Abstract
Free full text

Multiple Functions of Human Papillomavirus Type 16 E6 Contribute to the Immortalization of Mammary Epithelial Cells
Abstract
The E6 proteins from cervical cancer-associated human papillomavirus (HPV) types such as HPV type 16 (HPV-16) induce proteolysis of the p53 tumor suppressor protein through interaction with E6-AP. We have previously shown that human mammary epithelial cells (MECs) immortalized by HPV-16 E6 display low levels of p53. HPV-16 E6 as well as other cancer-related papillomavirus E6 proteins also binds the cellular protein E6BP (ERC-55). To explore the potential functional significance of these interactions, we created and analyzed a series of E6 mutants for their ability to interact with E6-AP, p53, and E6BP in vitro. While there was a similar pattern of binding among these E6 targets, a subset of mutants differentiated E6-AP binding, p53 binding, and p53 degradation activities. These results demonstrated that E6 binding to E6-AP is not sufficient for binding to p53 and that E6 binding to p53 is not sufficient for inducing p53 degradation. The in vivo activity of these HPV-16 E6 mutants was tested in MECs. In agreement with the in vitro results, most of these p53 degradation-defective E6 mutants were unable to reduce the p53 level in early-passage MECs. Interestingly, several mutants that showed severely reduced ability for interacting with E6-AP, p53, and E6BP in vitro efficiently immortalized MECs. These immortalized cells exhibited low p53 levels at late passage. Furthermore, mutants defective for p53 degradation but able to immortalize MECs were also identified, and the immortal cells retained normal levels of p53 protein. These results imply that multiple functions of HPV-16 E6 contribute to MEC immortalization.
Papillomaviruses induce benign epithelial cell expansions clinically referred to as warts or papillomas. There are >80 distinct human papillomavirus (HPV) types based on differences in their DNA sequences. These variations allow a diverse spectrum of disease: some HPV types are more frequent in skin, while others more commonly infect mucosa. These distinctions are particularly relevant in HPV infections of the lower genital tract and uterine cervix, where distinct types such as HPV type 16 (HPV-16) and HPV-18, referred to as high-risk HPVs, are associated with high-grade squamous intraepithelial lesions and invasive cervical carcinomas. In contrast to these types, low-risk HPVs such as HPV-6 and -11 are more often found in benign intraepithelial lesions (for a review, see reference 57). The HPV E6 and E7 genes are uniformly and selectively expressed in cervical carcinomas and derived cell lines, indicating that their expression is necessary for maintenance of the malignant state. E6 has multiple activities in tissue culture models that are thought to be linked to its oncogenic activities in vivo. These include malignant transformation of established cell lines, immortalization of primary human and rodent cells, resistance to calcium- or serum-induced terminal differentiation, and sensitization to or protection against apoptosis (for a review, see reference 41).
How does E6 alter the growth characteristics of the epithelial cell? The finding that high-risk, but not low-risk E6 proteins bind and target the tumor suppressor protein p53 for degradation suggested that this interaction accounts for its oncogenic properties (55). Transfection of high-risk E6 into cells results in enhanced turnover of p53 protein and inhibition of its transcriptional activation function (2, 3, 15, 16, 20, 26, 35, 37, 38). The interaction of E6 with p53 is mediated by the cellular factor E6-AP. E6 binds an 18-amino-acid peptide (amino acids 391 to 408) found near the center of E6-AP (27–29). The E6–E6-AP complex acts as a ubiquitin-protein ligase that results in the specific ubiquitination and degradation of p53 (46). This complex associates with the core DNA binding domain of p53, but stabilization of binding and degradation of the complexed p53 require sequences within the N terminus of p53 (37). Other investigators have reported that the E6 proteins bind to both the core domain and the C terminus of p53. However, the physiological relevance of the interaction with the C terminus of p53 remains to be determined (36). Although an E6-p53 complex in the absence of E6-AP has been reported (17, 34, 36), degradation of p53 occurs only when both E6 and E6-AP are expressed (17). Low-risk and cutaneous HPV E6 proteins bind p53 weakly if at all and are incapable of inducing its degradation.
Several cell culture systems have been employed to characterize the biological properties of E6. Keratinocytes are efficiently immortalized by high-risk E6 and E7 (for a review, see reference 57). We have previously reported that HPV-16 E6 alone can immortalize primary human mammary epithelial cells (MECs) and that the levels of p53 protein are drastically reduced in the immortal cells (2, 3). Mutational analysis of HPV-16 E6 demonstrated a strong correlation between p53 level reduction and immortalization (16). In contrast, low-risk HPV-6 E6 and bovine papillomavirus type 1 (BPV-1) E6 immortalized MECs with a much lower efficiency and only after a crisis period (2). Because HPV-6 and BPV-1 E6 do not induce p53 degradation in vitro or in early-passage MECs (2), immortalization may be p53 independent. However, the half-life of p53 in the postcrisis HPV-6 or BPV-1 E6-immortalized cells, and in all HPV-16 E6-immortalized cells, was found to be dramatically reduced (2). Spontaneous immortalization of MECs that express low p53 protein levels has never been observed in our experiments. Whether HPV-6 E6 binds p53 remains controversial. BPV-1 E6 does not bind or degrade p53, although it binds E6-AP, implying that E6-AP association is not sufficient for p53 binding and degradation. The reduced p53 levels in low-risk and BPV-1 E6-immortalized MECs imply that proteolysis of p53 may occur through more than one pathway in vivo. Furthermore, the majority of p53 mutations that are able to inhibit p53-activated transcription do not induce immortalization of MECs, although several p53 mutations do so with low efficiency (8, 21, 23). Therefore the ability of HPV-16 E6 to reduce p53 levels is not functionally equivalent to a trans-dominant p53 mutation.
Substantial evidence indicates that high-risk HPV E6 has p53-independent activities. In a search for additional targets, E6 proteins have been found to bind a variety of cellular factors, such as paxillin, AP-1, hDLG, IRF-3, Myc, hMCM7, Bak, and E6TP-1 (22, 24, 33, 43, 51); for a review, see reference 41). We have reported that E6 proteins from high-risk HPVs and BPV-1 bind E6BP (ERC-55) in vitro and in the yeast two-hybrid system. A strong correlation between in vitro E6BP binding and transformation of murine cells by a set of BPV-1 E6 mutants was observed (12). However, the biological relevance of the interaction between HPV-16 E6 and E6BP remains to be determined. Recently, we have mapped the region of E6BP that is necessary and sufficient for complex formation with HPV-16 E6 to a 25-amino-acid domain and found that the synthesized peptide comprising these 25 amino acids binds calcium and folds into a classical helix-loop-helix EF hand conformation (11). Further studies showed that 13 amino acids within the second α-helix mediated E6 association. Alignment of this α-helical E6-binding peptide with the E6-binding regions of other proteins, such as the 18-amino-acid E6-binding region of E6-AP and the first LD repeat of paxillin, revealed a consensus sequence required for E6 binding (11, 19).
While the regions in E6BP, E6-AP, and p53 necessary for association with HPV-16 E6 have been identified (11, 29, 36, 37), there is little information on the region of E6 necessary for these interactions. The C-terminal 11 amino acids in HPV-16 E6 are not required for p53 binding or degradation (reference 20 and our unpublished data). However, conflicting results regarding regions in E6 required for p53 binding and degradation have been reported (15, 20). To define the regions and amino acids in HPV-16 E6 required for interactions with E6-AP, p53, and E6BP, and to address the biological relevance of these interactions, we analyzed a series of HPV-16 E6 mutants for their in vitro and in vivo functions.
MATERIALS AND METHODS
Mutagenesis and DNA constructs.
