available cancer genomics data in the world (by size measured in terabytes). This article will (i) focus on the development and results of the CGHub repository in terms of its technical infrastructure, (ii) review the design and results of the GeneTorrent (GT) protocol for securely transferring large amounts of genomic data out of CGHub and (iii) seek to extrapolate the lessons learned from CGHub to future repositories for storing and computing over large genomics data.
Methods
In CGHub, we have brought together large amounts of raw research data (cancer genomes) and information technology best practices for storing nontrivial-sized data and querying the associated metadata, combining them into a single repository that complies with the NCI-required Federal Information Security Management Act (FISMA). CGHub is classified as ‘FISMA Low with Enhancements’. The technical design of CGHub addresses three key challenges in building the kind of repository that stores patient-related data:
Data organization and search (e.g. metadata)
Security (e.g. confidentiality of patients’ sequence data)
Efficient large data transfer (moving the bytes from the repository to the researchers)
Data organization and search (metadata)
The size of genomics data and the number of variables associated with any particular sequence file (e.g. disease, sample type, sequencing type) make managing and querying it challenging. A genomics data repository must therefore provide accurate and detailed metadata in a well-supported format that is easy to search and retrieve. We chose to leverage the Sequence Read Archive Metadata XML schema (currently at version 1.5) provided by the NCBI, which was already in use within the cancer genomics community. The schema is stored as a set of XML Schema Definitions (XSDs) that are updated periodically by the staff of the NCBI and referenced by CGHub via the Web.
A related objective to storing correct and useful metadata was a Web-accessible query interface for search and bulk retrieval. We addressed this by storing the metadata in a relational database, indexing it with the Apache Solr Web query engine and presenting it in XML format via a consistent set of Web service interfaces (WSIs). These Web services are a custom-built REST-based interface for public metadata search and retrieval (the metadata are not protected, as they are not patient-identifiable information). These WSIs make up the primary application programming interface of CGHub. To query and retrieve the metadata, we provide a command line script (cgquery) and a Web-based GUI, which supports searching and filtering via the Solr Web query engine (the CGHub data browser). The integration of these tools with the CGHub system is referenced in Figure 1.
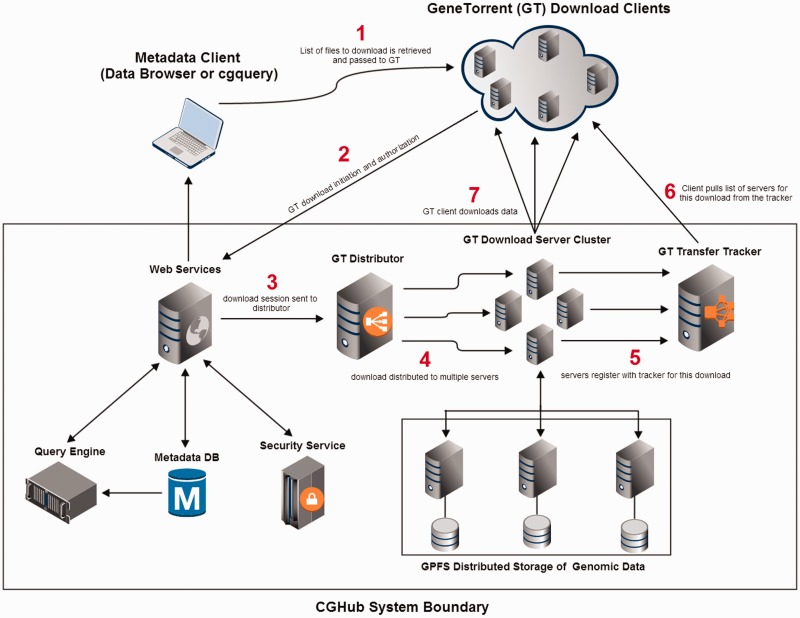
Figure 1.
CGHub and GeneTorrent conceptual system design and flow. (1) Client retrieves a list of downloadable files from the CGHub Web services. (2) Client uses GT to initiate a download. (3) Download is handed to GT distributor after proper security checks have passed. (4) Download is distributed to multiple transfer servers within a pool of available servers. (5) Servers announce themselves to the tracker as serving the requested file(s). (6) Client gets list of servers from the tracker. (7) Client downloads data directly from assigned transfer servers. All sequence data are read from the distributed GPFS.
