Abstract
Free full text

Simian Immunodeficiency Virus Infection of Chimpanzees
Simian immunodeficiency viruses (SIVs) are primate lentiviruses that infect no fewer than 36 different nonhuman primate species in sub-Saharan Africa (4, 9, 62). Two of these viruses, SIVcpz from chimpanzees (Pan troglodytes) and SIVsmm from sooty mangabeys (Cercocebus atys), have crossed species barriers on multiple occasions and have generated human immunodeficiency virus (HIV) types 1 and 2 (20, 24, 25, 94). AIDS, one of the most devastating infectious diseases to have emerged in recent history, is thus the result of cross-species infections of humans by lentiviruses of primate origin (33).
Identification of the primate source that spawned the HIV-1 pandemic is of scientific and public health importance. To this end, it is now well established that the immediate precursor of HIV-1 is a lentivirus that infects chimpanzees of the subspecies Pan troglodytes troglodytes in west central Africa (17, 24). What is also known but perhaps less widely appreciated is that SIVcpz strains have been transmitted to humans on at least three independent occasions and that the current HIV-1 group M pandemic, which has afflicted more than 60 million people and caused more than 20 million deaths, resulted from just one of these transmission events in the first half of the twentieth century (42, 77, 97). The reasons for HIV-1's sudden emergence, the adaptive changes that followed, and the mechanisms underlying its unique pathogenicity in humans have not yet been determined.
Primates naturally infected with SIV, including SIVcpz-infected chimpanzees, appear not to develop immunodeficiency or AIDS (31, 40, 49, 60, 70, 79). In contrast, HIV infection of humans is nearly always characterized by progressive loss of CD4+ T lymphocytes, chronic immune activation, and gradual destruction of an array of immune functions (82). The basis for this difference in pathogenicity is not understood, but deciphering which viral and/or host factors are responsible for the nonpathogenic course of natural SIV infections could well prove useful in developing more-effective treatment or prevention strategies for HIV/AIDS.
Chimpanzees and humans share more than 98% sequence identity across their genomes (91). This degree of relatedness provides a unique opportunity to search for key differences in virus-host interactions responsible for differences in viral pathogenicity, an approach that could prove complementary to current studies that focus primarily on human disease susceptibility (55). In this review, we summarize recent progress in four key scientific areas related to AIDS pathogenesis: (i) the origin of HIV-1; (ii) the origin of SIVcpz; (iii) natural SIVcpz reservoirs; and (iv) the natural history of SIVcpz infection. Scientific and public health implications of these data as they relate to human infections by the corresponding virus, HIV-1, will also be discussed.
THE ORIGIN OF HIV-1
The first suspicion that chimpanzees might be the source of HIV-1 arose when a wild-born chimpanzee from Gabon was found to harbor a lentivirus (SIVcpzGAB1) that was isogenic to HIV-1 (36). That is, the SIVcpzGAB1 genome carried the same nine reading frames as HIV-1, including an accessory gene termed vpu, which had thus far been identified only in HIV-1. In addition, phylogenetic analyses indicated that HIV-1 was more closely related to SIVcpzGAB1 than to any other SIV. But not all data supported the hypothesis that SIVcpz had given rise to HIV-1. First, of 50 wild-caught chimpanzees initially tested for SIVcpz infection, only two (GAB1 and GAB2) harbored HIV-1 cross-reactive antibodies, indicating an unexpectedly low SIVcpz infection rate compared with that of other naturally occurring SIV infections (1, 44, 63). This raised the possibility that chimpanzees and humans had acquired this virus from some still unidentified monkey species representing the true virus reservoir (36). Second, molecular characterization of an SIVcpz strain from a third chimpanzee (Noah) confiscated upon illegal importation to Belgium from Zaire revealed an unexpectedly high degree of genetic divergence compared with the other two SIVcpz strains (GAB1 and GAB2) (36, 37, 87). This virus, termed SIVcpzANT (64), also carried a vpu gene and clustered with SIVcpzGAB1 (and HIV-1 strains) in phylogenetic trees but was twice as distant from SIVcpzGAB1 and HIV-1 strains as they were from each other (87). This degree of sequence diversity had previously been seen only among SIV strains from different primate species, thus again raising the possibility that an unknown reservoir was the source of both SIVcpz infection in chimpanzees and HIV-1 infection in humans. Moreover, 43 other wild-caught chimpanzees from the same study were all antibody negative (64), raising further doubts about chimpanzees as the natural SIVcpz reservoir.