The construction of several HPV-16 E6 mutants has been described previously (16). Site-directed mutations were generated by a PCR-based mutagenesis method (9) with modifications to optimize PCR conditions. Mutations were subcloned into pSP65 (Promega) and the retroviral vector pLXSN (39) as PCR products by using oligonucleotides primers containing appropriate restriction enzyme sites. Plasmids were sequenced to confirm the presence of directed mutations. The L119R, H126C, and H126S mutations were also subcloned into pMSI · ref (14) by replacing the BsaBI-PinAI fragment of the wild-type E6 gene. Plasmids p16E6SP65 and pProp53SP65 (16), GST-E6BP-211 (12), GST-E6BPdlM (11), and GST-E6-AP and GST-E6-APΔ391-408 (29) were described previously. GST-E6BPFS−, which encodes glutathione S-transferase (GST) fused to E6BP lacking a signal sequence, was constructed by using a pGEX2T-derived vector, pGEX2TS (7). GST-E6BPFS−dlM was constructed by using GST-E6BPFS− as the backbone, in which the HindIII-SpeI fragment was replaced by that of GST-E6BPdlM.
In vitro p53 degradation and ubiquitination assays.
The E6 and p53 proteins were prepared in a rabbit reticulocyte lysate (RRL) transcription-translation system (TNT; Promega) in the presence of [35S]cysteine for E6 and [35S]methionine for p53. pSP65-based constructs were used as templates for the synthesis of E6, most of the E6 mutants, and p53, and SP6 RNA polymerase was used in these reactions. The L119R, H126C, and H126S E6 proteins were translated from the pMSI-based constructs (14), and T7 RNA polymerase was used. For in vitro p53 degradation assays, approximately equal amounts of each E6 protein and 1 μl of human p53 made in RRL were mixed in a 30-μl reaction mixture containing 9 μl of additional RRL in 25 mM Tris-HCl (pH 7.5)–100 mM NaCl–3 mM dithiothreitol (DTT). Samples were incubated at 25 or 37°C for 3 h, 1 volume of 2× sodium dodecyl sulfate (SDS) sample buffer was added, and the samples were boiled for 5 min. The reaction products were resolved on SDS–12% polyacrylamide gels, dried, and visualized with a phosphorimager (Bio-Rad). For the ubiquitination assays, equal amounts of in vitro-translated 35S-labeled E6 and p53 proteins were incubated under the conditions described above except that 4 mM ATPγ-S (Sigma) was included in the reaction mixture for 30 min. The reaction mixture was then incubated with the p53-specific monoclonal antibody pAb421 and rocked at 4°C for 30 min. The proteins were collected on protein A-Sepharose beads, washed with LSAB (100 mM NaCl, 100 mM Tris-HCl [pH 8.0], 1% Nonidet P-40 [NP-40], 2 mM DTT, 1 mM phenylmethylsulfonyl fluoride), and released from the beads by boiling in 60 mM Tris-HCl (pH 6.8)–2% SDS–10% glycerol–1% β-mercaptoethanol for 7 min. The beads were pelleted, the supernatant was removed and diluted with LSAB, and immunoprecipitations were performed with an antiubiquitin antiserum (Sigma). Proteins were collected on protein A-Sepharose beads, resolved on an SDS–15% polyacrylamide gel, and visualized by autoradiography.
In vitro binding assays.
The p53 and E6 binding assays were performed as described previously (16). Briefly, 275 ng of purified p53 protein expressed in baculovirus was incubated at 4°C with comparable amounts of in vitro-translated 35S-labeled E6 proteins in LSAB for 1 h. The mixtures were then allowed to react with the anti-p53 monoclonal antibody pAb421 and protein A-Sepharose beads for 1 h at 4°C. The beads were washed four times with LSAB, boiled in SDS sample buffer, and subjected to SDS–12% polyacrylamide gel electrophoresis. GST–E6-AP and GST–E6-APΔ391–401 proteins were made as described previously (29). GST–E6BP-211, GST-E6BPdlM, GST-E6BPFS−, and GST-E6BPFS−dlM fusion proteins were made in bacteria by standard procedures (49). Small aliquots of the fusion proteins were run on SDS-polyacrylamide gels and stained with Coomassie blue to confirm sizes and homogeneity. Protein concentrations were determined by the bicinchoninic acid assay (Pierce) with bovine serum albumin as a protein standard. For E6-AP and E6BP binding experiments, comparable amounts of each [35S]cysteine-labeled E6 protein made in RRL were incubated with 2 μg of the GST fusion protein coupled on glutathione-Sepharose beads in 250 μl of binding buffer. Lysis buffer (250 mM NaCl, 20 mM Tris-HCl [pH 7.4], 0.5% NP 40, 1 mM EDTA) (56) was used for GST-E6BP binding, and LSAB was used for GST–E6-AP binding. All binding buffers contained 2 mM DTT and 1 mM PMSF. The mixtures were subjected to rotary shaking at 4°C for 3 h, and the beads were washed four times with 1 ml of the corresponding binding buffer for each wash. The proteins associated with the beads were then released and subjected to SDS–12% polyacrylamide gel electrophoresis. The gels were dried and visualized with a phosphorimager.
Yeast two-hybrid assays.
The plasmid constructs and the yeast strain used for two-hybrid analysis have been described previously (12). A subset of the site-directed mutants of E6 were constructed in the yeast vector for the purpose of two-hybrid assays. For quantification of β-galactosidase activity in yeast, colonies were inoculated into selective medium containing glucose and grown as master cultures. A fraction of the master culture was used to inoculate a culture containing glucose or galactose. The cells used to inoculate the galactose culture were pelleted and then washed with medium lacking a carbon source to avoid glucose repression. The cultures were grown to an optical density at 600 nm of 0.9 to 1.2. The cells were then harvested and permeabilized, and the β-galactosidase activity was measured by using o-nitrophenyl-β-galactoside as described previously (25). Induction of β-galactosidase activity was determined as the ratio of β-galactosidase activity measured in galactose medium to that measured in glucose medium. Each mutant was assayed in one to three independent experiments. In each experiment, two representative transformants of each mutant were assayed.
Analysis of p53 and E6 in MECs.
The pLXSN-based constructs expressing E6 or mutants were introduced into the amphotrophic packaging cell line PA317 (kindly provided by D. Galloway and A. D. Miller) by calcium phosphate precipitation (10). The virus stocks were made in DFCI-1 medium, and titers were determined on the radiation-immortalized MEC line 76R-30 (53). The derivation and culture of the normal epithelial cell strain 76N from reduction mammoplasty in DFCI-1 medium has been described previously (4). 76N cells (105 to 106/100-mm-diameter dish) were plated in DFCI-1 medium 18 h before infection. Cells were infected with retroviral stocks, selected with G418, and expanded in DFCI-1 medium. Early-passage cells (passage 2 or 3) were divided for p53 protein analysis by Western blotting (53) with the anti-p53 monoclonal antibody pAb1801 or for selection of immortal clones by propagation in D2 medium (4, 5). Late-passage immortalized cells were also subjected to Western blot analysis as described above. For immunoprecipitation of the E6 proteins, the radiation-immortalized MEC line 76R-30 (53) was infected with retroviruses expressing each E6 gene. After selection in G418, the cells were labeled with [35S]cysteine, immunoprecipitations were performed as described previously (1), and the gels were analyzed with a phosphorimager.
For DNA sequencing, 1 μg of total cellular RNA from the HPV-16 E6 mutant-immortalized cell lines (F2V II or Y54H I) was used as a template to synthesize cDNA by using SuperScript II reverse transcriptase and an oligo(dT) primer (Gibco BRL). A pair of primers spanning the entire coding region of p53 and internal primers were used to generate p53-specific PCR products. The reverse transcription-PCR products were subjected to automatic DNA sequencing. The E6-coding region was amplified by PCR with genomic DNA isolated from E6 mutant-immortalized MECs as a template and primers spanning the entire coding region of E6. The PCR products were then subjected to automatic DNA sequencing.
Actinomycin D-induced responses in immortalized MECs.
