Abstract
Free full text
an Archive of life science journals
Determination of conifer age biomarker DAL1 interactome using Y2H-seq
Abstract
Age is a sophisticated physiological signal that ensures the sequence of different developmental stages in organisms. The regulation of ageing pathways appears to differ between gymnosperms and angiosperms. We previously identified DAL1 as a conserved conifer age biomarker that plays a crucial role in the transition from vegetative to reproductive life-history phases in pines. Therefore, elucidating the specific interaction events related to DAL1 is key to understanding how age drives conifer development. Large-scale yeast two-hybrid (Y2H) analysis followed by next-generation high-throughput sequencing (Y2H-seq) allowed us to identify 135 PtDAL1 interacting proteins in Pinus tabuliformis. Our study found that PtDAL1 interacting proteins showed an ageing-related module, with sophisticated interacting networks composed of transcription factors (TFs), transcriptional regulators (TRs), and kinases. These interacting proteins are produced in response to a variety of phytohormones and environmental signals, and are likely involved in wood formation, needle development, oleoresin terpenoids biosynthesis, and reproductive development. In this study, we propose a novel regulation model of conifer ageing pathways whereby PtDAL1 coordinates different environmental stimuli and interacts with corresponding proteins to regulate appropriate development.
INTRODUCTION
Ageing covers every developmental stage of plants and includes vegetative growth, maturity (beginning reproduction and continuingvegetative growth), senescence, and mortality[1]. Some species have short lifespans, while others may be extremely long-lived. Conifers are renowned for their longevity[2] and may even use a special strategy to respond to different environmental stimuli at different ages to ensure their survival and successful reproduction.
The miR156-miR172 module has long been recognized as the key player in ageing pathways in angiosperms[3−5]. The miR156 acts as an age biomarker in angiosperms and targets SPLs to repress AP1, LEAFY, SOC1, and miR172 in the shoot apical meristem[3]. The expression of miR156 is gradually downregulated with age and releases the expression of the target genes of miR172[4]. miR172 represses the expression of AP2-like genes to activate FT, AP1, and SOC1 further[5]. In addition, many transcription factors (TFs) also play an irreplaceable role in ageing regulation (e.g., the WRKY, NAC, and MYB gene families)[6−9]. DELLAs repress the expression of WRKY54, thereby significantly reducing the expression of its target gene senescence-associated genes to increase plant lifespan[8]. Likewise, the atnap knockdown mutant delays leaf senescence, while the overexpression of AtNAP causes precocious senescence[7]. The MYB-bHLH-TTG1 complex regulates the trichome development, which is a phenotype biomarker of age[9]. Although these miRNAs and their targets are also found in gymnosperms, they do not show the same age expression trend as they do angiosperms[10]. Therefore, whether the miR156-miR172 regulatory module plays a core role in the conifer ageing pathway remains to be confirmed.
In gymnosperms, AGL6-like genes have been shown to be involved in ageing pathways and reproductive development. For example, PaDAL1, LaAGL2-1, and CjMADS14 display an ascending expressive trend with age in Picea abies[11], Larix kaempferi[12] and Cryptomeria japonica[13]. In L. kaempferi, some traditional rejuvenating measures such as cutting and pruning[14] can significantly reduce the expression of LaAGL2-1[15]. In C. japonica, the expression of CjMADS14 can be induced by gibberellin treatment[13]. In Picea abies, PaDAL1 causes early flowering and abaxial trichomes initiation in transgenic Arabidopsis[11]. These results indicate that AGL6-like genes have a conserved regulatory function in gymnosperm ageing pathways. However, the regulation mechanisms of AGL6-like genes in these ageing pathways are largely unknown.
In our previous study, PtDAL1, an AGL6-like homologue in P. tabuliformis, also displayed a strict age-related expression pattern. Heterologous overexpression of PtDAL1 also caused extremely early flowering and abaxial trichomes initiating phenotype in transgenic Arabidopsis[16]. Moreover, we revealed that PtDAL1 interacted with PtMADS11 to transfer ageing signals to reproduction signals in mature tissues[16]. Thus, the formation of heterologous protein complexes with other proteins may be an important mechanism for DAL1 participation in regulating diversified age effects.
Plant ageing pathways are affected by phytohormones, signaling molecules, and TFs, and also respond to environmental cues[17]. As age increases, the biochemical substances in plants also experience tremendous changes. In Platycladus orientalis, soluble protein content reaches its highest level in 20-year-old trees[18] and protein variety declines with age[19]. Additionally, the carbonylation[17], glycosylation[17], and tyrosine phosphorylation[20] of proteins in plant cells increases with age. The accumulation of calcium, manganese, and even lignin and tannins in needles, also shows an age-dependent relationship[21]. The dynamic expression of TF coding genes has a more active contribution in transitions of biological processes at the senescence stage[22,23]. Although these previous studies are valuable for understanding key regulatory pathways, analyses were mostly limited to illustrating the effects of biological processes during the senescence phase at the chemical and protein levels.
Most biological processes are regulated by recruiting different proteins to form a protein complex. MADS-box family TFs also function by forming tetramers[23]. Screening interacting proteins is a technique that can be applied to understand the regulatory mechanism of PtDAL1 in ageing pathways. Immunoprecipitation, pull-down assays, and yeast two-hybrid (Y2H) assays are commonly used for the screening of interacting proteins[24]. Among them, the Y2H system is the most accessible method for large-scale screening[25,26]. Unfortunately, the traditional Y2H system has limitations including low plasmid extraction and high sequencing costs[25,27].
Here, we developed a Y2H-seq method that combined Y2H with next-generation sequencing techniques. With this optimized and efficient new technology, we screened the PtDAL1 interactome at the entire transcriptome scale. We show that PtDAL1 recruits relevant proteins to regulate growth or reproduction at different age phases. We propose a model whereby PtDAL1 responds to different environmental stimuli and interacts with the relevant proteins to regulate plant development through the ageing pathway. Our data provide the foundation for understanding ageing regulatory mechanisms in conifers.