The seemingly contradictory findings concerning SIVcpz diversity were ultimately explained when SIVcpz phylogeny was analyzed in the context of the subspecies origin of the infected chimpanzee host (24). Four different subspecies of chimpanzees are currently recognized on the basis of mitochondrial DNA (mtDNA) sequence differences (27, 30, 48): these include the western Pan troglodytes verus, the Nigerian P. t. vellerosus, the central P. t. troglodytes, and the eastern P. t. schweinfurthii (Fig. (Fig.1).1). When the genome sequence of a fourth SIVcpz strain (SIVcpzUS) was obtained from a chimpanzee (Marilyn) housed in an American primate center, mtDNA analysis was used to determine the subspecies of this ape as well as the three previously identified SIVcpz-infected apes (24). This analysis revealed that the chimpanzees (GAB1, GAB2, and Marilyn) who harbored viruses closely related to each other and to all groups of HIV-1 were all members of P. t. troglodytes (24). In contrast, the ape (Noah) who harbored the phylogenetically divergent SIVcpzANT strain was identified as a P. t. schweinfurthii. These data suggested that SIVcpz strains cluster in two divergent phylogenetic lineages (SIVcpzPtt and SIVcpzPts) according to the subspecies origin of their chimpanzee hosts. In addition, the same phylogenetic data argued that the central subspecies (P. t. troglodytes) was the source of HIV-1.

Natural range of chimpanzee subspecies and location of SIVcpz isolation (adapted from reference 75). The locations of study sites in the Budongo Forest Reserve and Kibale National Park (Uganda), the Nyungwe Forest Reserve (Rwanda), Gombe and Mahale Mountains National Parks (Tanzania), and the Kisangani area (eastern Democratic Republic of Congo [DRC]) are indicated, as are the rescue locations of five previously reported SIVcpz-infected captive chimpanzees from southern Cameroon and northern Gabon (51). Asterisks indicate the presence of natural SIVcpz infection. The four recognized chimpanzee subspecies are color-coded. International borders (black lines) and major rivers (blue lines) are shown. Kinshasa, which marks the area where the greatest diversity of HIV-1 group M strains has been found (89), is also shown.
Detailed phylogenetic analyses of HIV-1 diversity in west equatorial Africa provided further evidence for a P. t. troglodytes origin of HIV-1. It was shown that HIV-1 strains fall into three highly divergent clades, groups M, N, and O (32, 46, 65, 80), each of which was more closely related to SIVcpz from P. t. troglodytes than from P. t. schweinfurthii (33, 78). Remarkably, these three HIV-1 groups were not each other's closest relatives but instead were interspersed with SIVcpz within the HIV-1/SIVcpz radiation (Fig. (Fig.2).2). This finding indicated that there had been three separate introductions of SIVcpz into the human population, all from the central subspecies of chimpanzee (24, 78). Consistent with this interpretation was the observation that HIV-1 groups N and O are largely restricted to west equatorial Africa (group N has been reported only in Cameroon [2, 11]), coincident with the natural range of the central chimpanzee subspecies. HIV-1 group M, of course, has spread globally; yet the greatest extent of HIV-1 group M diversity has been reported in Kinshasa (89), suggesting that the epidemic had its beginnings in this geographic region (69). While wild chimpanzees are not found in the immediate vicinity of Kinshasa, this urban area is not far from the range of central chimpanzees (Fig. (Fig.11).

Evolutionary relationships of SIVcpz and HIV-1 strains based on maximum-likelihood phylogenetic analyses of full-length envelope protein sequences (adapted from ref. 10). SIVcpz strains from P. t. troglodytes and P. t. schweinfurthii are highlighted in red and blue, respectively. Representative strains of HIV-1 groups M, N, and O were included for comparison. Asterisks indicate internal branches with estimated posterior probabilities of 95% or higher. The scale bar denotes 10% replacements per site.
The close phylogenetic relatedness between HIV-1 and SIVcpz notwithstanding, the low frequency of SIVcpz detection in captive chimpanzees was still unexplained. Additional surveys of both wild-caught and wild-living chimpanzee populations, however, began to identify new SIVcpz strains, suggesting that natural infection is not as rare as originally thought. Four SIVcpz strains (CAM3, CAM4, CAM5, and CAM13) were obtained from orphaned chimpanzees in Cameroon (17, 50, 51), while additional cases of SIVcpz infection were documented in wild-living chimpanzee communities in Tanzania (TAN1 to TAN5) and the Kisangani area of the Democratic Republic of Congo (72, 73, 75, 93). Moreover, the existence of two separate clades of SIVcpz, infecting members of the central (SIVcpzPtt) and eastern chimpanzee (SIVcpzPts) subspecies, was confirmed. Figure Figure33 shows that SIVcpz strains from chimpanzees in Tanzania and the Democratic Republic of Congo, all of whom were identified as P. t. schweinfurthii by mtDNA analyses, are most closely related to SIVcpzANT (75, 93). Conversely, three newly identified Cameroonian viruses group most closely with other SIVcpzPtt strains, and their hosts were typed as P. t. troglodytes (10, 17, 50). Interestingly, one SIVcpz-infected ape (CAM4) was classified as a P. t. vellerosus, but on further investigation it was found that CAM4 had been housed with CAM3 and that their viruses were virtually identical (17). Thus, it is most likely that SIVcpzCAM4 does not represent a true P. t. vellerosus virus but rather a SIVcpzPtt strain acquired by cage transmission in captivity (17). Thus, all existing evidence points to naturally infected P. t. troglodytes apes as the natural reservoir for HIV-1.