For G1 arrest, late-passage MECs were seeded in D medium at 2.5 × 105 per well in a six-well plate. The next day, cells were left untreated or were treated with 0.5 nM actinomycin D. Twenty-four hours later, cells were harvested, fixed in 70% ethanol, stained with propidium iodide, treated with RNase A, and analyzed on a FACScan flow cytometer (Becton Dickinson). Cell cycle analysis was performed by using ModFit software (Becton Dickinson), and the G1-to-S ratios were calculated.
For the p53-specific response assay, late-passage MECs were seeded in D medium at 2 × 105 per 35-mm-diameter culture dish on the day before transfection. Duplicate cultures were transfected with 1 μg of p53-responsive luciferase reporter pPG, a modified version of PG13-CAT (30), along with 0.3 μg each of pGreenLantern (green fluorescence protein) and pcDNA3.1/His3/LacZ (β-galactosidase) reporters by calcium phosphate precipitation (10). At 40 h posttransfection, cells were left untreated or were treated with 0.5 nM actinomycin D for 24 h. Luciferase (Promega) and β-galactosidase (Tropix) assays were performed with cell extracts according to the manufacturer’s instructions, and enzymatic activities were measured with a luminometer (MGM Instruments, Inc.). β-Galactosidase activities were used to correct luciferase activities by normalizing transfection efficiency.
RESULTS
Mutagenesis of HPV-16 E6.
We have previously characterized a series of HPV-16 E6 mutants for p53 binding and degradation and for the ability to immortalize MECs (16). In the present study, 22 additional HPV-16 E6 mutants were generated and analyzed for E6-AP and E6BP binding. The majority of E6 mutants were generated by site-directed mutagenesis. Targeted amino acids were selected from those (i) conserved among genital HPV types, such as Arg 124 and Gly 134; (ii) conserved among genital HPV types and BPV-1, such as His 118; (iii) unique to high-risk HPV types, such as Phe 2, Tyr 54, Gln 107, Leu 119, Asp 120, His 126, Asn 127, Ile 128, and Gly 130; and (iv) common to multiple papillomavirus types listed by Cole and Danos (13), such as Glu 114 and Phe 125 (Table (Table1).1). In addition, a randomly mutagenized E6 library was screened by the yeast two-hybrid system to identify E6BP-binding-defective mutants. Two single missense mutations, L37S and L110Q, were identified from this screen. In this nomenclature, L37S and L110Q represent changes of the normally found leucines at amino acids 37 and 110 to serine and glutamine, respectively.
TABLE 1
Correlation of E6-AP and E6BP binding with HPV-16 E6 mutants
mutants
| HPV-16 E6 protein | Papillomavirus types in which the mutated amino acid is conserved | Bindinga
| |
|---|---|---|---|
| E6-AP | E6BP | ||
| Wild type | +++ | +++ | |
| F2V | High risk | +++ | +++ |
| F2L | High risk | +++ | +++ |
| H24L/I27Vb | None/none | +++ | +++ |
| K34Eb | Genital | +++ | +++ |
| Q35Rb | None | +++ | +++ |
| L37S | Multiple | +/− | +/− |
| Y54D | High risk | + | + |
| Y54H | High risk | +++ | +++ |
| C63R/Y70C/K72R/T86Sb | Multiple/none/genital/none | − | − |
| Y84Cb | Genital | +++ | +++ |
| I101Vb | Multiple | ++ | ++ |
| Q107R | High risk | +/− | +/− |
| L110Q | Multiple | − | − |
| H118D | Genital and BPV-1 | +/− | +/− |
| H118N | Genital and BPV-1 | ++ | ++ |
| Δ118-122b | Genital and BPV-1 | + | +/− |
| L119R | High risk | +++ | +++ |
| D120A | High risk | +++ | +++ |
| D120T | High risk | +++ | +++ |
| R124T | Genital | +++ | +++ |
| F125V | Multiple | +/− | +/− |
| F125L | Multiple | +++ | +++ |
| N127K | High risk | +++ | +++ |
| I128T | High risk | +/− | +/− |
| G130V | High risk | − | − |
| W132Rb | Multiple | +/− | +/− |
| G134V | Genital | +/− | +/− |
Association of E6 with E6-AP and E6BP.
While the regions in E6-AP, E6BP, and p53 necessary for association with HPV-16 E6 have been identified (11, 29, 36, 37), little is known about the region of E6 necessary for these interactions. To gain this information, a large number of previously described and newly generated E6 mutants were examined for the ability to associate with E6-AP and E6BP in vitro. GST–E6-APΔ391-408, which lacks the 18-amino-acid E6-binding region, was used as the negative control for GST–E6-AP. Two versions of E6BP were expressed as GST fusions. GST–E6BP-211 expresses the C-terminal 211 amino acids, the original yeast two-hybrid isolate of E6BP (11, 12). We have previously mapped the E6-binding region to the fourth EF hand of E6BP. GST-E6BPdlM, in which the 24-amino-acid EF hand IV is deleted and which does not bind E6 (11), was used as the negative control for GST–E6BP-211. GST-E6BPFS− expresses E6BP beginning at amino acid 25 and lacks the N-terminal signal sequence. Similarly, GST-E6BPFS−dlM was constructed as a negative control for GST-E6BPFS−.
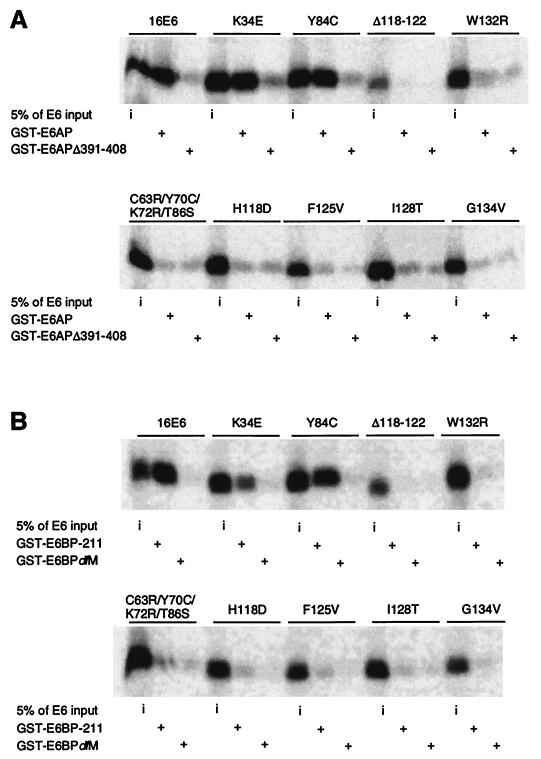
The results of the in vitro association experiments are summarized in Table Table1.1. Notably, the binding patterns of the E6 mutant proteins were very similar for E6-AP and E6BP. As shown by representative mutants (Fig. (Fig.1),1), mutants K35E and Y84C exhibited high-affinity binding to GST–E6-AP (Fig. (Fig.1A),1A), E6BP-211 (Fig. (Fig.1B),1B), and E6BPFS− (data not shown). In contrast, other E6 mutants showed very poor binding to these proteins, barely distinguishable from that for the respective negative controls (Fig. (Fig.1).1). The deletion mutant Δ118-122 showed undetectable binding in this experiment (Fig. (Fig.1),1), although low-level binding was detected in other experiments. H118D, F125V, I128T, and G134V showed low-level binding (<10%) to E6-AP under less stringent binding and washing conditions (0.2 versus 1% NP-40) or with Sepharose bead-coupled GST-16E6 mutant fusion proteins used to capture radiolabeled, in vitro-translated E6-AP (data not shown). Among the deletion mutants (15), only Δ143-147 retained substantial binding to E6-AP and E6BP; the others had greatly reduced associations with a similar pattern (data not shown). To confirm the abilities to bind to E6BP, a subset of E6 mutants was also analyzed in vivo by yeast two-hybrid analysis with E6BP-211 as the bait. H118D and I128T showed marginal interaction with E6BP (5% of the wild-type E6 level). F125V and G134V did not display significant interaction with E6BP (data not shown). In summary, a correlation between the associations of E6-AP and E6BP was observed for E6 mutants. However, no cellular protein binding domains could be localized in any short linear region of E6.