Both of these client query interfaces produce XML-formatted ‘manifest’ files, which can initiate a multi-genome download via GT, our data transfer tool. The manifest file provides basic information on each entry, such as file name, size and MD5 checksum. Given this manifest file, GT will download all selected genomic sequence data files. Because all steps in this process can be scripted, a user can construct automated pipelines to query and download genomics data from CGHub.
Security: identity management
Confidentiality of patient-derived genomics data is a core requirement of CGHub. Only authorized researchers are allowed access to ∼98% of the CGHub genomics data. Thus, before allowing downloads to be initiated, the system must ensure proper authentication and authorization. For simplicity, we decided to use a preexisting solution: the NIH InCommon identity provider via the Internet2 Shibboleth middleware package. This allows for a single-sign-on (SSO) architecture to be leveraged from the existing technology and infrastructure. This process is illustrated in Figure 2. As a part of this, CGHub stores a list of login names authorized for various NCI projects. The actual authorization for these logins is handled by the NCI-appointed Data Access Committee (DAC), which is distinct from CGHub.
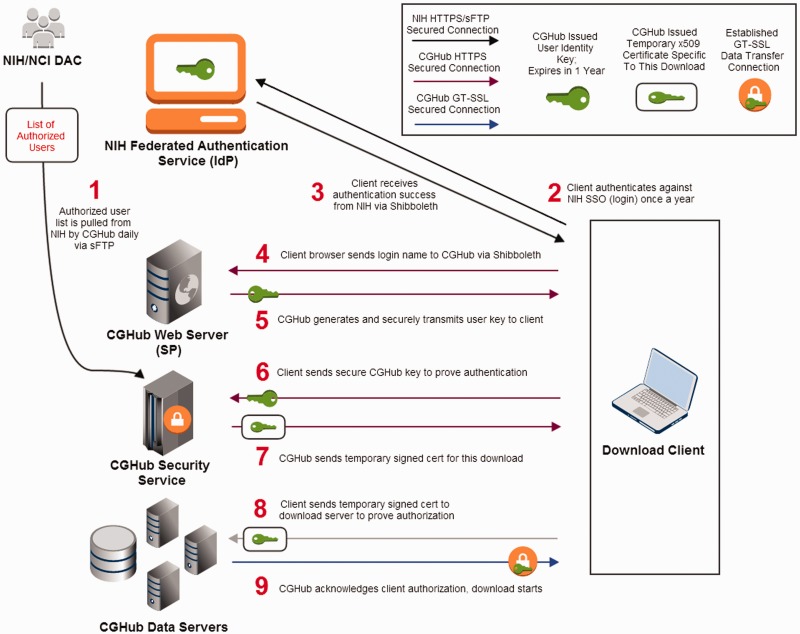
Figure 2.
CGHub client security transaction: (1) CGHub retrieves the authorized list of users from the NCI DAC multiple times a day. (2) The client accesses CGHub’s Web page to login and is redirected to NIH’s InCommon federated SSO authentication page over HTTPS where the client logs in using previously issued NIH credentials. (3) A successful authentication is sent back to the client via Shibboleth. (4) The client then sends the authentication via Shibboleth to CGHub’s Web server. (5) CGHub’s Web server then generates a unique cryptographically encoded key for the client, which has a 1-year expiration from the date of issuance, and sends it securely to the client over HTTPS. (6) The client then sends his\her file request with its encrypted identity key from Step 5 over HTTPS to CGHub’s security service. (7) If the client’s key is recognized and authorized to download the requested file, CGHub’s security service will send a signed temporary x509 certificate specific to the requested file back to the client over HTTPS. (8) The client then sends the x509 certificate from Step 7 to the CGHub data server to prove the client’s identity and authorization to download the requested file. (9) The CGHub data server verifies the certificate and establishes an SSL connection using AES-256 for encryption for the file transfer to the client.