RESULTS
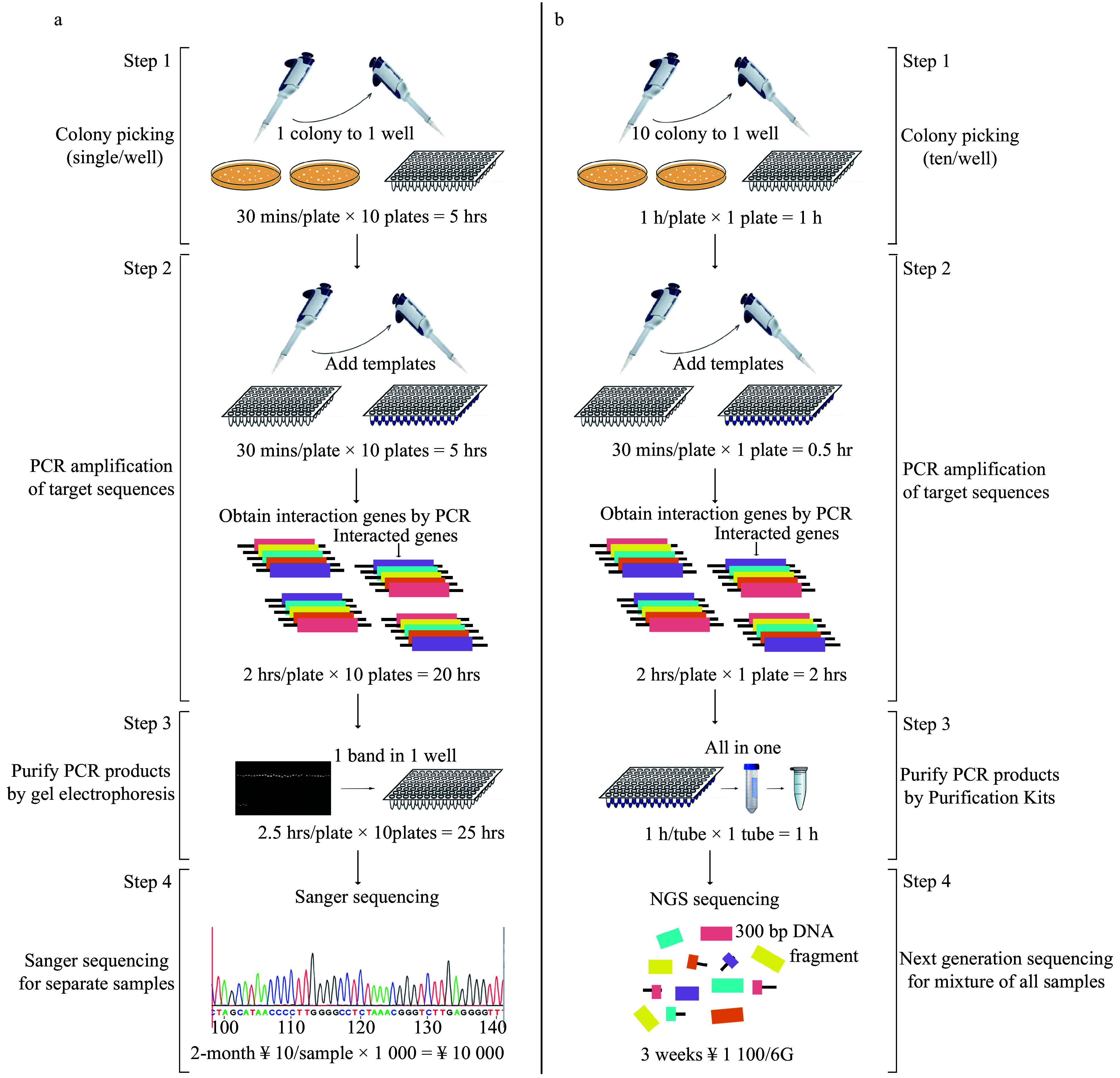
The efficiency of Y2H-seq compared to traditional Y2H
To obtain global PtDAL1 interacting proteins, the Y2H system was applied to screen a cDNA library from different age samples of P. tabuliformis. In a traditional Y2H assay (Fig. 1a), single clones would need to be individually selected to extract plasmids for PCR; this process would need to be repeated several times. Then, we would need to individually purify the PCR products and sequence the purified products using Sanger sequencing. These procedures are highly inefficient to complete a large-scale project and are very uneconomical due to the high redundancy of the results. We optimized this technology by combining the Y2H and next-generation sequencing (NGS) as an Y2H-seq assay (Fig. 1b) that requires carrying out PCR one time only. We picked 10 single clones and mixed them in 50 μl ddH2O, omitting the plasmid extraction. Next, all the PCR products were mixed in one tube for purification. This change decreased our experimental time and cost significantly. Finally, we sequenced the purified products using NGS sequencing[28]; this was chosen instead of Sanger sequencing, as Sanger cannot use a mixed sample. The optimized system reduced the whole experimental time by 66% and reduced cost by 89%. The optimized technology allowed us to identify the interacting proteins thoroughly.
Global interactome of age biomarker PtDAL1 in Pinus tabuliformis
Based on Y2H-seq, we screened the PtDAL1 interactome at the whole-transcriptome scale. We identified 135 PtDAL1 non-redundant interacting proteins from 1000 events (Table 1), including 21 TFs, two TRs, and two kinases, as well as enzymes, heat shock proteins, and RNA binding proteins.
Table 1
| Gene_ID\Symbol | At_homo | e-value | At_gene_symbol | TF/TR/PK | EST count |
| Pt0G09550 | AT1G74310 | 1.32E-39 | AtHSP101 | 33,639 | |
| Pt0G15520 | AT4G38510 | 0 | AtVAB2 | 34,522 | |
| Pt0G21450 | AT5G08640 | 2.8E-125 | AtFLS1 | 28,899 | |
| Pt0G22130 | AT4G37480 | 6.7E-137 | 22 | ||
| Pt1G04610\PtDAL9 | AT2G45660 | 3.41E-65 | AGAMOUS-LIKE 20 (AtSOC1) | TF | 66,859 |
| Pt1G04670\PtDAL19 | AT2G45660 | 8.53E-57 | AGAMOUS-LIKE 20 (AtSOC1) | TF | 573 |
| Pt1G04680 | − | 31 | |||
| Pt1G04770\PtMADS11 | AT2G45660 | 2.34E-53 | AGAMOUS-LIKE 20 (AtSOC1) | TF | 688 |
| Pt1G04910\PtMADS13 | AT2G45660 | 7.91E-61 | AGAMOUS-LIKE 20 (AtSOC1) | TF | 257 |
| Pt1G04940\PtMADS26 | AT2G45660 | 1.37E-61 | AGAMOUS-LIKE 20 (AtSOC1) | TF | 78 |
| Pt1G21750 | AT5G42270 | 0 | VARIEGATED 1 (VAR1) | 116,184 | |
| Pt1G21760 | AT5G42270 | 0 | VARIEGATED 1 (VAR1) | 40 | |
| Pt1G26900 | AT1G01090 | 0 | PDH-E1 ALPHA | 330 | |
| Pt1G31120 | AT2G05790 | 5.67E-74 | 57 | ||
| Pt1G38270 | AT2G05790 | 1.95E-81 | 13,837 | ||
| Pt1G38280 | AT2G05790 | 1.95E-81 | 7,453 | ||
| Pt1G42680 | AT5G63480 | 1.18E-37 | MEDIATOR 30 (MED30) | TR | 210,880 |
| Pt1G50870 | AT5G55160 | 6.87E-45 | AtSUMO2 | 26 | |
| Pt1G53330 | AT4G31200 | 1E-178 | 156,474 | ||
| Pt1G54050 | AT2G21660 | 8.22E-40 | GLYCINE RICH PROTEIN 7 (AtGRP7) | 11 | |
| Pt1G56320 | − | 52 | |||
| Pt1G57520 | AT3G05530 | 0 | RPT5A | 201,789 | |
| Pt1G58710 | AT5G55160 | 5.72E-33 | AtSUMO2 | 48,291 | |
| Pt1G70570 | − | 13 | |||
| Pt2G09370 | AT1G09070 | 4.32E-36 | AtSRC2 | 18,394 | |
| Pt2G09380 | − | 11,074 | |||
| Pt2G09430 | − | 62,709 | |||
| Pt2G12420 | AT2G46210 | 0 | AtSLD2 | 388 | |
| Pt2G22810\PtLEAFY | AT5G61850 | 2.7E-115 | LEAFY 3 (LFY3) | TF | 10 |
| Pt2G28740\PtbHLH143 | AT1G59640 | 1.27E-38 | BIG PETAL (BPE); ZCW32 | TF | 97 |
| Pt2G29110 | AT5G14040 | 0 | MPT3 | 52,616 | |