Binding to E6-AP and p53 is not sufficient for stimulation of p53 degradation by E6.
Next, we compared the E6-AP binding and p53 binding abilities of the mutants. As summarized in Table Table2,2, mutants that were impaired for E6-AP binding had a corresponding reduction in p53 association. However, E6-AP binding could be separated from p53 association. For example, E6 mutants F2V, F2L, and Y54H bound E6-AP at wild-type levels (Fig. (Fig.2A)2A) but were severely compromised for p53 association, showing 2, 10, and 7% of wild-type levels, respectively (Fig. (Fig.2B,2B, top panels; Table Table2).2). Similarly, mutant Y84C bound E6-AP at the wild-type level but showed very low binding to p53 (Fig. (Fig.1A;1A; Table Table2).2).
TABLE 2
Summary of E6 mutant studies
studies
| HPV-16 E6 protein | Mutant class | Bindinga
| p53 degradationa at:
| p53 levels in MECs
| MEC immortalizationb | ||||
|---|---|---|---|---|---|---|---|---|---|
| E6-AP | E6BP | p53 | 25°C | 37°C | Early passage | Late passage | |||
| Wild type | +++ | +++ | +++ | +++ | +++ | Low | Low | 6 (6) (6) | |
| H24L/I27Vc | I | +++ | +++ | +++ | +++ | +++ | Low | Low | 3 (3) (3) |
| K34Ec | I | +++ | +++ | +++ | +++ | +++ | Low | Low | 3 (3) (3) |
| Q35Rc | I | +++ | +++ | +++ | +++ | +++ | Low | Low | 3 (3) (3) |
| Y84Cc | I | +++ | +++ | +/− | +++ | +++ | Low | Low | 3 (3) (3) |
| I101Vc | I | ++ | ++ | +++ | +++ | +++ | Low | Low | 3 (3) (3) |
| E114A | I | + | NDd | + | +++ | +++ | Low | Low | 2 (2) (2) |
| H118D | I | +/− | +/− | + | +++ | ++ | Low | Low | 1 (1) (1) |
| H118N | I | ++ | ++ | ++ | +++ | +++ | Low | Low | 1 (1) (1) |
| Δ118-122c | I | + | +/− | − | − | − | Low | Low | 3 (3) (3) |
| F125V | I | +/− | +/− | ++ | +++ | − | Low | Low | 2 (2) (2) |
| I128T | I | +/− | +/− | +/− | +++ | − | Intermediate | Low | 3 (3) (3) |
| G134V | I | +/− | +/− | + | +++ | − | Low | Low | 2 (2) (2) |
| L37S | II | +/− | +/− | − | ++ | − | Normal | Low | 1 (2) (2) |
| Q107R | II | +/− | +/− | +/− | +++ | + | Normal | Low | 3 (3) (3) |
| L110Q | II | − | − | − | − | − | Normal | Low | 1 (2) (2) |
| W132Rc | II | +/− | +/− | − | − | − | Normal | Low | 1 (4) (4) |
| F2V | III | +++ | +++ | +/− | − | − | Normal | Normal | 3 (4) (4) |
| Y54H | III | +++ | +++ | + | +++ | − | Normal | Normal | 4 (4) (4) |
| C63R/Y70C/K72R/T86Sc | − | − | − | − | − | Normal | NAe | 0 (5) (5) | |
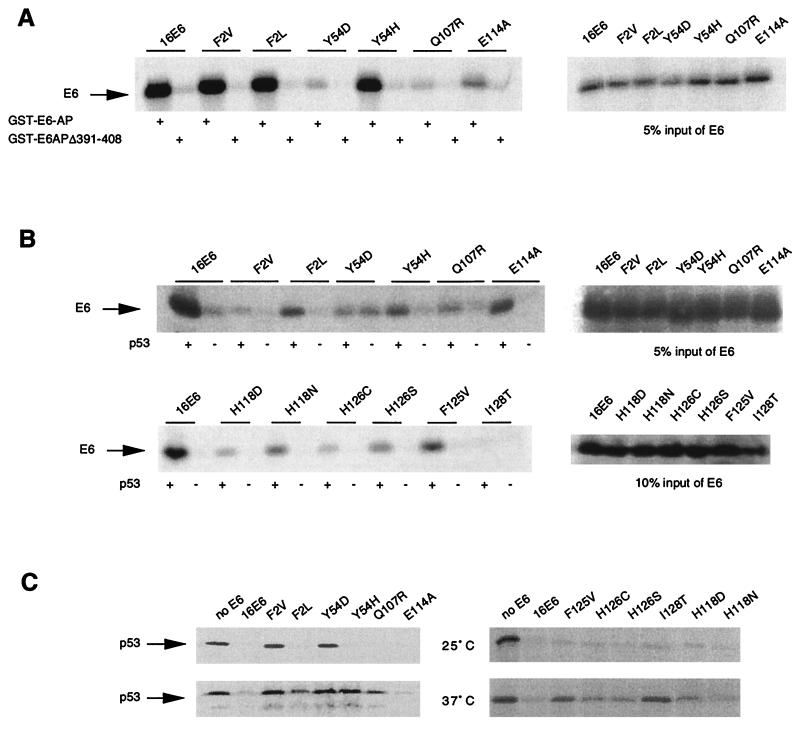
E6 interaction with E6-AP and p53. (A) E6-AP binding by E6 mutants (same as for Fig. Fig.1A1A except that different E6 mutants were tested for binding). The 5% input levels for the indicated E6 mutants are shown in the right panels. (B) p53 binding by E6 mutants. Immunoprecipitations were performed in the absence (−) or presence (+) of purified p53 protein as described in Materials and Methods. Input levels of E6 proteins are shown in the right panels. (C) p53 degradation by E6 mutants. The input levels of E6 are shown in the right panels in panel B.
These data imply that E6-AP binding is not sufficient for p53 association. Next we asked whether p53 binding by the E6–E6-AP complex was sufficient for induction of p53 degradation. Many E6 mutants, such as E114A, H118D, H118N, F125V, H126C, H126S, and I128T, which displayed reduced p53 binding abilities (Fig. (Fig.2B;2B; Table Table2),2), were able to induce p53 degradation at 25°C in vitro (Fig. (Fig.2C,2C, top panels) and in MECs (see Fig. Fig.4,4, top panel). The fact that low levels of p53 binding were sufficient for inducing p53 degradation by these mutants can be explained by the enzymatic nature of E6–E6-AP-mediated p53 degradation. However, mutants such as F2V and Y54H, which retained p53 binding ability at reduced efficiency (Fig. (Fig.2B),2B), failed to induce p53 degradation at 37°C in vitro (Fig. (Fig.2C)2C) and in MECs (see Fig. Fig.4).4). This indicates that binding to p53 is not sufficient for stimulation of p53 degradation and that a function in addition to p53 binding may be required for efficient p53 degradation stimulated by E6. This putative function may be more severely impaired in F2V than in other mutants.
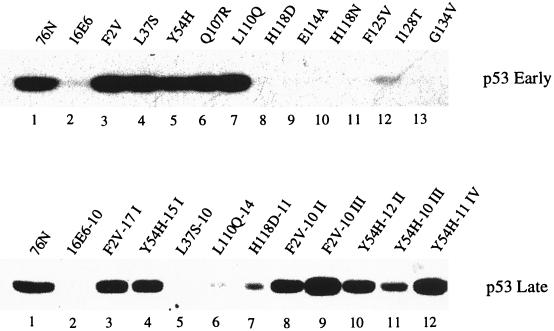
p53 degradation by the HPV-16 E6 mutants in vivo. Cell extracts of MECs infected with retroviruses expressing E6 or mutants as indicated were analyzed by Western blotting. The p53-specific monoclonal antibody pAb1801 was used to probe p53 of MECs at either early (top panel) or later (bottom panel) passages after infection in the immortal clones. 76N, normal MECs. The numbers following each E6 designation in the bottom panel indicate passage numbers; I, II, III, and IV indicate cell lines derived from separate experiments.