Security: confidentiality and integrity
Cancer data confidentiality and integrity must be secured both in storage and transfer. Other genomics databases (EBI and NCBI) have chosen, at least in the past, to encrypt these data independent of the network transfer. These encrypted genomics files—mainly encrypted binary alignment/map (BAM) files (4)—are sent over unencrypted network transfer protocols, and then decrypted offline by the end user. If this decryption process fails after the file has been downloaded, the transfer must be repeated, at least partially, incurring significant cost in time to the researcher. Although this approach keeps the data protected from unauthorized parties, it burdens the researcher with the task of managing post-transfer decryption and validation. Although GT does not necessarily reduce overall computational costs, because decryption and validation still occur on the client end, it reduces administrative overhead by running these operations automatically as part of the transfer.
File integrity checking is accomplished through the use of SHA-1 (160-bit) (5) hashing of each ‘piece’ of a genome sequence file. These are precomputed on upload to CGHub and are checked by the GT client during download. If any piece fails the hash check, it is re-transferred automatically. Encryption is implemented through a custom extension to the underlying BitTorrent (BT) library using the well-known OpenSSL libraries for securing Transmission Control Protocol (TCP) connections. GT uses the asymmetric RSA algorithm for establishing a connection and confirming the identity of the CGHub servers/clients. The AES-256 (6) symmetric encryption algorithm is then used for securing the data transfer. We chose the combination of RSA with SHA-1 (5) hashing and AES (6) using 2048-bit and 256-bit key strengths, respectively, to achieve a reasonable bit strength within a recognized set of security protocols (7).
Transfer
Transferring large amounts of data over wide area networks (WANs) remains a challenge (8). Additionally, security requirements exacerbate the problem owing to the computational demand of encryption/decryption and file integrity hashing. Commonly used applications such as Rsync-over-SSH and secureFTP (sFTP) suffer from the computational demands of encryption/decryption (9) and from TCP protocol inefficiencies (10) when used for high volume, and secure transfers over high-bandwidth networks with long round trip times (‘long-fat networks’ or LFNs).
To address these limitations, we selected BT as a foundational transfer protocol. BT is a common protocol for data transfer over WANs. A major goal of BT was efficient and robust transfer of large files to multiple nodes concurrently (11). BT has held a top position in its share of the world’s bandwidth usage in the recent past with 27% of total network usage in Asia and a nontrivial share in Europe (12) and has been the subject of various research papers (13, 14).
The CGHub repository makes use of the GT extension of BT, developed by Annai Systems and based on the open-source, TCP-based libtorrent-rasterbar version of BT developed by Rasterbar Software. GT includes support for SSL encrypted data channels, preauthentication and authorization keys and genomic metadata integration. The latter two components are integrated with Annai’s proprietary Genomic Network Operating System (GNOS), which also provides the aforementioned WSI in CGHub. The CGHub was the first large-scale repository to use Annai’s GT and provided input on its ongoing design and development.
There are two additional factors that are critical in using GT/BT to support high-performance transfers: BT’s inherent parallelization and its provision for relatively painless horizontal scaling (adding more concurrent downloads) to support a growing number of simultaneous users. Parallelization allows more efficient utilization of the previously mentioned LFNs, over which our clients connect to CGHub. These networks are provided by third-party organizations such as Internet2 and are provisioned with links often ≥1 Gbps but with round-trip times >50 ms, depending on the client’s location.
The CGHub supports parallelization through both hardware and software. The hardware supports highly parallelized data storage and transfer with many internal 10 Gbps Ethernet interconnects for both storage and transfer nodes and a 10+ Gbps outbound connection to research and public backbone networks. The GT software supports parallelization intrinsically at multiple layers of the transfer. Following on both of these features of CGHub, horizontal scaling can be achieved with minor additional administration cost.