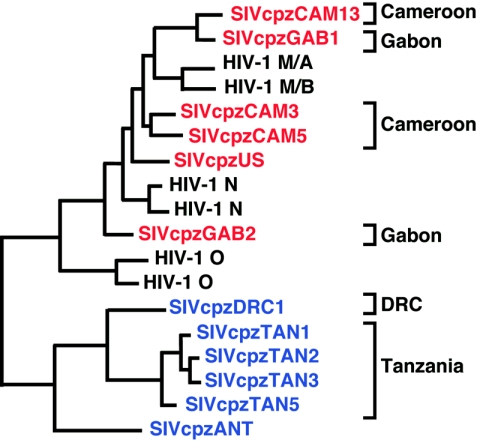
Phylogeography of SIVcpz (adapted from ref. 51). The phylogenetic relationships of available SIVcpzPtt (red) and SIVcpzPts (blue) strains are shown in a partial envelope region (180 amino acids). Brackets on the right indicate the country of origin of the various SIVcpz strains; SIVcpzUS and SIVcpzANT are of unknown geographic provenance.
THE ORIGIN OF SIVcpz
But if HIV-1 arose from cross-species transmission of SIVcpz, where did SIVcpz originate? To answer this question, investigators had to look to the 70 other species of primates inhabiting sub-Saharan Africa, more than half of which are known to harbor SIV-related lentiviruses (4, 9, 62). All primate lentiviruses so far identified fall within a single phylogenetic lineage distinct from lentiviruses from other species of mammals (4). Within this primate lentivirus radiation, each primate species generally has its own type of SIV, such that multiple strains from any one host species form a monophyletic clade. An exception is the mandrill, Mandrillus sphinx, which harbors two phylogenetically divergent types of SIV (81, 83). In some cases, closely related monkey species harbor closely related SIVs. Examples include L'Hoest's and suntailed monkeys, mandrills and drills, and the four species of African green monkeys (genus Chlorocebus) (1, 7, 38, 83). This has led to the hypothesis that some SIVs may have been coevolving with their hosts for an extended period of time, such that ancestors of today's monkey species were infected with ancestors of their respective SIVs (1, 7, 38). However, this hypothesis is controversial, largely because attempts to estimate the time depth of the primate lentivirus phylogeny using molecular clocks have produced time estimates several orders of magnitude more recent than the comparable ancestry of the monkey species (34, 77). Moreover, there are many examples of cross-species transmission and recombination, indicating that many SIVs have spread between different primate species (38, 81, 83). Thus, current data indicate that the evolutionary history of primate lentiviruses is complex, involving virus/host coevolution as well as interspecies transmission, with the timelines and directions of the latter remaining to be deciphered (3, 9, 76).
While certain SIVs may have coevolved with their primate hosts, this appears not to be the case for SIVcpz. Two chimpanzee subspecies, the central P. t. troglodytes and eastern P. t. schweinfurthii, are naturally infected by SIVcpz, but a third one, the western P. t. verus, is not (Fig. (Fig.1).1). Extensive studies of over 1,500 captive apes of either known P. t. verus (by mtDNA analysis) or west African origin (based on capture records) failed to uncover any evidence of SIVcpz infection (68, 84), and the absence of SIVcpz was further confirmed in 28 members of a wild-living P. t. verus community (72). Chimpanzees thus appear to have acquired SIVcpz by cross-species transmission sometime after their divergence into different subspecies. That SIVcpz infection occurred relatively more recently is also supported by the fact that the other great apes, including the gorilla (Gorilla gorilla) and the chimpanzee's closest relative, the bonobo (Pan paniscus), seem to be free of SIV infection (44, 88) (M. Peeters, B. Keele, and B. H. Hahn, unpublished data). Thus, existing evidence suggests that chimpanzees acquired SIVcpz long after the divergence of Pan troglodytes from the other ape species and even long after the split of the western P. t. verus subspecies from the central and eastern subspecies, estimated to have occurred about 1.5 million years ago (48).