We have previously reported a temperature-sensitive (ts) phenotype for induction of p53 degradation in vitro (16). Here we observed more E6 mutants showing this ts phenotype, including Y54H, F125V, and I128T (Fig. (Fig.2C)2C) and G134V (Table (Table2).2). Some mutants, such as F2L and Q107R, showed an intermediate ts phenotype (Fig. (Fig.2C).2C). Since E6-AP and p53 binding assays are routinely performed at 4°C, we suspected that these E6 mutants might be unable to degrade p53 at 37°C because of an inability to bind E6-AP and/or p53 at this temperature. We thus carried out binding reactions for the representative mutants F2L and Y54H at 0, 25, and 37°C simultaneously. Both mutants bound E6-AP at all temperatures with an efficiency similar to that for wild-type E6, although background binding to GST–E6-APΔ391-408 increased in the case of Y54H (data not shown). These E6 mutants also bound p53 at all temperatures with reduced efficiency compared to the wild type and showed increased background binding, especially in the case of F2L (data not shown). Therefore, these mutant E6 proteins bind E6-AP and p53 at 37°C but are unable (Y54H) or have a reduced ability (F2L) to induce p53 degradation at this temperature (Fig. (Fig.22C).
These observations suggest that specific amino acids in HPV-16 E6 may be necessary for a critical step in the targeted proteolysis of p53 in vitro. It has been demonstrated that a unique cysteine residue in the C terminus of E6-AP can be covalently ligated to ubiquitin (46), which is subsequently transferred to p53 in the presence of HPV-16 E6 (42). To investigate whether these p53 degradation ts mutants were impaired for the ubiquitination of p53 at 37°C, we performed p53 ubiquitination assays at the permissive (25°C) and nonpermissive (37°C) temperatures. As expected, wild-type HPV-16 E6 induced p53 ubiquitination at both temperatures compared to the control water-primed lysate (Fig. (Fig.3).3). F2L induced p53 ubiquitination at 25°C with an efficiency similar to that for wild-type E6 and at 37°C at lower levels than the wild type (Fig. (Fig.3).3). In contrast, Y54H failed to induce ubiquitination of p53 at 37°C but retained this activity at 25°C (Fig. (Fig.3).3). Clearly, ubiquitination correlated well with degradation. The inability of Y54H to induce p53 ubiquitination at 37°C indicates that binding to p53 is not sufficient for ubiquitination of p53. Taken together, these results suggest that E6 has multiple roles in the ubiquitin-mediated degradation of p53.
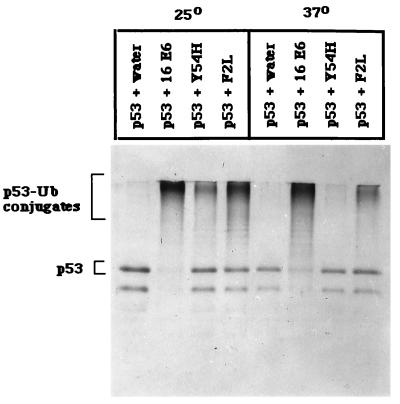
Ubiquitination of p53 by E6 and E6 mutants. In vitro-translated, 35S-labeled p53 was incubated with E6 proteins or water-primed lysate in the presence of ATPγ-S at 25 and 37°C. After incubation, the mixture was immunoprecipitated with a monoclonal antibody to p53 (pAb421), collected on protein A-Sepharose, and boiled, and then the supernatant was reimmunoprecipitated with antiubiquitin antiserum, absorbed on protein A-Sepharose, and run on an SDS-polyacrylamide gel (see Materials and Methods).
Correlation of in vitro and in vivo p53 degradation by E6 mutants.
Normal human MECs in culture exhibit readily detectable wild-type p53 protein. Infection of these cells with retrovirus expressing wild-type HPV-16 E6 induced a rapid decrease in the p53 half-life and resulted in low p53 levels (16). This represents an in vivo model to analyze the HPV-16 E6 mutants independent of immortalization. MECs were infected with recombinant E6-expressing retroviruses and analyzed for p53 protein by Western blotting. Parallel cultures were transferred to D2 medium to select for immortal cells. Consistent with in vitro results, wild-type HPV-16 E6 and mutants E114A, H118D, and H118N, which induced p53 degradation in vitro (Fig. (Fig.2C),2C), also showed low p53 levels at early passage (Fig. (Fig.4,4, top panel). F2V, L37S, Y54H, Q107R, and L110Q, which were unable to induce p53 degradation at 37°C in vitro (Fig. (Fig.2C2C and Table Table2),2), also failed to reduce p53 levels in early-passage MECs (Fig. (Fig.4,4, top panel). Similarly, I128T was greatly impaired for inducing p53 degradation at 37°C in vitro (Fig. (Fig.2C)2C) and was slightly compromised for reducing p53 levels in early-passage MECs (Fig. (Fig.4,4, top panel, lane 12). An exception to the general correlation between in vitro and in vivo phenotypes was observed with mutants F125V and G134V. Analogous to Δ118-122 (16, 20), these mutants were severely compromised for inducing p53 degradation in vitro at 37°C (Fig. (Fig.2C2C and Table Table2)2) yet were able to reduce p53 levels in vivo in early-passage MECs (Fig. (Fig.4,4, top panel).
Immortalization of MECs by E6 mutants.
We have reported the correlation between the abilities of E6 mutants to promote p53 degradation and to immortalize MECs (16). Those studies analyzed a limited number of mutants. In the present study, we extended the characterization to include additional E6 mutants, with an emphasis on those mutants with defective or greatly reduced interaction with E6-AP, E6BP, or p53. MECs infected with the E6 mutant C63R/Y70C/K72R/T86S, which was defective in interaction with all three cellular proteins, senesced along with the control LXSN-infected cells after several passages in the D2 selective medium (Table (Table2).2). Mutants E114A (Fig. (Fig.2A),2A), H118D (Fig. (Fig.1),1), and H118N are compromised for binding with E6-AP and E6BP yet exhibited competence for inducing p53 degradation in vitro (Fig. (Fig.2C)2C) and in vivo (Fig. (Fig.4,4, top panel) and for immortalization of MECs (Table (Table2).2). Another group of mutants (L37S, Q107R, F125V, I128T, and G134V), which showed greatly reduced binding ability with all three cellular proteins and a ts phenotype for p53 degradation in vitro (Fig. (Fig.11 and and22 and Table Table2),2), also immortalized MECs (Table (Table2).2). While F125V, I128T, and G134V immortalized MECs as efficiently as wild-type E6 did, L37S and Q107R showed a lag period. Notably, mutants Δ118-122, L110Q, and W132R, which are highly defective for E6-AP and E6BP associations (Fig. (Fig.11 and Table Table2)2) and completely defective for p53 binding and degradation in vitro, also immortalized MECs (Table (Table2).2). The immortalization by L110Q or W132R exhibited a slow period and yielded immortal cells in only a subset of experiments. Δ118-122 induced immortalization as efficiently as wild-type HPV-16 E6 (Table (Table2).2). Interestingly, we identified a novel group of mutants that immortalized MECs. Mutants F2V and Y54H were competent for association with E6-AP (Fig. (Fig.2A)2A) and E6BP (Table (Table1)1) but had a greatly reduced ability to bind p53 (Fig. (Fig.2B,2B, top panels). While F2V was defective in p53 degradation in vitro (Fig. (Fig.2C)2C) and in vivo (Fig. (Fig.4,4, top panel), Y54H was ts for p53 degradation in vitro (Fig. (Fig.2C)2C) and defective in vivo (Fig. (Fig.4,4, top panel). Nevertheless, these mutants immortalized MECs without a lag period. Immortal clones emerged in three of four experiments from MECs infected with F2V-expressing retrovirus, while infection with Y54H-expressing retrovirus yielded immortal cells in all of the four experiments (Table (Table22).
p53 levels in early- and late-passage MECs.