| Pt2G29160 | AT5G55180 | 6.73E-42 | 30 | ||
| Pt2G29170 | − | 36 | |||
| Pt2G29410 | − | 133 | |||
| Pt2G36780 | AT3G09970 | 3.2E-138 | RHIZOBIALE-LIKE PHOSPHATASE 2 (RLPH2) | 72 | |
| Pt2G41770\PtDAL10 | AT1G26310 | 4.84E-47 | CAULIFLOWER (CAL) | TF | 4 |
| Pt2G48290 | − | 10 | |||
| Pt2G58590 | AT1G63000 | 0 | UER1 | 56,441 | |
| Pt2G66560 | − | 16,151 | |||
| Pt2G71640 | AT1G75290 | 2.28E-97 | 42,597 | ||
| Pt2G72330 | − | 669 | |||
| Pt2G72340 | − | 33 | |||
| Pt2G73010 | − | 16,500 | |||
| Pt3G00960 | − | 16 | |||
| Pt3G26100 | AT2G29630 | 0 | THIAMINC (THIC) | 136,084 | |
| Pt3G52020 | AT1G47710 | 2.7E-133 | AtSERPIN1 | 853 | |
| Pt3G52520 | − | 30 | |||
| Pt3G54180\PtMADS49 | TF | 1 | |||
| Pt3G61960 | AT4G32460 | 9.17E-63 | AtHA1-1 | 119 | |
| Pt4G01770\PtTPS_di41 | AT4G16730 | 1.72E-72 | TERPENE SYNTHASE 02 (TPS02) | 25 | |
| Pt4G02100 | − | 91 | |||
| Pt4G06200 | − | 551,090 | |||
| Pt4G06880 | AT4G32470 | 4.21E-52 | 2,220,400 | ||
| Pt4G23680 | − | 575 | |||
| Pt4G35470 | AT5G21060 | 2.6E-50 | 554 | ||
| Pt4G38090 | − | 31 | |||
| Pt4G39400 | AT5G09760 | 1.6E-155 | 117 | ||
| Pt4G40470 | AT3G05530 | 1.46E-88 | RPT5A | 21,709 | |
| Pt4G40480 | AT3G05530 | 3.5E-127 | RPT5A | 21,180 | |
| Pt4G42010 | AT1G74310 | 9.99E-92 | AtHSP101 | 1,847 | |
| Pt4G47930 | − | 49 | |||
| Pt4G64210 | AT2G04160 | 4.49E-46 | AUXIN-INDUCED IN ROOT CULTURES 3 (AIR3) | 10 | |
| Pt5G00580 | AT1G21980 | 4E-146 | AtPIP5K1; AtPIPK1 | PK | 98,948 |
| Pt5G06310 | AT3G46740 | 0 | TOC75-III; MAR1 | 349 | |
| Pt5G19560 | AT1G75030 | 2.04E-95 | AtLP-3 | 17,623 | |
| Pt5G20480\PtGATA5 | TF | 1 | |||
| Pt5G40090 | − | 12 | |||
| Pt5G50950 | AT3G05530 | 5.4E-143 | RPT5A | 85,346 | |
| Pt5G54180 | AT5G43060 | 0 | ESPONSIVE TO DEHYDRATION 21B (RD21B) | 27 | |
| Pt5G57130 | AT5G43060 | 5.3E-131 | ESPONSIVE TO DEHYDRATION 21B (RD21B) | 41 | |
| Pt5G62470\PtDAL4 | AT2G45660 | 1.96E-67 | AGAMOUS-LIKE 20 (AtSOC1) | TF | 1,003,270 |
| Pt5G62640\PtDAL35 | AT2G45660 | 2.36E-67 | AGAMOUS-LIKE 20 (AtSOC1) | TF | 182,559 |
| Pt6G17610 | AT2G21660 | 7.65E-43 | AtGRP7 | 2,751 | |
| Pt6G34620 | AT2G21660 | 7.48E-41 | AtGRP7 | 518 | |
| Pt6G35050\PtDAL1 | AT2G45650 | 1.44E-81 | AGAMOUS-LIKE 6 (AGL6) | TF | 126 |
| Pt6G37350\PtMADS5 | AT2G22540 | 3.69E-59 | SHORT VEGETATIVE PHASE (SVP) | TF | 9 |
| Pt6G39740 | AT3G07320 | 8.7E-174 | 290,696 | ||
| Pt6G41700 | AT4G25200 | 3.8E-36 | ATHSP23.6-MITO | 157 | |
| Pt6G44930 | − | 23 | |||
| Pt6G51040 | AT4G26830 | 7E-145 | 202,392 | ||
| Pt6G51050 | AT2G05790 | 0 | 4,185,410 | ||
| Pt7G03490 | AT3G04880 | 2.3E-137 | DNA-DAMAGE-REPAIR/TOLERATION 2 (DRT102) | 224 | |
| Pt7G14660 | AT5G42800 | 9.3E-164 | DFR | 2,048 | |
| Pt7G15050 | AT5G06570 | 1.48E-37 | 21,889 | ||
| Pt7G15060 | AT1G68620 | 3.89E-50 | 73,081 | ||
| Pt7G20560 | AT3G23900 | 4.2E-120 | IRR | 4,284 | |
| Pt7G21710 | − | 6,227 | |||
| Pt7G24630 | − | 14 | |||
| Pt7G32610 | AT3G05500 | 3.24E-70 | SRP3; LDAP3 | 12,253 | |
| Pt7G48580 | AT5G55180 | 1.85E-85 | 53 | ||
| Pt7G50230 | AT3G45310 | 1.3E-174 | 32 | ||
| Pt7G50300 | AT3G45310 | 2E-174 | 4,816 | ||
| Pt7G58880 | AT5G43060 | 9.3E-122 | RD21B | 8,270 | |
| Pt8G00820 | AT1G18250 | 4.7E-107 | ATLP-1 | 7,596 | |
| Pt8G00840 | AT1G18250 | 4.5E-107 | ATLP-1 | 94,972 | |
| Pt8G04720 | AT5G55180 | 8.1E-82 | 135 | ||
| Pt8G04770 | AT5G55180 | 1.27E-88 | 5,885 | ||
| Pt8G15270\PtDPL1 | AT1G14920 | 6.5E-170 | RESTORATION ON GROWTH ON AMMONIA 2 (RGA2) | TF | 9 |
| Pt8G15340\PtDPL2 | AT2G01570 | 1.1E-128 | REPRESSOR OF GA1-3 1 (RGA1) | TF | 64 |
| Pt8G16740 | − | 27 | |||
| Pt8G19120 | AT5G46290 | 0 | 3-KASI | 101 | |
| Pt8G35940 | AT5G47000 | 4.2E-125 | 75,498 | ||