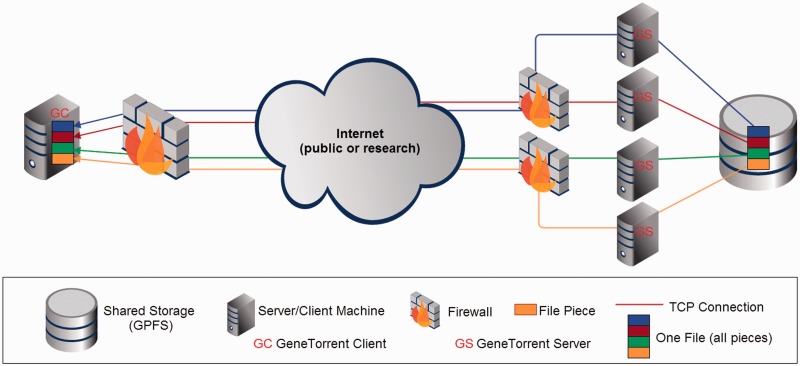
GT’s parallel features are built on the piece-wise nature of the underlying BT protocol, which allows for large files to be handled as collections of independent segments that can be transferred and verified via hashing individually. In addition, BT leverages concurrent TCP connections to maximize bandwidth usage for any given file transfer, and it provides the ability to efficiently scale to support increasing numbers of clients in aggregate with relatively minimal system changes. The high aggregate transfer rate is further supported at the storage system level by CGHub’s use of IBM’s General Parallel File System (GPFS). Figure 3 shows the distributed nature of the system with the file storage subsystem (GPFS) using commodity near-line SAS 7200 RPM hard drives attached to storage servers (not shown), which are interconnected with all the file servers by 10 Gbps Ethernet links.
Figure 3.
CGHub’s multi-layered parallelized transfer architecture using GT.
GT provides multiple dynamically allocated download servers and multiple concurrent client transfer processes—each of which initiates a concurrent TCP connection to all download servers—per download. Additionally, the GT download servers are fronted by multiple 10 Gbps connected firewalls, which are also part of the parallelization, a requirement in the presence of the packet processing load that 10 Gbps traffic places on a network filter device such as a firewall. A single encrypted download is spread across these layers of parallelization (Figure 3) to achieve a more efficient use of network bandwidth and CPU processing power than would be likely with a single encrypted TCP stream.
Based on this, a certain amount of horizontal scaling can be achieved with minor changes, simply by using commodity hardware for additional download servers and requiring only the GT server software to be installed. The dynamic GT server distribution component (part of GNOS), shown in Figure 1, is easily updated to handle the additional servers in the pool of download machines. A potential limiting factor to scaling horizontally is the storage back end. However, adding additional storage servers with attached disk arrays extending both the available storage and theoretically the parallel performance of the system is feasible and has been done in practice without extensive re-tooling in the CGHub system.
Results
Transfer performance comparison
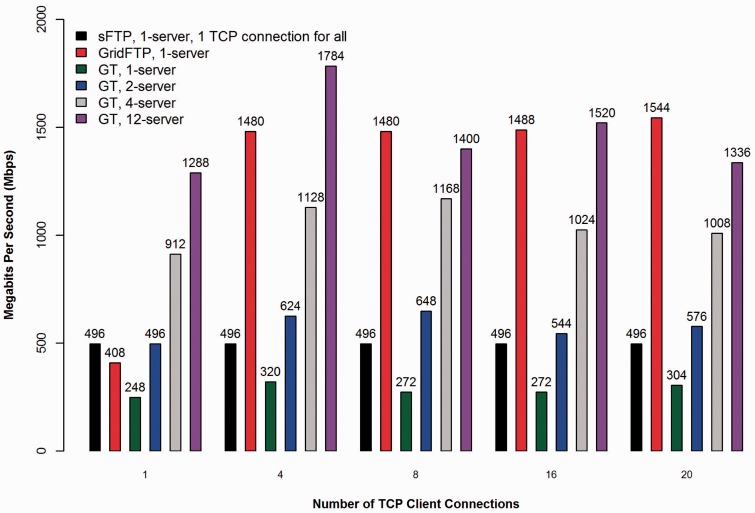
To test network transfer performance, we ran GT with varying numbers of TCP connections per server and the number of servers using encrypted data channels via OpenSSL as shown in Figure 4. We compared GT against the nearly ubiquitous SSH secure File Transfer Protocol (sFTP) program and the open-source GridFTP transfer program (8) used as part of the Globus system that supports multiple parallel encrypted TCP connections. For our tests, all transfers were run reading an 8.88 GB cancer BAM file from GPFS with server-side caching enabled. The sFTP program supports only a single encrypted TCP connection to a single server but is present in all the TCP connection groups in the graph for consistency and comparison. GridFTP was run as a single client connecting to a single server for all tests, as GridFTP’s multi-server striping mode requires the same number of servers and clients participating in the transfer. Technical details such as hardware specifications and software versions used, as well as links to guides covering GT installation and firewall configuration, are available in the Supplementary Data. In addition, we have included in the Supplementary Data a second performance comparison graph to demonstrate the effect encryption has on GT transfer performance.