The comparison of p53 levels in early- and late-passage MECs revealed three distinct classes of E6 mutants (Fig. (Fig.44 and Table Table2).2). Consistent with our previous findings (16), the majority of E6 mutants (classes I and II) showed low levels of p53 in late-passage immortal cells (Table (Table2).2). Class I, represented by E114A, H118D, H118N, F125V, and G134V, showed greatly reduced p53 levels at both early and late passages (Fig. (Fig.4;4; Table Table2).2). Mutant I128T falls into this class by showing moderately reduced p53 levels at early passage (Fig. (Fig.4,4, top panel, lane 12) and low p53 levels at late passage (Table (Table22 and data not shown). Mutants of this class were able to immortalize MECs as efficiently as wild-type HPV-16 E6, except that I128T showed a lag period in one experiment (see below). Class II mutants (L37S, Q107R, L110Q, and W132R) failed to reduce p53 levels at early passage but displayed low p53 levels in the immortalized cells (Fig. (Fig.44 and Table Table2).2). This class of mutants immortalized MECs with a lower efficiency than wild-type E6. For these mutants, immortal clones emerged in only a subset of independent experiments with a lag period not observed with the wild type or with class I mutants (Table (Table2).2). Mutants F2V and Y54H represent a new phenotype (class III) that showed normal p53 levels at both early and late passages (Fig. (Fig.4)4) yet immortalized MECs. These mutants yielded immortal cells in multiple experiments (Table (Table2).2). We examined p53 levels in all individual cell lines. As shown in Fig. Fig.4,4, bottom panel, from the three F2V-derived lines (lanes 3, 8, and 9) and three of the four Y54H-derived lines (lanes 4, 10, and 12) normal p53 levels were detected at late passage, except for one line (lane 11) that showed an intermediate level of p53. To carefully evaluate the efficiency of immortalization by various mutants, viruses with a relatively low titer (2 × 104 PFU) were used to infect MECs in one of the experiments. In this experiment, class III mutants F2V and Y54H immortalized MECs as efficiently as wild-type HPV-16 E6, while class II mutants L37S, L110Q, and W132R failed to immortalize and Q107R and I128T underwent a lag period before yielding immortal cells.
p53 is wild type in E6 mutant-immortalized MECs.
One possibility was that the normal p53 levels observed in F2V- and Y54H-derived late-passage cells were due to selection of a p53 mutation(s) during the immortalization process. To evaluate this, the immortalized cells F2V II and Y54H I (Fig. (Fig.4,4, bottom panel) were subjected to reverse transcription-PCR. Sequencing of the PCR products across the entire coding region revealed a normal p53 sequence in both cell lines. To verify that p53 is functionally competent, late-passage cells immortalized by wild-type HPV-16 E6, mutant F2V, or mutant Y54H were exposed to the DNA-damaging agent actinomycin D, and cell cycle profiles were assessed by flow cytometric analysis of DNA content. Consistent with previous findings, wild-type E6-immortalized cells exhibited analogous cell cycle profiles in the presence or absence of actinomycin D (Fig. (Fig.5A).5A). This indicated abrogation of the DNA damage-induced G1 arrest by E6. In contrast, F2V- and Y54H-derived cells (F2V II and Y54H IV) displayed a G1 arrest in response to actinomycin D treatment (Fig. (Fig.5A)5A) that was similar to that for the normal 76N cells (data not shown). This result indicates that the p53-mediated DNA damage response is retained in these cells. The same experiment was also performed with F2V III and Y54H I cells, and similar G1 arrest cell cycle profiles were observed (data not shown). To further test the function of the p53 protein, cells were transfected with a p53-responsive luciferase reporter, and luciferase activities in response to actinomycin D treatment were measured. As shown in Fig. Fig.5B,5B, the normal 76N cells and the 76N cells immortalized by F2V or Y54H displayed nearly twofold activation of the reporter when exposed to actinomycin D. By contrast, the MECs immortalized by wild-type E6 showed a twofold reduction, and the radiation-immortalized, p53-null 76R-30 (53) MECs showed virtually no change in luciferase activity. These results argue that MECs can proliferate with an unlimited life span yet maintain normal levels of functional p53 protein in the presence of E6.
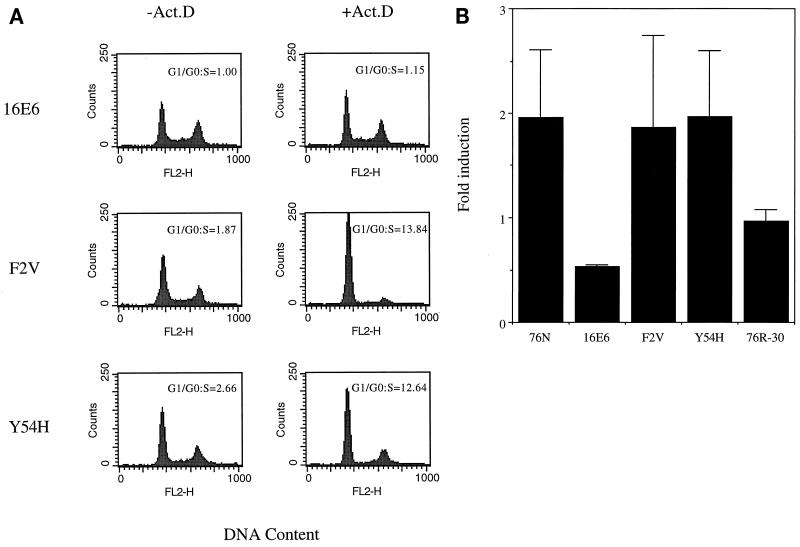
Characterization of p53 in E6 mutant-immortalized MECs. (A) G1 arrest in response to DNA damage. Exponentially growing cells (16E6, F2V II, and Y54H IV) were treated with 0.5 nM actinomycin D (Act.D) for 24 h or left untreated. DNA content was analyzed by propidium iodide staining followed by flow cytometric analysis. The G1/S ratios determined with ModFit software (Becton Dickinson) from a representative experiment are shown. (B) Activation of a p53-responsive reporter in response to DNA damage. Immortal MECs (16E6, F2V II, and Y54H IV), normal 76N cells, and p53-null 76R-30 cells were transfected with a p53-responsive luciferase reporter construct, pPG, along with β-galactosidase and green fluorescence protein reporter constructs. At 40 h posttransfection, the cells were changed to medium containing or lacking 0.5 nM actinomycin D. Twenty-four hours later, cell extracts were made and luciferase and β-galactosidase activities were measured. Luciferase activities were corrected by normalizing β-galactosidase activities. Values shown are the means and standard deviations from three independent experiments, each performed in duplicate.
Immortalization of MECs by E6 mutants that were compromised for p53 binding and/or degradation might have resulted from contamination by wild-type or other mutant E6 during retroviral infection. To exclude this possibility, we PCR amplified the E6-coding sequences from the cellular DNAs isolated from MECs immortalized by F2V, L37S, Y54H, Q107R, L110Q, E114A, F125V, Δ118-122, I128T, W132R, and G134V. DNA sequencing of these PCR products confirmed the presence of the expected mutations and the absence of wild-type or other mutant E6 sequences. Furthermore, we examined representative mutants for E6 expression levels by immunoprecipitation. Wild-type and mutant E6 proteins were confirmed to be expressed at similar low levels in immortal 76R-30 cells (data not shown).