| Pt8G46580 | AT5G43060 | 0 | RD21B | 14,683 | |
| Pt8G46590 | AT5G43060 | 0 | RD21B | 186,050 | |
| Pt8G48720 | AT3G05530 | 1.5E-143 | RPT5A | 13,449 | |
| Pt9G05110 | − | 415 | |||
| Pt9G05120 | − | 28 | |||
| Pt9G10750 | − | 11,338 | |||
| Pt9G10800 | − | 8,111 | |||
| Pt9G21000 | AT5G38660 | 1.37E-61 | APE1 | 87,565 | |
| Pt9G28260 | − | 372 | |||
| Pt9G36230 | AT4G23160 | 1.7E-116 | CYSTEINE-RICH RLK 8 (CRK8) | PK | 17 |
| Pt9G39600 | AT4G12070 | 1.1E-151 | 95,337 | ||
| Pt9G40380 | AT1G55000 | 4.51E-87 | 26,718 | ||
| Pt9G46960 | AT2G17420 | 0 | NTR2; AtNTRA | 61,919 | |
| PtJG05540\PtNEEDLY | AT5G61850 | 1.22E-88 | LEAFY 3 (LFY3) | TF | 24 |
| PtJG13360 | AT1G54320 | 7.1E-173 | 12 | ||
| PtJG38830 | AT1G24510 | 0 | CCT5 | 30,744 | |
| PtJG40190 | AT3G04880 | 1.16E-53 | DRT102 | 271 | |
| PtQG03450/PtLWD1 | AT1G12910 | 0 | AtAN11 | 41,358 | |
| PtQG06810\PtSMU | AT2G26460 | 2.8E-113 | SUPPRESSORS OF MEC-8 AND UNC-52 2 (SMU2) | 1 | |
| PtQG22390 | AT4G27670 | 1.21E-64 | HEAT SHOCK PROTEIN 21 (HSP21) | 22,925 | |
| PtXG01260 | AT1G18640 | 2.4E-115 | PSP1 | 66,835 | |
| PtXG01380 | AT1G18640 | 1.6E-115 | PHOSPHOSERINE PHOSPHATASE 1 (PSP1) | 49 | |
| PtXG01810 | AT5G21060 | 1.92E-53 | 30 | ||
| PtXG11800 | − | 43 | |||
| PtXG23600\PtbZIP59 | AT4G38900 | 1.05E-89 | BASIC LEUCINE-ZIPPER 29 (BZIP29) | TF | 1 |
| PtXG26610\PtLUG5 | AT4G32551 | 0 | LEUNIG (LUG) | TR | 7 |
| PtXG33640\PtRGA | AT2G01570 | 2.2E-172 | REPRESSOR OF GA1-3 1 (RGA1) | TF | 5 |
| PtXG33750\PtDPL3 | AT2G01570 | 2.2E-172 | REPRESSOR OF GA1-3 1 (RGA1)) | TF | 133 |
| PtXG35990 | − | 56 | |||
| PtXG36010 | AT4G26830 | 6.01E-90 | 37 | ||
| PtXG37060 | AT3G44620 | 3.69E-86 | 106,593 | ||
| PtXG44840\PtDAL21 | AT1G24260 | 4.83E-48 | AGAMOUS-LIKE 9 (AGL9); SEPALLATA3 (SEP3) | TF | 13 |
| PtXG49760 | AT4G29040 | 0 | RPT2a | 92,990 |
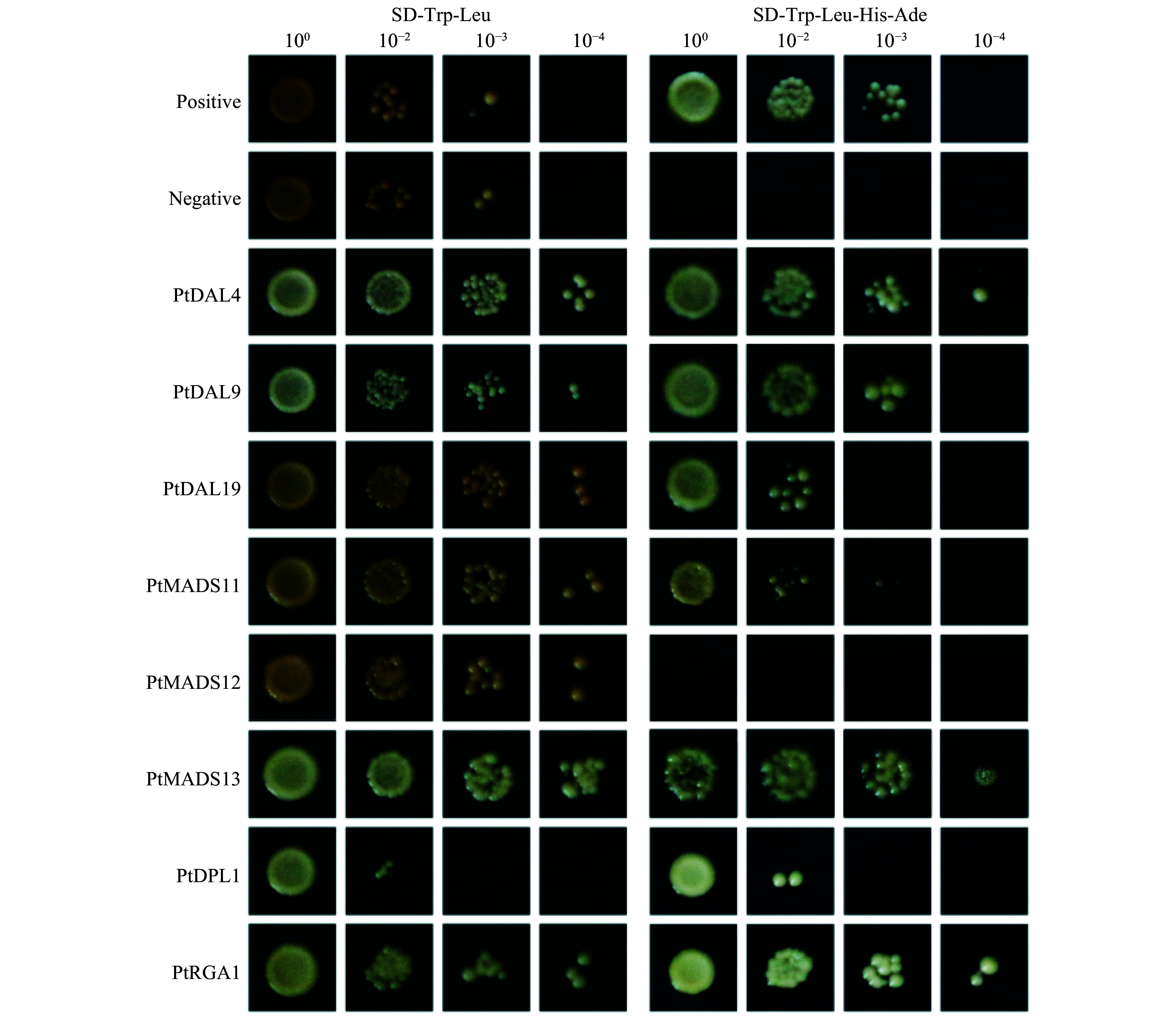
A total of 12 MADS-box proteins and four DELLA proteins were screened out as PtDAL1 interacting proteins (DIPs). Due to their important role in reproductive development and hormone signal transduction, we verified the interaction between them and DAL1 point-to-point through Y2H assay (Fig. 2). In Y2H, the majority of the point-to-point results were consistent with the screening results. Only the PtMADS12 produced a negative result, which suggests that Y2H-seq has comparable accuracy to the traditional Y2H method (Fig. 2). These results suggest that Y2H-seq is a valid technique, but it requires further verification and research.
Conifers and Arabidopsis AGL6 homologs have different interaction networks
PtDAL1 is a homolog of AtAGL6 in Arabidopsis. To understand the different functions of conifers and Arabidopsis AGL6 homologs, we obtained the AtAGL6 interactome from protein interaction databases to compare with the PtDAL1 interactome (Fig. 3a). Interestingly, we found that all of the AtAGL6 interacting proteins belong to the MADS-box family (Fig. 3a). Although the MADS-box proteins are the major components of DIPs, our data showed a more diverse DIP network, including DELLAs (PtRGA, PtDPL1, PtDPL2, and PtDPL3), a bHLH family member (PtbHLH143) and bZIP family member (PtbZIP59), glycosyl hydrolase (Pt6G51050 and Pt2G29160), and floral meristem identity factor (PtNEEDLY) (Fig. 3a). AtAGL6 and PtDAL1 interactomes share some common subfamily members, such as AGL42, SOC1, SVP, and AGL16 (Fig. 3b). This result indicates that the AGL6 homolog interactomes were partially conserved during the seed plant evolution, and these MADS-box proteins may be the core module in the AGL6-mediated regulatory network.