Until recently there has been no indication as to what might have been the source of chimpanzees' newly acquired infection, because in evolutionary trees SIVcpz strains formed a lineage that was not particularly closely related to any of the other known SIVs (78). However, this changed with the isolation of viruses from red-capped mangabeys (Cercocebus torquatus) and greater spot-nosed monkeys (Cercopithecus nictitans), each of which exhibited similarity to SIVcpz but in different parts of their genomes. SIVgsn was the first monkey virus identified to contain a vpu gene, similar to SIVcpz. Moreover, SIVgsn was found to be closely related to SIVcpz but only within the 3′ half of its genome (19). In the 5′ region of the genome, SIVcpz was found to be most closely related to SIVrcm (6). Such discordant phylogenetic relationships are generally indicative of recombination during the evolution of viruses (38, 76, 81, 83). Both SIVrcm and SIVgsn were initially believed to have been the product of recombination events involving SIVcpz and other as yet unidentified SIV lineages (6, 19). But since the regions of similarity between SIVcpz and the two other viruses did not overlap and since SIVrcm and SIVgsn were only distantly related throughout their genomes, the more plausible interpretation was that SIVcpz is the product of recombination between ancestors of SIVrcm and SIVgsn. Extensive phylogenetic analyses of all major SIV lineages, including a comprehensive analysis of topologies obtained for four genomic subdivisions, ultimately demonstrated that SIVcpz is indeed the most likely recombinant (3).
More recent studies of SIVs from mustached monkeys (Cercopithecus cephus) and mona monkeys (Cercopithecus mona) have shown that both form a phylogenetic clade together with SIVgsn (5, 18). In the 3′ region of their genome (including the env gene), the three Cercopithecus strains are closely related to each other, but they also cluster with SIVcpz (Fig. (Fig.4A).4A). In the 5′ region of their genome (including the pol gene), SIVmus, SIVmon, and SIVgsn are similarly closely related, but SIVcpz again clusters with SIVrcm. Thus, the phylogenetic positions of SIVcpz in Fig. Fig.4A4A are consistent with a comparatively recent origin of this virus by recombination between (i) an ancestor of the SIV now infecting red-capped mangabeys and (ii) an ancestor of the SIVs found in greater spot-nosed, mustached, and mona monkeys. Within the region between pol and env, the vif and vpr genes of SIVcpz are SIVrcm-like, whereas the vpu gene must have originated from the SIVgsn/SIVmus/SIVmon ancestor (Fig. (Fig.4B).4B). Thus, one recombination crossover likely occurred in the short region between vpr and vpu (the 5′ exons of tat and rev lie within this region but are too short to be phylogenetically informative). However, the recombination event that generated SIVcpz may have involved more than just exchanging genome halves, since SIVrcm encodes a vpx gene 5′ of its vpr gene, whereas no remnant of this gene is apparent in SIVcpz (Fig. (Fig.4B4B).

Recombinant origin of SIVcpz. (A) Maximum-likelihood phylogenies of primate lentivirus Pol and Env sequences (adapted from ref. 9). The close genetic relationship of SIVcpz to SIVrcm from red-capped mangabeys in Pol, and to SIVgsn, SIVmus, and SIVmon from greater spot-nosed, mustached, and mona monkeys in Env, are highlighted in green and magenta, respectively. (B) Schematic diagram of the genomic organization of SIVcpz. Genomic regions are colored according to their genetic relationship to SIVrcm (green) or the SIVgsn/SIVmus/SIVmon lineage (magenta). Grey areas in SIVcpz are of unknown origin. Vpu and vpx genes are highlighted.
Given the complex evolutionary history of SIVcpz, the question arises as to how this virus emerged as a new infectious agent in chimpanzees. One explanation could be that chimpanzees acquired both ancestral viruses, which then recombined in that host. Chimpanzees are known to prey on monkeys (Fig. (Fig.5).5). Goodall was the first to document that wild chimpanzees hunt smaller monkeys for food during her pioneering field work in Gombe National Park (28), and this behavior has since been reported in ape communities throughout equatorial Africa (12, 47, 86, 92). It is not difficult to envisage that such hunting provided an opportunity for transmission of blood-borne viruses from monkeys to chimpanzees. It is possible that the two monkey SIVs that gave rise to SIVcpz were transmitted independently to different chimpanzees and that each spread separately in the chimpanzee population before they had an opportunity to recombine in an ape coinfected with both viruses. Alternatively, only one of the monkey viruses may have established itself as a chimpanzee virus and the recombinant may have arisen when a chimpanzee infected with that virus acquired a second SIV, again through predation. In either case, the recombinant eventually replaced any initial monkey SIV infection(s), since the genomes of all known SIVcpz strains exhibit an identical mosaic structure, regardless of their geographic origin. Still another possibility is that the recombinant virus was generated in a monkey host and was subsequently transmitted to chimpanzees; however, this explanation is less likely, since there is no evidence of such a recombinant in today's Cercopithecus monkeys or red-capped mangabeys.

Chimpanzees hunt and eat other primates. An adult male chimpanzee from the Ngogo community (Kibale National Park, Uganda) feeds on the arm of a female red colobus monkey (photograph courtesy of John Mitani).