Figure 4.
Comparison of transfer performance between sFTP, GridFTP and GT with varying numbers of TCP connections and GT server instances.
GT showed differences in performance with varying number of TCP connections and servers simultaneously serving the same download to the same client, where 12 is the current maximum number of download servers possible in the CGHub production system. From the graph we see that GT is fastest when running a medium number, four, of concurrent TCP connections to the maximum number of server instances (12 servers). This is most likely owing to the CPU-bounded nature of encryption, as the transfer rates are significantly lower than the line speed of 10 Gbps and the client disk IO has little or no consistent deleterious performance impact. Spreading the transfer across additional servers attains faster rates in all tests, as more CPU cores are harnessed to perform encryption on the server side for a single download. Conversely, maximizing the number of concurrent TCP connections without raising the number of servers shows diminishing returns with more than four connections and in some cases it significantly decreases the performance of the transfer (8 and 20 connections).
The above result is most likely due to the increasing overhead of additional concurrent TCP connections to a fixed number of GT servers. GridFTP in constrast continues to achieve better performance as the number of TCP connections is raised to a single server, demonstrating one potential difference in how the two protocols operate. However, even GridFTP should eventually see diminishing returns as more concurrent TCP connections are added, according to its authors (8). Further research would be required to say for certain what the actual root cause for the difference is, but it is likely that BT does not attempt to optimize any specific TCP connection, whereas GridFTP is most likely using each TCP connection more efficiently. The added benefit of GT is then primarly in its inherent ability to scale to an arbitrary number of servers (within some limit), which GridFTP cannot currently do for a single client. In any case, there is clearly a ‘sweet spot’ for the configuration of the number of transfer servers and the number of concurrent TCP connections controlled from the GT client. These two variables represent the primary tuning parameters of GT.
In practice, the performance impact of SSL/TLS encryption for Web sites has been largely alleviated by advances in CPU speed and architecture (15). However, there is a significant mismatch between the many-users-small-data transfers of a typical Web application such as Gmail and the few-users-huge-data transfers from a data repository like CGHub. A single load of a Gmail page transfers on the order of 1 MB of data, and even in the presence of attachments, is limited to 25 MB per attachment. An average user of CGHub will attempt to transfer 14 GB in a single download. Our results therefore indicate that this negation of the computational effect of encryption in the context of much larger data than Web applications still requires careful configuration and tuning on both the server and the client side, as the scale of data being transferred per user is approximately three orders of magnitude greater.
In the near future, we are not likely to see significant improvement in general-purpose CPU performance for cryptographic functions on a single core because chip design is limited by power consumption and heat buildup to current clock frequencies, and chip manufacturers have multiplied the number of cores rather than raising individual core frequencies (16, 17). Special instruction sets, such as Intel’s support of the symmetric cipher block algorithm AES (AES-NI), require software support (e.g. newer versions of OpenSSL), but are one option for alleviating the encryption burden. In general, GT, regardless of the CPU architecture, and building off of BT’s parallelism, load shares the computational work of a single encrypted download across multiple systems, lowering the bottleneck effect of encryption on transfer speeds. In light of the above results, this appears to be an effective answer to the limitations of current architectures for securing large transfers efficiently.