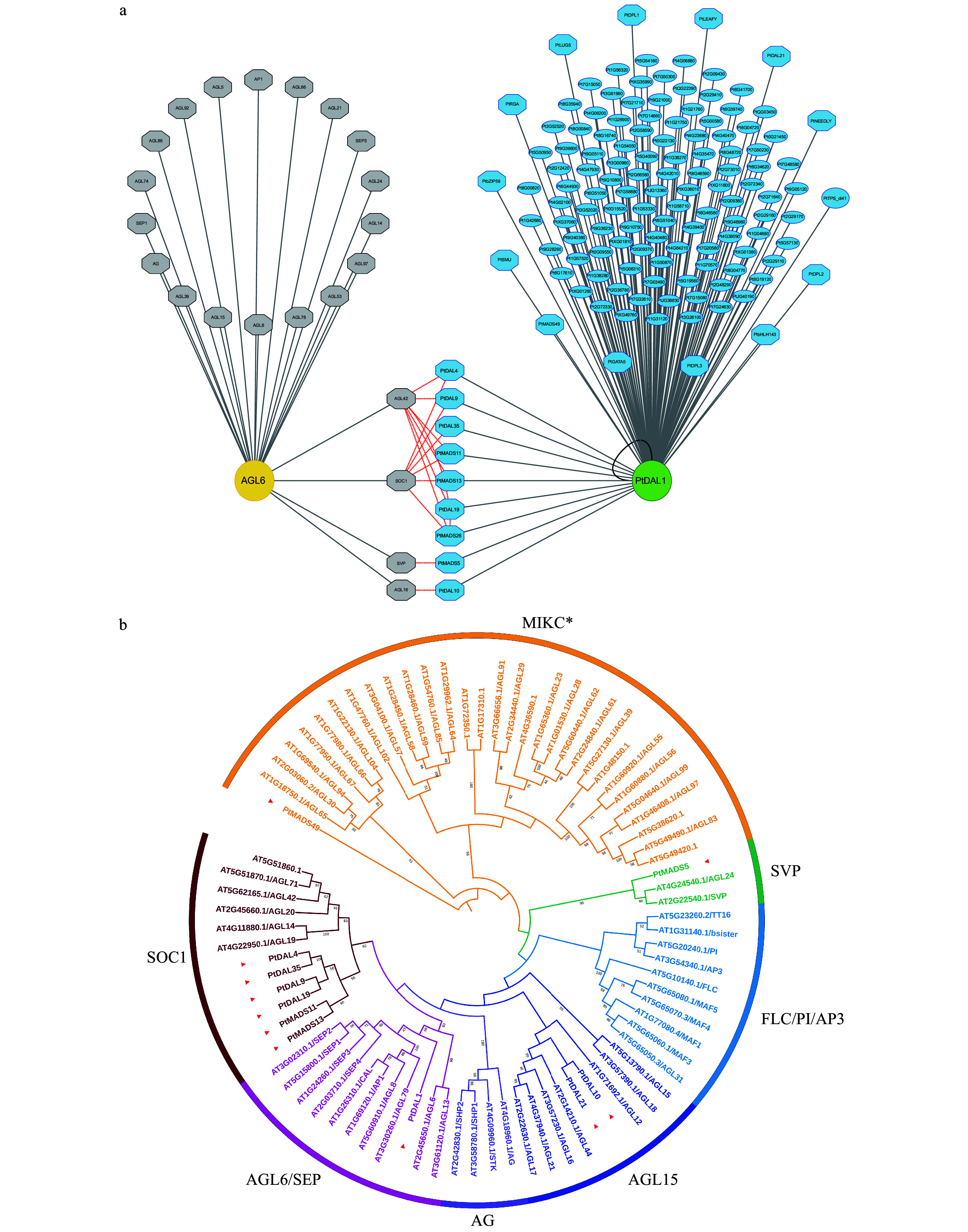
The interactome of PtDAL1 and its Arabidopsis homologue AtAGL6. (a) Gray, AtAGL6 interacted proteins in Arabidopsis. Blue, PtDAL1 interacted proteins in P. tabuliformis. Red lines indicate the homologs between Arabidopsis and P. tabuliformis. Black lines indicate homologous dimer. Octagon, transcription factors. (b) The phylogenetic analysis of DIPs in MADS-box gene family. Red inverted triangle, DIPs of MADS-box family in P. tabuliformis.
The expression profiles of PtDAL1 interacting protein coding genes
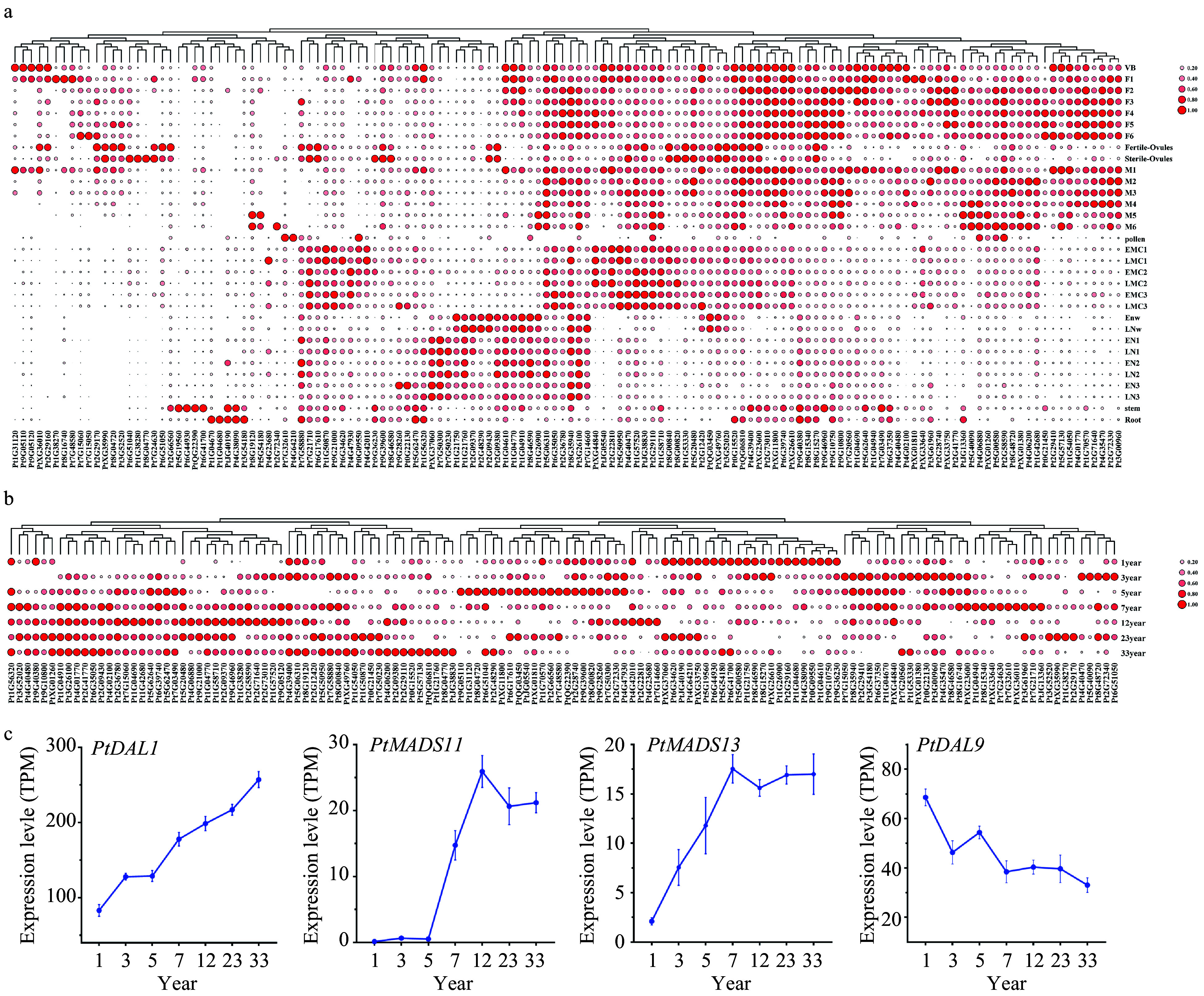
To reveal the potential biological functions of DIPs, we analyzed the spatial and temporal expression patterns of all DIP coding genes. The results show that most of the DIP genes were expressed in the tissues most similar to PtDAL1 in P. tabuliformis (Fig. 4a, Supplemental Fig. 1). This suggests that PtDAL1 serves as a global regulatory factor that participates in developing most tissues through interactions with different proteins at different ages. The expression of DIP genes has a significant ageing-related module (Fig. 4b, Supplemental Fig. 2). For example, there are 22 DIP genes that accumulate highly in 1-year-old trees. Similarly, there are 22 DIP genes that accumulate highly in 3-year-old trees, 21 DIP genes that accumulate highly in 5-year-old trees, 18 DIP genes that accumulate highly in 7-year-old trees, and 47 DIP genes that accumulate highly in 12, 23, and 33-year-old trees. These findings suggest that PtDAL1 establishes an age-related genetic program that recruits different proteins corresponding with the developmental stage. PtMADS11, PtMADS13, and PtDAL9 all have significant ageing effects similar to PtDAL1 (Fig. 4c), implying that MADS-box proteins probably play an important role in balancing ageing effects at different stages.