Today the ranges of red-capped mangabeys and the three Cercopithecus monkey species overlap each other, as well as that of P. t. troglodytes, in west central Africa (30). While these ranges may have been different in the past, the current geographical relationships would suggest that the early stages of SIVcpz evolution, i.e., the transmission of two different monkey SIVs to chimpanzees and their subsequent recombination, occurred in the central subspecies. It is not known whether this happened before the split of the eastern subspecies, which has been inferred from chimpanzee mtDNA data to have arisen comparatively more recently by eastward expansion of P. t. troglodytes (23, 26). Thus, it is not clear whether SIVcpz infected central chimpanzees prior to this event or whether SIVcpz spread later from the central subspecies to an already established eastern subspecies. Under either scenario, one might expect SIVcpzPts strains from eastern chimpanzees to form a clade within the radiation of SIVcpzPtt strains from central chimpanzees, mirroring the mtDNA phylogeny of their hosts. However, SIVcpz isolates from central and eastern chimpanzees form two distinct monophyletic clades (Fig. (Fig.22 and and3),3), indicating that these two subspecies have been effectively isolated in recent times. It is quite possible that the current lack of SIVcpz strains that outflank existing SIVcpzPtt and SIVcpzPts lineages is the result of sampling bias, since all SIVcpzPtt strains of known geographic origin were identified in either Gabon or Cameroon (Fig. (Fig.1).1). Alternatively, some degree of lineage sorting may have taken place, occurring more rapidly in SIVcpz than in the mtDNA of the chimpanzee hosts. While this sorting could have been the result of a selective sweep within SIVcpz-infected central chimpanzees, this explanation is not required. Coalescence theory indicates that, in the absence of selection, the time back to a single common ancestor is a function of the effective population size (35). The effective population size of SIVcpz in central chimpanzees is likely to have been much lower than that of the host, simply because the prevalence of SIVcpz is much less than 100%. Thus, it need not be surprising that there is a mismatch between the phylogeny of SIVcpz strains and that of the hosts. In conclusion, current evidence indicates that not only HIV-1 but also its chimpanzee precursor are of west central African origin.
NATURAL SIVcpz RESERVOIRS
Of the four subspecies of chimpanzees, only two are known to harbor SIVcpz naturally, and even then, unexpectedly few apes were initially found to be virus positive. These observations have continued to raise questions as to whether chimpanzees represent a natural SIV reservoir. Much of the problem in evaluating chimpanzees as a potential reservoir relate to their endangered status, remote and fragmented habitat, reclusive nature, and declining numbers. Wild chimpanzees live in communities of 5 to 150 individuals in isolated forest regions, and they avoid human contact except for the rare instances when they have been habituated to human observers (12, 28, 52). Chimpanzee populations are also rapidly declining due to relentless poaching, habitat destruction, and infectious diseases (14, 90). Finally, it is unethical and unsafe (for both apes and humans) to tranquilize wild chimpanzees for investigative purposes. In response to these challenges, our laboratory has developed methods to identify SIVcpz-specific nucleic acids and antibodies in chimpanzee fecal and urine samples collected noninvasively from the forest floor (72, 75, 93).
Noninvasive SIVcpz testing entails the screening of chimpanzee fecal and urine samples for the presence of virus-specific antibodies, using an enhanced chemiluminescence Western blotting approach followed by virion RNA extraction and reverse transcriptase PCR (RT-PCR) amplification of viral sequences to confirm and molecularly characterize SIVcpz infection (72, 75). In a pilot study of SIVcpz-infected captive chimpanzees, antibodies were found (by Western immunoblot analysis) in 63% of fecal and 100% of urine samples, suggesting that this approach was of sufficient sensitivity and specificity for field testing (72, 75). The mucosal antibodies were of the immunoglobulin G class and were directed against multiple viral proteins; samples from uninfected chimpanzees did not exhibit nonspecific or false-positive Western blotting reactivities (Fig. (Fig.6).6). A proportion (56%) of fecal samples also contained viral nucleic acids in the form of virion RNA (vRNA). Although the sensitivity of fecal vRNA detection was lower than that for antibodies, the fact that all SIVcpz-infected chimpanzees studied had at least one RT-PCR-positive fecal sample suggested that vRNA analyses could be useful for molecular confirmation of infection as well as for screening, especially if multiple samples from the same individuals could be collected and tested. In contrast, vRNA detection in urine was rare and thus not useful for diagnostic purposes: only 10% of urine samples were vRNA positive, suggesting that virus particles are not normally passed through intact kidney epithelium.