CGHub usage statistics
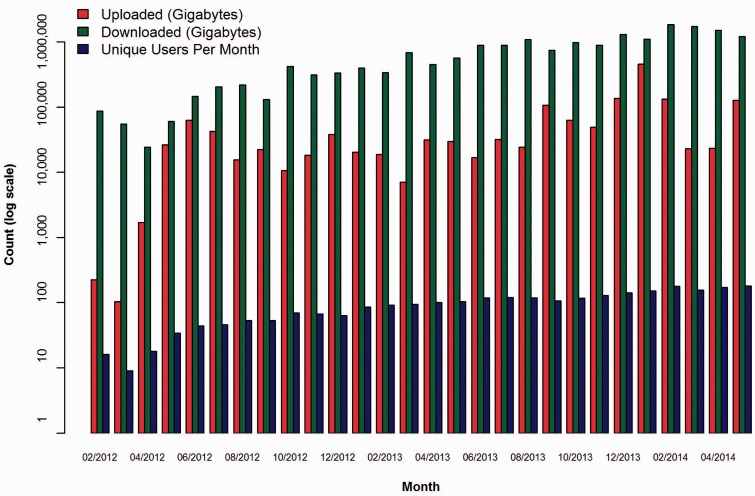
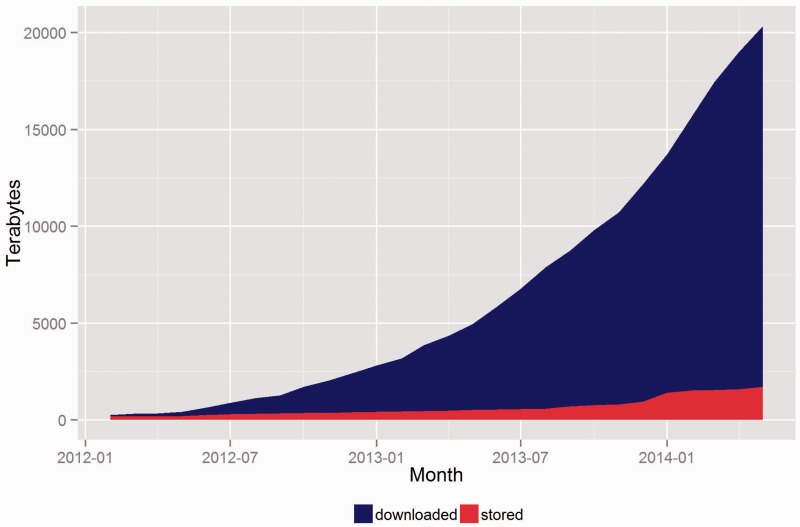
The following graphs demonstrate the growth of CGHub’s data footprint in terms of size and usage. All download numbers are estimates based on records of initiated downloads, including tests but excluding retries. This evidence demonstrates that CGHub is not only operational but also provides an ongoing source of cancer genomics data to many research groups. Figure 5 shows that every metric has increased by nearly a factor of 10 since CGHub’s inception. This demonstrates a substantial increase in CGHub submissions and downloads. Figure 6 shows the total size and download usage cumulatively by month. Downloads totaling in aggregate of ~20 PB of data have been initiated while the repository has grown to >1.4 PB in size with hundreds of terabytes projected to still be uploaded.
Figure 5.
CGHub growth measured by size (GB) of uploads, downloads and number of users per month. The y-axis is in log scale.
Figure 6.
CGHub cumulative growth: downloaded vs. stored data.
Discussion
We have presented an overview of the CGHub data repository for storing and transferring petabytes of cancer genomics data. We have emphasized our approach to security and analyzed the file transfer performance of GT for accessing multi-gigabyte files over WANs via authenticated and encrypted channels. The highly redundant nature of the system at both the storage and the server level combined with the scalability of parallel transfer algorithms such as GT provides a working base to build on in the quest to support the ever-growing amount of cancer-related genomics data that need to be stored, accessed and ultimately reasoned over in a secure manner. What follows is a discussion of the limitations of the current approach primarily for smaller less-built-out groups accessing the data we store and the possible solution of an integrated data storage repository with a computational analysis grid in the same geographical location.