Promoter cis element analysis of PtDAL1 interacting protein coding genes.
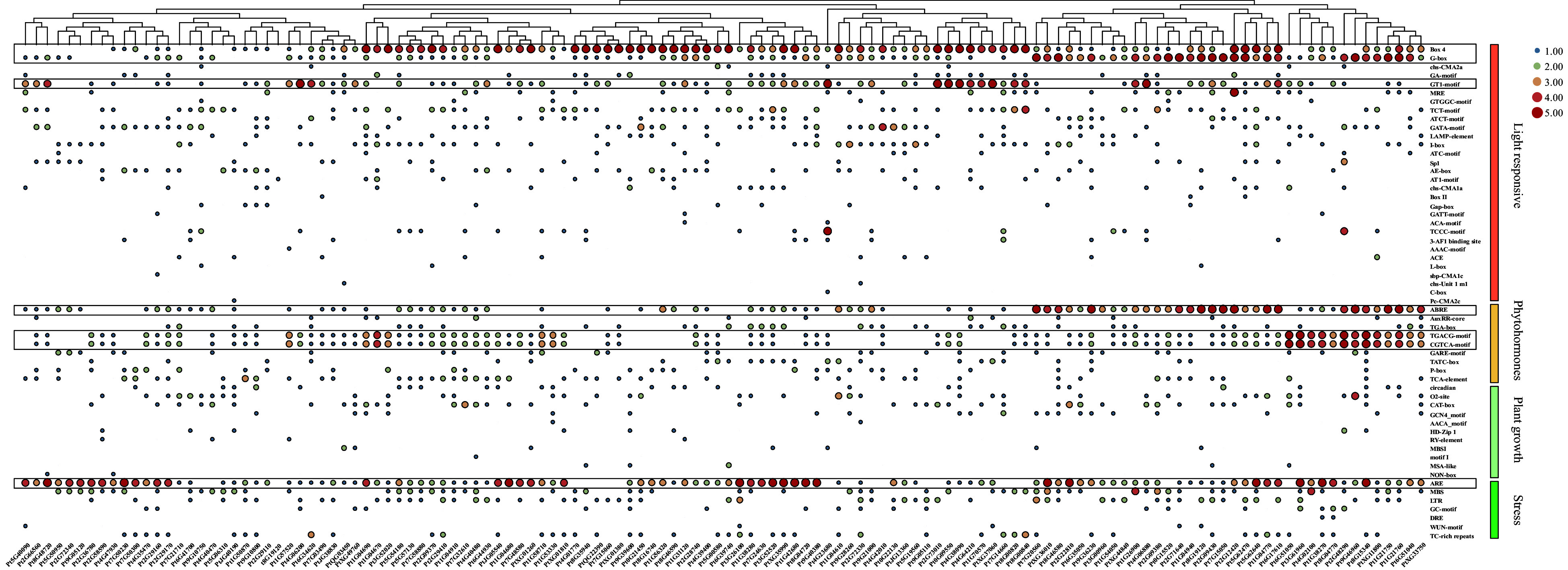
To explore the potential regulatory mechanism of DIP genes, the cis-elements were identified from their promoters. We found 57 types of cis-elements, including 30 light response, 9 phytohormone response, 11 plant growth-related response, and 7 stress response elements (Fig. 5). Among these cis-elements, light-responsive elements accounted for the largest proportion, and Box-4, G-box, and GT1-motif were critical elements of light response. For phytohormone response, the ABRE motif related to abscisic acid (ABA) as well as the TGACG motif and CGTCA motif related to the methyl jasmonate (MeJA) were mainly enriched. For stress response, only the ARE motif related to anaerobic induction was enriched. These results indicate the potential role of DIP genes in the circadian rhythm, photosynthesis, ABA-mediated stomatal regulation, MeJA-mediated anthocyanin biosynthesis, and stress response.