DISCUSSION
The E6–E6-AP complex specifically binds p53 and induces its proteolysis by using components of the ubiquitin system present in RRL (27–29, 44–47). The molecular mechanisms underlying E6-mediated p53 degradation in vivo have not been characterized. Recent experiments using antisense inhibition of E6-AP expression have supported its involvement in E6-mediated, but not E6-independent, p53 degradation in vivo (6). One aim of our investigations was to identify and discriminate the potential roles of HPV-16 E6 in p53 degradation. In addition to the in vitro system with RRL, we employed cultured human MECs as an in vivo system to examine the ability of a series of E6 mutants to induce p53 degradation (this study and reference 16). In addition, we addressed the importance of interactions of E6 with cellular proteins E6-AP, p53, and E6BP in the immortalization of MECs.
A systematic evaluation of E6 mutants for E6-AP binding has not been reported. A limited peptide domain within E6 could not be identified, nor did the E6-AP binding-defective mutations cluster in a specific region. Consistent with previous observations (20), we found that small deletions in the central region and the second zinc finger of the E6 protein, such as Δ73-77, Δ78-82, Δ101-105, Δ106-110, Δ118-122, Δ123-127, Δ128-132, Δ133-137, and Δ138-142, resulted in a dramatic reduction in the ability of HPV-16 E6 to bind E6-AP, E6BP, and p53 and to target p53 degradation in vitro (data not shown). We also identified mutations in the N terminus (F2V) or the first zinc finger of E6 (L37S and Y54D) that greatly reduced binding to E6-AP, E6BP, and/or p53. This suggests that the N-terminal region and the first zinc finger are also important for E6 function, consistent with previous observations (15, 20, 40). The strong correlation between E6-AP and E6BP binding in this series of small deletion and point mutations of E6 was not surprising given that E6-AP and E6BP share an α-helical motif (11, 19). It is highly conceivable that these α-helical motifs make similar contacts with E6.
It is controversial whether E6-AP is absolutely required for the association of E6 with p53. Some investigators (27, 28) but not others (17, 18, 34, 36) have found that E6-AP is necessary for the complex formation between E6 and p53. However, it has recently been demonstrated that the presence of E6-AP is required for E6-mediated degradation of p53 (17, 50). Our genetic and biochemical studies extend these observations by implying that E6-AP association is necessary but not sufficient for p53 binding and degradation. We identified two point mutants of HPV-16 E6, F2V and Y54H, which bound E6-AP at wild-type levels but had greatly reduced binding to p53. Another interesting mutant, Y54D, bound E6-AP at reduced levels but did not bind p53. These results suggest a direct contribution of E6 to the coordination of p53. One likely scenario is that binding to E6-AP exposes the p53 interaction region of E6. This conformational change may be facilitated in GST-E6 fusions, which were reported to bind p53 in the absence of E6-AP (17, 34, 36). The Phe 2 and Tyr 54 mutants may be unable to respond to E6-AP binding or may affect amino acids in E6 that are involved in p53 contact. Another possibility is that HPV-16 E6 may induce a conformational change in E6-AP that reveals its p53 binding domain. This appears to be unlikely, since E6-AP is not thought to be normally involved in p53 catabolism (6, 50).
The separation of p53 binding and degradation properties of HPV-16 E6 has not been clearly established (15, 20). In this study, we demonstrated that binding to p53 is necessary but not sufficient for inducing p53 degradation, in agreement with the conclusions of others (15). Specifically, F2L and Y54H bound p53 at all temperatures tested yet exhibited diminished or defective stimulation of p53 degradation at 37°C. F2V and I128T showed similar p53 binding properties, although F2V was defective while I128T was competent for p53 degradation at 25°C. Furthermore, mutant F125V retained substantial p53 binding ability but was less efficient in promoting p53 degradation at 37°C in vitro than other mutants that bound less p53. Mutants Y84C and E114A, which exhibited low levels of p53 binding, retained the ability to efficiently direct p53 degradation in vitro and in vivo. These observations document that p53 binding is not directly proportional to p53 degradation and can in part be rationalized as a series reactions that mediate the enzymatic degradation of p53 by the E6–E6-AP complex. Our mutational analyses imply that HPV-16 E6 participates in at least one of these steps in addition to p53 binding. This putative function(s) is impaired by mutations of Phe 2 and Tyr 54.
With these mutants and an earlier series of E6 mutants (16), we have observed a correlation between E6-induced p53 degradation in vitro and reduced p53 levels in MECs (Table (Table2).2). The general concordance between the in vitro and in vivo p53 degradation assays suggests that the latter is also an E6-AP-dependent process. However, we have also observed a few exceptions. In addition to mutant Δ118-122 (16, 20), mutants F125V, I128T, and G134V showed reduced levels of p53 in MECs at early passage although they were defective for p53 degradation at 37°C in vitro (Table (Table2).2). These exceptions imply that additional functions may be involved in E6-enhanced p53 turnover in MECs. Alternatively, the in vivo assay may be more sensitive than the in vitro assay. It is conceivable that mutant E6 proteins may be partially misfolded when synthesized in RRL but assume their proper conformation in vivo. In contrast to these mutants, Y54H and F2L showed an absent or reduced ability to induce p53 degradation at 37°C in vitro, and Y54H failed to reduce p53 levels in early-passage MECs, although both mutants bound E6-AP and p53. Consistent with the reduced ability to induce p53 degradation, F2L and Y54H are impaired in their ability to stimulate p53 ubiquitination. In the process of ubiquitination, the C terminus of E6-AP is modified by an activated ubiquitin peptide that is transferred to p53 in the presence of HPV-16 E6 (42). Phe 2 and Tyr 54 may be necessary to catalyze the ubiquitination of p53 by the E6–E6-AP complex or the presentation of the ubiquitinated p53 to the proteasome pathway.
Based on the comparison of p53 levels in early- and late-passage MECs, we have observed three classes of E6 mutants that are able to immortalize MECs (Table (Table2).2). Class I mutants resemble wild-type HPV-16 E6 in causing low p53 levels at both early and late passage and efficient immortalization of MECs. Class II mutants include L37S, Q107R, L110Q, and W132R and show low p53 levels at late passage but do not reduce p53 levels at early passage. This class of mutants immortalizes MECs with lower efficiency, resembling the phenotype observed with low-risk and BPV-1 E6 (2). The reduced p53 half-life observed in the immortal cells that eventually grew out with this class of E6 mutants may occur through selection for loss of p53. Another intriguing possibility is that all E6 proteins may be capable of inducing enhanced p53 turnover, perhaps through activation of a normal cellular pathway. The third possibility is that the E6 proteins of class II mutants are completely misfolded in vitro, but a fraction is properly folded in vivo. This may explain why some mutants of this class are impaired in p53 degradation in vitro and in early-passage MECs but eventually diminish p53 in the immortalized MECs. Class III is the new phenotype observed in the present study. Mutants of this class, i.e., F2V and Y54H, do not reduce p53 levels at early or late passage yet efficiently immortalize MECs. We confirmed the presence of the E6 mutations and wild-type p53 in the immortal cells by DNA sequencing. The p53 protein retained the ability to activate G1 checkpoint arrest and a p53-responsive reporter in response to DNA damage. Given that these two class III mutants bind to p53, although with reduced affinity, it is possible that they alter p53 function through binding in a way leading to MEC immortalization without reducing p53 levels. Interestingly, in contrast to class II mutants, which do not or barely bind E6-AP yet lead to reduced p53 levels in late-passage MECs, class III mutants retain binding to E6-AP but fail to target p53 for degradation in the immortalized MECs. This phenomenon points to the possible existence of a potential cellular target(s) of E6 other than p53; E6–E6-AP-mediated targeting of this cellular factor(s) may be critical for MEC immortalization independent of p53 inactivation. It has recently been reported that HPV-16 E6 induces degradation of Myc proteins through the ubiquitin pathway (24), suggesting that Myc may be a relevant target of E6 in MEC immortalization. However, opposing effects of E6 on Myc through both transcription (31) and posttranscription (24, 52) mechanisms have been reported. Recently a mutant of E6 analogous to class III mutants was reported by other investigators. This mutant, 8S9A10T, efficiently immortalized human MECs that retained normal p53 levels. Interestingly, this mutant retained the ability to activate telomerase, leading to the model that telomerase activation is required for the immortalization of MECs by E6 (32). Paradoxically, another mutant, Δ118-122, was reported to have lost the ability to activate telomerase and immortalize MECs (32). Δ118-122 displayed full immortalization of MECs in our experiments (16) (Table (Table2).2). Sequencing of the E6-coding region from the immortalized MECs revealed the expected deletion and the absence of wild-type contamination. Expression of the Δ118-122 protein in the immortalized MECs was also confirmed (data not shown). We have not tested telomerase activity and telomere length with this mutant in our MECs. Experiments to evaluate the class II and class III E6 mutants described here for telomerase activation will be of interest. In addition to Δ118-122, Kiyono and colleagues (32) described an E6 mutant with a C-terminal truncation, Δ140-151, which was competent for promoting p53 degradation yet failed to immortalize MECs and was defective in telomerase activation.