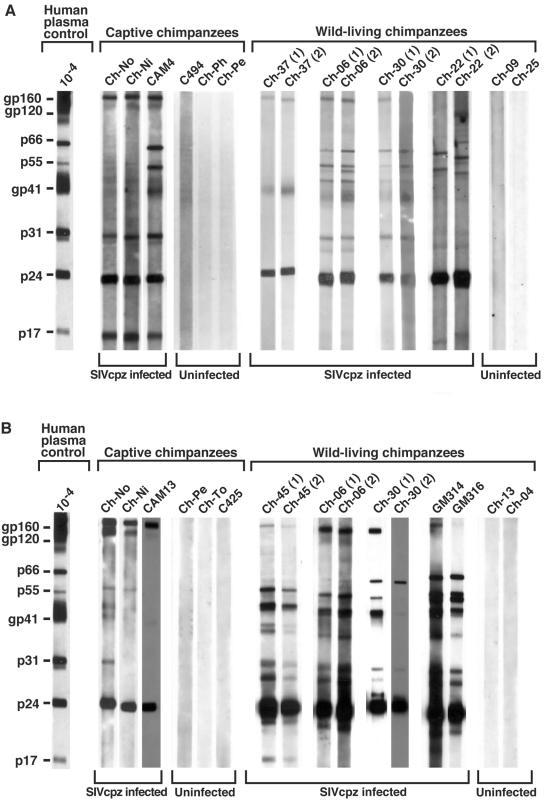
Detection of SIVcpz-specific antibodies in chimpanzee urine and fecal samples obtained by noninvasive methods. Urine (A) and fecal (B) specimens from captive and wild-living chimpanzees were tested by ECL Western immunoblot analysis for the presence of virus-specific antibodies. (Data for captive chimpanzees was adapted from Fig. Fig.22 in reference 75.) Chimpanzees are identified by code number, with serial samples identified in parentheses. All wild-living chimpanzees are from Gombe National Park. Ch-37 and Ch-45 are members of the Mitumba community, and Ch-06, Ch-30, and Ch-22 are members of the Kasekela community. Fecal samples GM314 and GM316 are from unknown members of the nonhabituated Kalande community. Molecular weights of HIV-1-specific proteins are indicated. The banding patterns of plasma from an HIV-1-infected human (analyzed at a dilution of 1:10,000) is shown as a positive control.
Noninvasive testing of chimpanzee fecal and urine samples for virus-specific antibodies and nucleic acids thus made possible, for the first time, field studies of SIVcpz prevalence and molecular epidemiology (72, 73, 75, 93) and has even been extended to SIV detection in other primate species (43, 74). The first SIVcpz-infected wild-living chimpanzee (P. t. schweinfurthii) was identified in Gombe National Park (72), which in turn led to additional surveys of resident communities in this same area (Fig. (Fig.6)6) (75). Gombe National Park is located on the shores of Lake Tanganyika (Fig. (Fig.1)1) and is home to three chimpanzee communities (28). The largest community, Kasekela, has been studied by Goodall and colleagues since the 1960s and currently comprises about 55 apes. The neighboring Mitumba and Kalande communities are smaller, each comprised of only about 20 apes. An initial survey of 76 of the 95 (or so) Gombe chimpanzees yielded prevalence estimates of 17, 5 and 30% for Mitumba, Kasekela, and Kalande, respectively (75). SIVcpz infection could be confirmed by urine antibody and/or fecal vRNA analysis in seven of these individuals, and two additional apes exhibited urine antibody profiles suggestive of infection (75). More recent studies of the entire Gombe chimpanzee population with more sensitive methods identified 11 additional infections, suggesting an overall SIVcpz prevalence throughout Gombe of about 20% (B. Keele, M. L. Santiago, and B. H. Hahn, unpublished data). Most of the new infections were identified in the Kalande community. Since these chimpanzees are not habituated, it was necessary to characterize host DNA by using highly polymorphic microsatellite loci to determine which samples came from the same individual. Thus far, 8 of 18 chimpanzees in this community have been found to harbor SIVcpz (Keele, Santiago, and Hahn, unpublished). These field studies in Gombe thus reveal, for the first time, a prevalence of SIVcpz infection that begins to approximate that found in other primates naturally infected by SIVs (61). Moreover, with approximately half of the Kalande community members infected, it would appear as if hotspots of SIVcpz infection might exist. The dynamics of SIVcpz infection in this and other endemically infected ape communities requires further study.