The functions of PtDAL1 interacting proteins
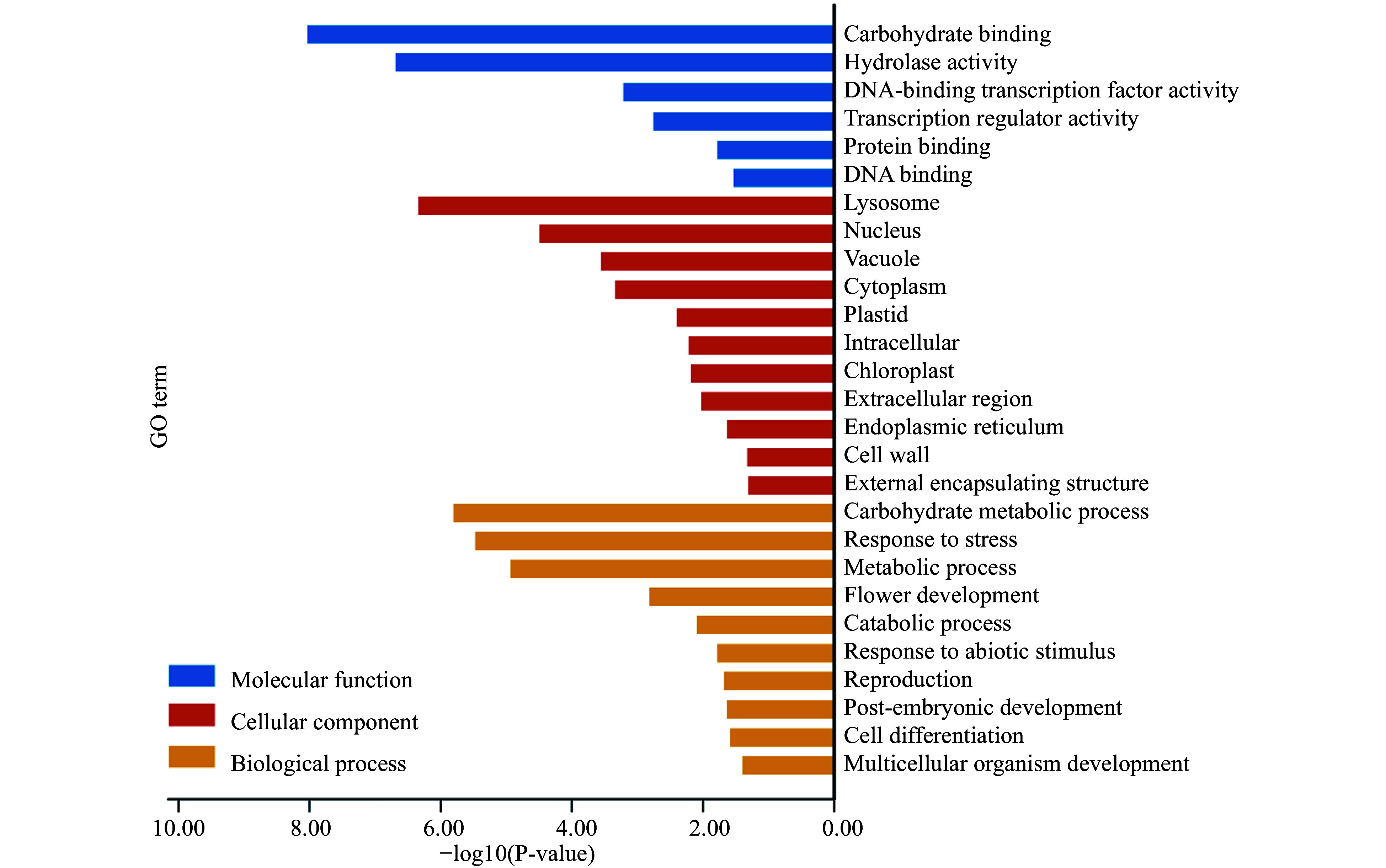
The GO enrichment analysis categorized 135 PtDAL1 interacting proteins into three GO subgroups. These interacting proteins were involved in stress response (BP), flower development (BP), abiotic stimulus response (BP), post-embryonic development, lysosome development (CC), and DNA-binding transcription factor activity (MF) (Fig. 6). Considering promoters of DIP genes enriched stress response motif and light-responsive elements, it is not surprising that they participate in stress response and abiotic stimulus response. These results indicate that DIPs play an important role in several growth stages. In addition, the results show that DIPs have a function in flower development, which is consistent with the main functions of the MADS-box gene family[29].
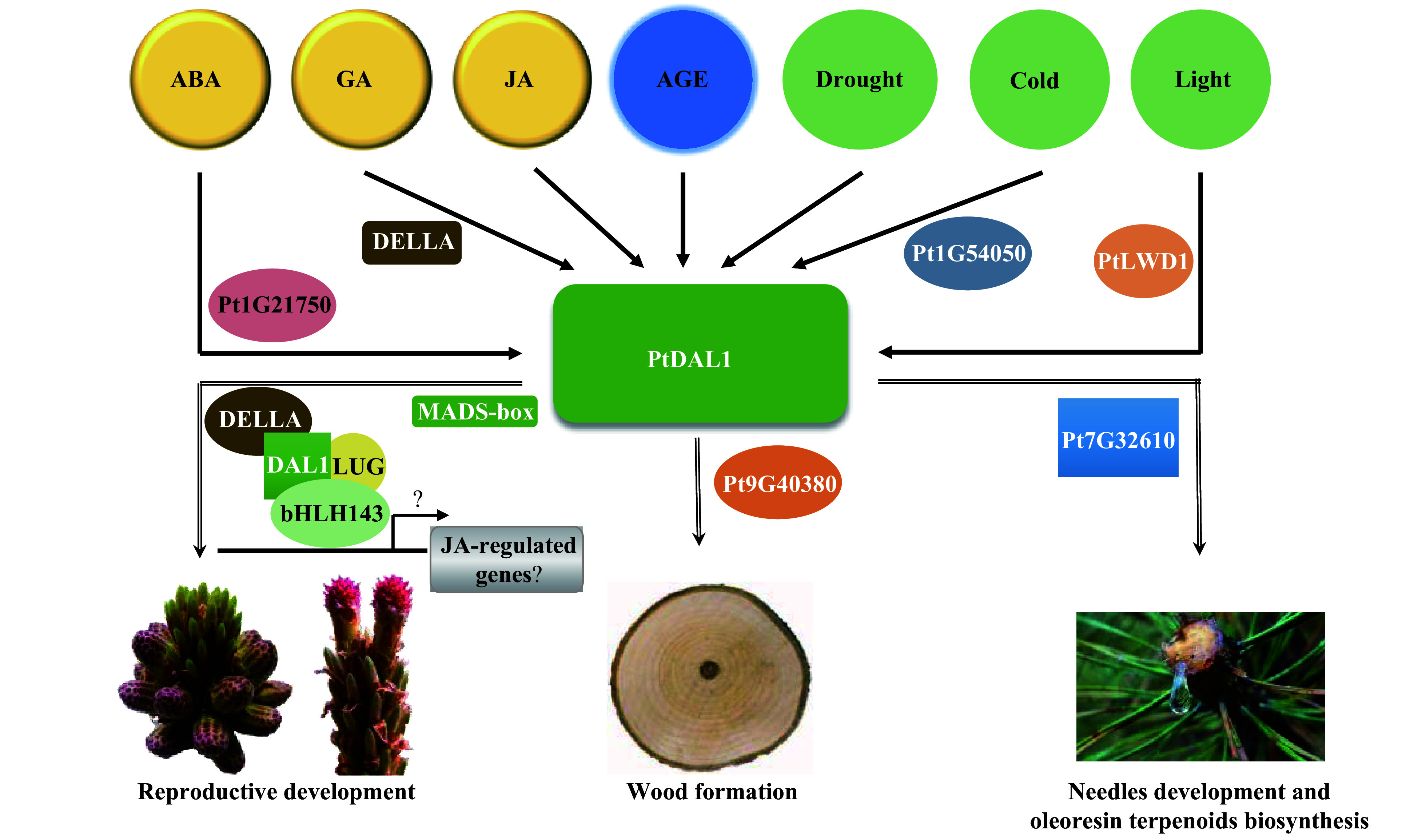
According to the results described above, we present a working model for PtDAL1 as a core node in ageing networks. Our results suggest that PdDAL1 interacts with proteins selectively under phytohormone, light, and stress conditions to regulate plant vegetative growth, reproductive development, and oleoresin terpenoid biosynthesis (Fig. 7). In summary, PtDAL1 is an ageing signal integrator, combining environmental signals and endogenous signals to mediate the balance of vegetative and reproductive development at different growth stages.
DISCUSSION
The conifer undergoes a series of developmental processes throughout its whole lifespan, requiring the coordination of phytohormones, TFs, and a variety of environmental cues. In this study, we explored which proteins PtDAL1 interacted with to play a phased regulatory role in the ageing pathway of P. tabuliformis. Interrogation of the PtDAL1 interactome in different ages of P. tabuliformis provided a comprehensive resource for understanding transcriptional programs underlying the age-dependent utilization of diverse compositions of protein polymers.