Based on our data (this study and reference 16) and the data from others (32), HPV-16 E6 appears to utilize multiple functions to immortalize MECs. These include a putative function of growth stimulation in addition to p53 inactivation and telomerase activation. E6 either inactivates p53 or utilizes another function to immortalize MECs when the p53 inactivation function is impaired. The function that bypasses p53 inactivation may be telomerase activation or growth stimulation or both. From the panel of E6 mutants presented here, we have not identified any mutant that binds and degrades p53 but is unable to immortalize MECs.
We also investigated the potential contribution of E6-E6BP association in the immortalization of MECs. The abilities of the E6 mutants to bind E6BP in vitro and in the yeast two-hybrid system were compared with the ability to induce MEC immortalization. We observed a good correlation between E6BP binding and MEC immortalization for the majority of E6 mutants examined. However, several mutants, such as Δ118-122, F125V, I128T, and G134V, showed marginal binding to E6BP yet immortalized MECs with high efficiency. These data argue against the importance of E6BP association in the immortalization of MECs induced by E6. However, we cannot completely rule out the E6BP-binding function of E6 in MEC immortalization, for the following reasons. First, these mutants also showed greatly reduced E6-AP as well as p53 binding ability. The E6–E6-AP complex has an identified cellular target, p53, whereas the target of the E6-E6BP complex, if any, has not been identified. Mutants such as Q107R and H118D, which retained very low levels of binding to E6-AP and E6BP (Fig. (Fig.11 and and2A),2A), induced p53 degradation with high efficiency at 25°C and modest efficiency at 37°C in vitro (Fig. (Fig.2C).2C). Both mutants efficiently immortalized MECs (Table (Table2).2). Analogous to the situation for E6-AP–p53, E6 mutants showing marginal E6BP binding may still retain substantial function of a putative target of E6-E6BP that results in immortalization of MECs. Second, none of the mutants, e.g., F125V, I128T, and G134V, which efficiently immortalized MECs showed an absolute loss of the ability to bind to E6BP. However, mutants L37S and L110Q showed no binding to E6BP in all of the assays employed, yet they immortalized MECs with low efficiency (Table (Table2).2). MECs may not be a suitable system to assess E6BP function. Given that it is a calcium-binding protein (11, 12, 54), E6BP may play a role in HPV-16 E6-mediated resistance to calcium- and serum-induced differentiation of keratinocytes. Previous studies have implied the importance of p53 in this cellular process (14), but MDM-2 and a trans-dominant p53 mutant cannot substitute for E6 for this calcium resistance function, suggesting that activities other than p53 inactivation are also involved (48). Finally, in addition to E6-AP, p53, and E6BP, it should be interesting to address the biological relevance of other E6-binding proteins (22, 24, 33, 43, 51; for a review, see reference 41) by using MECs as a model system. In summary, our results demonstrate that HPV-16 E6 is a multifunctional protein. Interactions with E6-AP, E6BP, or p53 do not account for all of the abilities of E6 to immortalize MECs; other pathways may exist.
ACKNOWLEDGMENTS
Y. Liu and J. J. Chen contributed equally to this work.
We thank S. Fawell for p53 protein; D. Galloway and A. D. Miller for retroviral vectors and the packaging cell line; P. Howley, J. Huibregtse, and K. Vousden for plasmids; and S. Chan, N. Doshi, and C. Lorsen for technical assistance and advice on certain experiments.
This work was supported by NIH grant F32 CA69738-03 to Y.L., NIH grants CA70195 and CA64823 and grants from the Massachusetts Department of Public Health to V.B., NIH grant CA73558 to E.J.A., NIH grant AR01952 to C.P.M., and a Dermatology Foundation Dermik Laboratories Career Development Award to J.J.C.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.73.9.7297-7307.1999
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/73/9/7297.full.pdf
Free after 4 months at jvi.asm.org
http://jvi.asm.org/cgi/content/full/73/9/7297
Free to read at jvi.asm.org
http://jvi.asm.org/cgi/content/abstract/73/9/7297
Free after 4 months at jvi.asm.org
http://jvi.asm.org/cgi/reprint/73/9/7297
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.73.9.7297-7307.1999
Article citations
HPV upregulates MARCHF8 ubiquitin ligase and inhibits apoptosis by degrading the death receptors in head and neck cancer.
PLoS Pathog, 19(3):e1011171, 03 Mar 2023
Cited by: 8 articles | PMID: 36867660 | PMCID: PMC10016708
HPV-Associated Breast Cancer: Myth or Fact?
Pathogens, 11(12):1510, 09 Dec 2022
Cited by: 11 articles | PMID: 36558844 | PMCID: PMC9786769
Review Free full text in Europe PMC
A Drosophila model of HPV16-induced cancer reveals conserved disease mechanism.
PLoS One, 17(12):e0278058, 12 Dec 2022
Cited by: 0 articles | PMID: 36508448 | PMCID: PMC9744332
HPV E6 regulates therapy responses in oropharyngeal cancer by repressing the PGC-1α/ERRα axis.
JCI Insight, 7(18):e159600, 22 Sep 2022
Cited by: 7 articles | PMID: 36134662 | PMCID: PMC9675449
Human Papillomavirus 16 E6 and E7 Oncoproteins Alter the Abundance of Proteins Associated with DNA Damage Response, Immune Signaling and Epidermal Differentiation.
Viruses, 14(8):1764, 12 Aug 2022
Cited by: 6 articles | PMID: 36016386 | PMCID: PMC9415472
Go to all (135) article citations
Protocols & materials
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mutational analysis of human papillomavirus type 16 E6 demonstrates that p53 degradation is necessary for immortalization of mammary epithelial cells.
J Virol, 70(2):683-688, 01 Feb 1996
Cited by: 57 articles | PMID: 8551603 | PMCID: PMC189867
hAda3 degradation by papillomavirus type 16 E6 correlates with abrogation of the p14ARF-p53 pathway and efficient immortalization of human mammary epithelial cells.
J Virol, 82(8):3912-3920, 06 Feb 2008
Cited by: 17 articles | PMID: 18256148 | PMCID: PMC2293003
Human papillomavirus type 16 E6-induced degradation of E6TP1 correlates with its ability to immortalize human mammary epithelial cells.
J Virol, 75(9):4459-4466, 01 May 2001
Cited by: 56 articles | PMID: 11287601 | PMCID: PMC114197
Mechanism of HPV E6 proteins in cellular transformation.
Semin Cancer Biol, 7(6):317-326, 01 Dec 1996
Cited by: 38 articles | PMID: 9284524
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: CA64823
Grant ID: F32 CA69738-03
Grant ID: F32 CA069738
Grant ID: CA70195