To determine the prevalence of SIVcpz infection in other areas of East Africa, field surveys were conducted in wild-living ape communities north and south of Gombe National Park (75, 93; M. L. Santiago, M. Emery, N. Gross-Camp, V. Reynolds, M. Huffman and B. H. Hahn, unpublished data). These included the Kanyawara and Ngogo communities, both in Kibale National Park in western Uganda; habituated chimpanzees in the Budongo Forest, in northern Uganda; and members of the M group in Mahale Mountain National Park in southern Tanzania (Fig. (Fig.1).1). Surprisingly, this screening effort, which involved the collection and examination of samples from more than 180 known individuals, failed to uncover a single SIVcpz-infected chimpanzee. In addition, more than 300 fecal samples collected from nonhabituated chimpanzees in the Nyungwe Forest Reserve in western Rwanda were also found to be negative for SIVcpz antibodies and virion RNA. In contrast, SIVcpz infection was documented in wild apes in the eastern Democratic Republic of Congo (Fig. (Fig.1).1). A limited survey of forested areas in the vicinity of Kisangani documented the presence of SIVcpz infection in two of three field sites (93). In one instance, the infecting viral strain (DRC1) was characterized by vRNA analysis and shown, as expected, to fall within the phylogenetic radiation of other SIVcpzPts strains (Fig. (Fig.3).3). Thus, SIVcpz seems to be broadly, but unevenly, distributed across East Africa, with endemic foci of infection in some communities and rare or absent virus in others. In this context, it should be noted that the communities seemingly free of SIVcpz infection (Budongo, Kibale, Nyungwe, and Mahale Mountains) are all located at the easternmost limits of the P. t. schweinfurthii range (Fig. (Fig.1),1), where habitat loss and fragmentation are particularly severe. This finding raises the possibility that SIVcpz may have been present there in the past but may have since become extinct. Alternatively, the eastward spread of SIVcpz may have been influenced by biogeographical barriers, such that certain communities were never exposed to this virus. More-extensive sampling of chimpanzee communities in the Democratic Republic of Congo will be necessary to address these questions.
Noninvasive testing for SIVcpz infection in wild-living chimpanzees from west central Africa are only beginning, primarily because of a lack of established field sites with appropriate research infrastructure. However, over the past decade more than 100 wild-borne P. t. troglodytes and 50 P. t. vellerosus apes have been screened in wildlife centers and sanctuaries (50, 51, 63, 64). Although only a few naturally infected P. t. troglodytes apes were identified, molecular characterization of their viruses has yielded important insight into the evolutionary history of SIVcpz in this geographic area. First, SIVcpz infection has been identified in captive members of the P. t. troglodytes but not the P. t. vellerosus subspecies (51). While this finding does not exclude the possibility that P. t. vellerosus apes harbor SIVcpz, the data suggest that natural infection, if it occurs, is rare. Second, SIVcpzPtt strains from geographically neighboring areas in southern Cameroon or northern Gabon were found to exhibit a surprising degree of genetic diversity, suggesting a longstanding infection in this area (10, 51). Finally, phylogenetic analyses provided evidence of ancient recombination among members of the SIVcpzPtt lineage, at a time when the precursors of HIV-1 groups M, N, and O were still infecting chimpanzees (10, 51). This evidence implies that divergent SIVcpz strains must have circulated at sufficiently high prevalence rates in P. t. troglodytes populations that individual apes became coinfected, since that would be required for recombination to occur. Taken together, these data suggest that P. t. troglodytes represent a long-standing SIVcpz reservoir. The scarcity of SIVcpz in captive members of this subspecies does not argue against this conclusion, since the majority of wild-caught apes are acquired as infants at a very young age (51, 63). Thus, such apes likely acquire their infection by mother-to-infant transmission (51). The frequency of vertical transmission in chimpanzees is not known, but data from one naturally SIVcpz-infected breeding female (Marilyn) suggests that it may be rare (84). This is also true for other primate species that are naturally infected by SIV, e.g., African green monkeys and sooty managbeys (22, 66), and even for humans: only about one-third of HIV-1-infected women transmit virus to their infants in the absence of antiretroviral therapy (67, 71). Thus, studies of captive orphans are unlikely to yield accurate estimates of SIVcpz prevalence rates in the wild (51).
In summary, natural SIVcpz infection has been documented in wild (or wild-born) chimpanzees from Cameroon, Gabon, the Democratic Republic of Congo, and Tanzania, indicating that infection of both P. t. troglodytes and P. t. schweinfurthii subspecies is endemic across equatorial Africa. P. t. verus does not seem to be infected for reasons outlined above, and the infection status of P. t. vellerosus is unknown. SIVcpz prevalence rates are only beginning to be determined in wild populations, but in the eastern communities that have been studied, prevalence estimates range from 0 to 30% (75). In addition, there is some evidence to suggest that SIVcpz prevalence rates may be higher in west central than in East Africa; however, a systematic and comprehensive analysis of the molecular epidemiology of SIVcpz will require additional field surveys of nonhabituated chimpanzee populations, especially in west central Africa. Such studies are now feasible, since new technologies have been developed especially for this purpose. These include genotyping methods for species confirmation and individual identification (utilizing highly polymorphic microsatellite loci) as well as methods to preserve fecal antibodies in the absence of refrigeration (B. Keele and B. H. Hahn, unpublished data). These technical innovations will permit more-extensive field studies than have been possible in the past, especially at remote field sites.