Identifying interacting proteins by traditional Y2H technique requires extracting plasmids from yeast to PCR for sequencing[25,27]. In our study, we found that the difficulty of sample identification and the cost of sequencing will rise exponentially as hundreds to thousands of interacting events are screened. Many technological improvements have enabled the next generation of high-throughput sequencing technology[28]. In the Y2H-seq we present in this study, we omitted several costly and time-consuming steps, reducing experimental time and cost significantly. We selected TFs with important functions among the DIPs, verifying the reliability of the screening results. Most of the tested DIPs interacted with PtDAL1, except PtMADS12 (Fig. 2). This false positive result may be caused by short reads in the building of the sequencing library because the sequences of PtMADS11, PtMADS12, and PtMADS13 are too similar to distinguish. In conclusion, Y2H-seq is a convenient and reliable method that can be used to supplement common Y2H.
Comparing the interactome between PtDAL1 and AtAGL6, we found that there are more DIPs than interacting proteins of AtAGL6, indicating that PtDAL1 may be associated with more biological functions than AtAGL6. In addition, the PtDAL1 and AtAGL6 interactome share some common interactors belonging to MADS-box subfamilies, including AGL42, SOC1, SVP, and AGL16 (Fig. 3a). This result suggests that the function of PtDAL1 is probably conservative in reproductive development. AGL42 and SOC1 are positive regulators to accelerate flowering times[30−34], while SVP and AGL16 have a negative impact on reproductive development[35−38]. PtDAL1 may also interact with AGL42/SOC1-like proteins or AGL16/SVP-like proteins to balance vegetative growth and reproduction. However, there is not enough conclusive evidence showing the function of AtAGL6 in ageing pathways in Arabidopsis. Moreover, age-related genes and microRNA in angiosperms do not show conspicuous expression differences with age[10], such as WRKY[6], NAC[39], and MYB[22] family genes as well as miR156[3,4], and miR172[3]. These findings indicate that PtDAL1 is probably the core regulator of the unique ageing regulatory system in gymnosperms.
Phytohormones are the prominent factors in ageing pathways. In angiosperms, salicylic acid (SA), jasmonic acid (JA), abscisic acid (ABA), and ethylene are all involved in ageing regulation[1,22]. JA response decays with plant age, but defensive substances are accumulated[40−42]. In conifers, researchers also found that the expression levels of ethylene increased with age[18]. In our study, we found that JA and ABA were also involved in ageing regulation (Fig. 5), indicating that the effects of some phytohormones in ageing are conserved. We also found many light-related proteins in DIPs, as TOC1 connects the circadian and ageing pathways in angiosperms[43], suggesting light may be a factor in ageing regulation. Thus, further exploration of the mechanisms of ageing pathways and biological processes is warranted at the molecular level.
In Arabidopsis, DELLAs-WD40-bHLH-MYB protein polymer plays a key role in JA-mediated regulation of trichome development[9]. LEUNIG (LUG) is a repressor having the WD40-repeat domain, which interacts with different proteins to regulate critical developmental processes in angiosperms. Some of these processes include promoting adaxial cell identity in leaves and embryonic shoot apical meristem initiation[44] as well as flower development[45]. In addition, LUG is also involved in JA response by regulating MYELOCYTOMATOSIS 2 (MYC2) targets[40]. bHLH family genes also regulate flower size[46,47] and trichome development through JA response[48]. In our data, PtLUG5, PtbHLH143, and DELLAs were highly expressed in the 5-year-old tree. Thus, we suggest PtDAL1 will interact with DELLAs, PtLUG5, and PtbHLH143 to form polymers that regulate reproductive development. This proposition requires further verification.
Various biological processes of the interactome resource of PtDAL1 in P. tabuliformis can be analyzed to address a broad spectrum of questions on conifer lifespan. For example, the conifer ageing effect is the most important limitation of somatic embryogenesis and organogenesis[49]. Thus, a thorough understanding of biological networks that delineate conifer ageing is essential to achieve plant regeneration. Further studies to characterize the detailed function of each DIP will provide a more comprehensive network that further extends our understanding of age-associated pathways in gymnosperms. The global DIPs revealed in this study will serve as fundamental resources and facilitate future studies on perceiving the age-dependent processes in perennial forest trees.
MATERIALS AND METHODS
Plant materials
We collected new shoots from 1, 3, 5, 7, 12, 23, and 33-year-old trees of P. tabuliformis Carr. from a primary clonal seed orchard located in Pingquan City, Hebei Province, China to construct the yeast library.
For tissue-specific expression analysis, we collected needles[50], cones[51], pollen, new shoots, roots, stems[52], and ovule[53]. For ageing-specific expression analysis, we collected vegetative shoot apexes[16].
Yeast two-hybrid (Y2H) and Y2H-seq assay
For the Y2H assay, PtDAL1 CDS was cloned into the pGBKT7 (BD) (Clontech), and PtDAL4, PtDAL9, PtDAL19, PtMADS11, PtMADS12, and PtMADS13 were inserted into pGADT7 (AD)(Clontech). We transferred recombinant plasmids into the yeast strain using Y2H (Weidi Biotechnology Co. Ltd (Shanghai, China)). Transformants were placed on SD-Leu-Trp plates and incubated for 2 d at 30 °C. The interactions were tested on SD-Trp-Leu-His-Ade plates and incubated for 3–4 d at 30 °C.
For the Y2H-seq assay, we constructed the yeast library in OEbiotech company (OEbiotech Co., Ltd (Shanghai, China)). Here, RNA was extracted from new shoots, and then reverse transcription was applied to obtain the whole cDNA in new shoots. Next, the whole cDNA was inserted in pDONR222 and pGADT7-DEST vectors. Finally, the converted expression vectors in the Y187 strain to obtain working liquid. In the meantime, we used pGBKT7-PtDAL1 plasmids to verify self-activation and screen PtDAL1 interacting proteins using the Y2H mating protocol (OEbiotech Co. Ltd (Shanghai, China)). Then, we selected 10 events in one PCR tube with 50 μl ddH2O and mixed them. Then, we added 1 μl of mixed liquid to every tube as the template for PCR analyses. To extract as much DNA fragments of DAL1 interacting proteins as possible, we extended the extension time to 3 min if the extension efficiency was 1 kb/min. All PCR products were mixed in a 10 ml tube to purify them. Finally, we used the purified products to sequence by NGS sequencing.
Construction of interaction network
The AtAGL6 interactome were obtained from the database of EMBL-EBI (http://www.ebi.ac.uk) and BAR (http://bar.utoronto.ca/interactions2). We listed the interacted proteins of PtDAL1 and then constructed the interaction network using Cytoscape 3.6.1 (National Institute of General Medical Sciences (NIGMS)).
Cis-elements analysis
The 2000 bp upstream sequencing of CDS of PtDAL1 interacting proteins in P. tabuliformis genome was obtained using TBtools[54]. Then, the cis-elements of DIP promoters were screened using the PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html).
Heatmap analysis and GO enrichment analysis
We obtained data annotations from the GO database (http://geneontology.org), and functional annotation of these sequences was performed by running BLAST against protein sequences from Arabidopsis thaliana. GO enrichment analysis and heatmap of cis-elements, tissue-specific expression, and ageing-specific expression were carried out using TBtools[54]. Due to the large amounts of data, we used the mean for ageing and tissue-specific expressions (Fig. 4). The entire raw data are provided as supplementary material.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
Acknowledgements
This work was supported by 'The National Natural Science Foundation of China (NO. 31870651)' and 'The Fundamental Research Funds for the Central Universities (NO. 2021ZY55)'. www.letpub.com
Supplementary Information
accompanies this paper at (http://www.maxapress.com/article/doi/10.48130/FR-2021-0012)
References
Articles from Forestry Research are provided here courtesy of Maximum Academic Press

 1,*
1,*