The current paradigm is to store genomics data centrally and distribute them in large volumes to satellite centers and labs for analysis. There are concerns about the sustainability of this paradigm, and these concerns are important in light of the large volumes of genomics data currently being generated and expected in the future. Here we provide additional insight into this matter by estimating our capacity for handling future growth based on our 2+ years of experience running CGHub, discussing the ongoing feasibility of researchers transferring these data to their local systems, and suggesting a possible solution.
Future usage projections
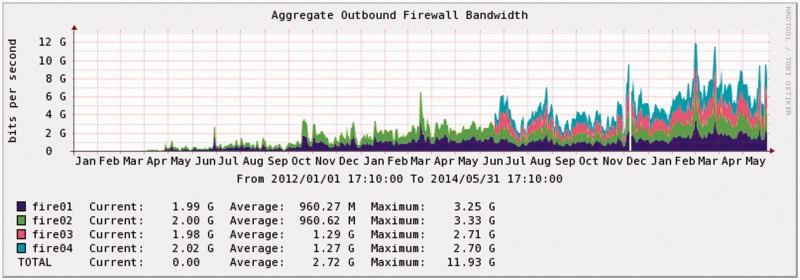
We estimate an upper limit of CGHub’s aggregate outbound bandwidth to be between 20 and 30 Gbps given the current hardware—CPU processing capacity and storage internetworking—and GT transfer capabilities, assuming upgraded network link(s) and additional firewalls, which are currently limited to an aggregate of 15–20 Gbps. We believe this upper limit could be realized with some software optimization but without an architecture redesign. Averaged bandwidth usage—overall peaks of ∼15 Gbps and a 5 Gbps average in the past year—suggests that we will not hit the 30 Gbps ceiling in the next 6–12 months (Figure 7 shows averages over 2+ years).
Figure 7.
CGHub’s outbound firewall usage 1/2012–5/2014 (averaged 24-h intervals, some precision is lost).
In our performance comparison results, we showed that a baseline secure transfer using sFTP would run no faster than ∼496 Mbps. By current estimates, downloading the entire CGHub repository at that speed would take ∼8 months (1.4 PB in 261 days) given that ∼98% of the data would require encrypted transfers. Such a download would still take ∼73 days of constant downloading at 1784 Mbps even when using a highly optimized GT client and servers, as presented in the Results section. Many users would run multiple downloads in parallel but this would still be dependent on their hardware and available network bandwidth (496 Mbps is almost half of a 1 Gbps network link).
That said, we currently have users running parallel jobs with GT. One client on the east coast of the USA reported a download rate from CGHub’s location at the San Diego Super Computing Center (SDSC) of ∼2 TB an hour (∼4444 Mbps) spread across ∼50 concurrent instances of GT. In contrast, many other clients are limited to rates much <496 Mbps due, at least in part, to constrained network and IO capacity. It is also important to state here that CGHub’s use of the BT-using GT is currently in a client/server mode, not in the more common peer-to-peer mode, thus partially obviating the potential performance benefit of a many-to-many transfer scheme (there is still some benefit in being able to scale the server ‘peers’ controlled by CGHub). This limitation is in part because of the restrictive data-sharing policy surrounding the protected genomic data CGHub stores. If the data continue to grow at the rates we have observed—and possibly even at the present size of the repository—small-to-medium-sized research groups will need to pursue alternative methods of accessing and computing on these data.
An immediate solution to this problem is to couple the data in CGHub with colocated automated analysis pipelines backed by high-performance compute cluster(s) at the SDSC. This would allow a community involved in high-throughput cancer genomics analysis to develop around CGHub. There are already groups subscribing to colocalized compute farms positioned near CGHub at the SDSC. Longer-term solutions include the creation of a few centers, each hosting a research computational cloud made up of persistent high capacity storage, joined with an extensive compute and networking infrastructure. These centralized groups operating the clouds could then optimize the sharing of large volumes of data between themselves via an automatable tool such as GT using its swarm approach to file transfer for rapid synchronization of data sets. This consolidated approach would then bring genomics back in line with the prevailing wisdom in the big data world, which is to move computation to the data rather than data to the computation (18).
Supplementary Data
Supplementary data are available at Database Online.
Acknowledgements
The authors thank the research community using CGHub who have continuously encouraged our work and inspired us to do better. The authors also thank the University of California, specifically staff at the Santa Cruz and San Diego campuses for their continued support of this project. The management of Leidos Biomedical Research, Inc has been excellent partners in our efforts to assure that CGHub meets the needs of our community. Diagrams were created using the Gliffy Online service.
Funding
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Funding for open access charge: US NCI (HHSN2611200800001E).
Conflict of interest: The core software for CGHub uses the GNOS network repository software and is a commercial product provided by Annai Systems. Cardinal Peak is a contract engineering services company.
References
- 1.Kent W.J., Haussler D. (2001) Assembly of the working draft of the human genome with GigAssembler. Genome Res., 11, 1541–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilks C., Maltbie D., Diekhans M., et al. (2013) CGHub: Kick-starting the Worldwide Genome Web. Proc. Asia Pac. Adv. Netw., 35, 1–13 [Google Scholar]
- 3.Moore G. (1998). Cramming more components onto integrated circuits. Proc IEEE, 86, 82–85 [Google Scholar]
- 4.Li H., Handsaker B., Wysoker A., et al. (2009) The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics, 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Institute of Standards and Technology. (2012) Secure Hash Standard, FIPS180-4. NIST, Gaithersburg, MD [Google Scholar]
- 6.National Institute of Standards and Technology. (2001) Advanced Encryption Standard (AES), FIPS197. NIST, Gaithersburg, MD [Google Scholar]
- 7.Al Hasib A., Haque A.A.M.M. (2008) A comparative study of the performance and security issues of AES and RSA cryptography. In: Convergence and Hybrid Information Technology, 2008. ICCIT'08. Third International Conference on, Vol. 2 IEEE Computer Society, Washington, DC, USA, pp. 505–510 [Google Scholar]
- 8.Allcock W., Bresnahan J., Kettimuthu R., et al. (2005) The Globus Striped GridFTP Framework and Server. In: Proceedings of the 2005 ACM/IEEE conference on Supercomputing. IEEE Computer Society, Washington, DC, USA, p. 54 [Google Scholar]
- 9.Khalil-Hani M., Nambiar V.P., Marsono M.N. (2010) Hardware Acceleration of OpenSSL cryptographic functions for high-performance Internet Security. In: Intelligent Systems, Modelling and Simulation (ISMS), 2010 International Conference on. IEEE Computer Society, Washington, DC, USA, pp. 374–379 [Google Scholar]
- 10.Gu Y., Grossman R.L. (2007) UDT: UDP-based data transfer for high-speed wide area networks. Comput. Netw., 51, 1777–1799 [Google Scholar]
- 11.Cohen B. (2003) Incentives build robustness in BitTorrent. In: Workshop on Economics of Peer-to-Peer Systems, Vol. 6, pp. 68–72 [Google Scholar]
- 12.Sandvine Incorporated. (2012) Global Internet Phenomena Report: 1H 2012Sandvine Incorporated ULC, Ontario, Canada [Google Scholar]
- 13.Liogkas N., Nelson R., Kohler E., et al. (2006) Exploiting bittorrent for fun (but not profit). In Proceedings of the 5th International Workshop on Peer-to-Peer Systems (IPTPS'06). [Google Scholar]
- 14.Zhang C., Dhungel P., Wu D., et al. (2011) Unraveling the BitTorrent ecosystem. Parallel Distrib. Syst. IEEE Trans., 22, 1164–1177 [Google Scholar]
- 15.Langley A., Modadugu N., Chang W.T. (2010) Overclocking ssl. In: Velocity: Web Performance and Operations Conference. [Google Scholar]
- 16.Dongarra J., Gannon D., Fox G., et al. (2007) The impact of multicore on computational science software. CTWatch Q., 3, 1–10 [Google Scholar]
- 17.Sutter H. (2005) The free lunch is over: a fundamental turn toward concurrency in software. Dr. Dobb's J., 30, March 2005 [Google Scholar]
- 18.Schadt E.E., Linderman M.D., Sorenson J., et al. (2010) Computational solutions to large-scale data management and analysis. Nat. Rev. Genet., 11, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.