THE NATURAL HISTORY OF SIVcpz INFECTION
Little is known about the natural history of SIVcpz infection since only very recently has it become possible to identify and observe infected apes in the wild. Even in captive chimpanzees, only seven naturally infected individuals have been identified (17, 24, 51, 63, 64), and among these, only one (Noah) has been subjected to virological and immunological analyses (13, 40, 41, 54, 56-58). Observational studies of both captive and wild-living apes seem to suggest that SIVcpz-infected chimpanzees do not become noticeably immunodeficient or succumb prematurely to opportunistic infections or neoplasms, although an obvious caveat here is that it is difficult in field studies to distinguish between these and other natural causes of death.
Whether chimpanzees, like humans, experienced an outbreak of an AIDS-like disease when they first acquired SIVcpz from their prey species is not known but has been a subject of speculation. In a recent study, de Groot and colleagues found that chimpanzees have a restricted array of major histocompatibility complex (MHC) class I alleles in comparison to that of humans; that is, the MHC class I loci (HLA-A, HLA-B, and HLA-C) in humans exhibit a much higher degree of allele diversity than do the orthologous loci in chimpanzees (Patr-A, Patr-B, and Patr-C) (21). Since this loss of allelic diversity is not present at other loci within the chimpanzee genome (39, 95), de Groot and colleagues argued that it likely reflected a “selective sweep” (i.e., elimination) of chimpanzees caused by an ancient infection with a pathogenic virus. The authors went on to speculate that this pathogen was SIVcpz (or a closely related SIV) and that today's chimpanzees are immune to SIV/HIV disease because apes who were inherently susceptible to disease on the basis of particular MHC (or linked) genes were eliminated during the initial devastating infection with this virus. If this scenario were true, it would not only begin to explain the apparent lack of SIVcpz pathogenicity in chimpanzees today but it could also foreshadow potential events to come for human populations infected by HIV-1 (16). But there are problems with the de Groot hypothesis. First, reduced MHC diversity was identified in western (P. t. verus) as well as central and eastern (P. t. troglodytes and P. t. schweinfurthii) chimpanzees. If such a sweep were caused by SIVcpz, all chimpanzee subspecies would be expected to show evidence of infection, not just P. t. troglodytes and P. t. schweinfurthii. Contrary to this prediction, there is no evidence of natural SIVcpz infection in over 1,500 P. t. verus apes (68, 72, 84). Second, the reduced MHC diversity observed in the common chimpanzee (Pan troglodyte) also appears to be present in bonobos (Pan paniscus), placing the timing of the selective sweep even before chimpanzee speciation; again, there is no evidence that bonobos are naturally infected with SIVcpz or a related SIV (88). Thus, while it is possible that some pathogen led to a widespread infection, followed by the elimination of certain MHC-linked loci in the ancestors of present day chimpanzees and bonobos 2 to 3 million years ago, it is highly unlikely that this agent was SIVcpz or a closely related SIV.
We are thus left with little information regarding the natural history and pathogenesis of SIVcpz infection in chimpanzees. What insight we do have, by necessity, has come from laboratory analyses of SIVcpz-infected captive chimpanzees and ex vivo analyses of their viruses and lymphoreticular tissues. For example, studies of several SIVcpz isolates indicate that, like HIV-1, they use CD4 and CCR5 as their main receptor and coreceptor for entry into target cells (10, 50, 58). For reasons that remain to be explained, it appears that chimpanzee peripheral blood mononuclear cells (PBMCs) are considerably less susceptible to SIVcpz (and HIV-1) infection in vitro than are human PBMCs (8, 57). In this context, it is of interest that chimpanzee and human CD4 and CCR5 molecules differ at several potentially significant amino acid residues (15, 85, 96). In vivo, SIVcpz establishes a persistent infection and replicates to relatively high titers (104 to 105 viral RNA copies/ml) (58) but does not seem to be associated with CD4 cell decline or loss of CD4 cell function (40), increased apoptosis of CD4+ T-lymphocytes (29), or extensive trapping of virus particles in germinal centers and degenerative changes in lymph node architecture (41). In this way, SIVcpz seems to resemble other nonpathogenic lentiviral infections, e.g., those of sooty mangabeys and African green monkeys (49, 60, 70, 79), although an obvious caveat of these studies is that the in vivo chimpanzee data are derived from only a single naturally infected individual. Many more chimpanzees will need to be studied to characterize the biology and natural history of this virus. Of note, chimpanzees experimentally infected by HIV-1 are not equivalent to apes who acquired SIVcpz by natural routes. In most experimentally infected chimpanzees, HIV-1 replicates poorly, frequently at undetectable levels (40, 53). Serial passage in chimpanzees can serve to increase this in vivo replication potential, but chimpanzee-adapted strains of HIV-1 cause CD4 T-cell decline and clinical AIDS, symptoms not observed in naturally infected apes (53, 59). Thus, meaningful data of SIVcpz natural history will require analyses of chimpanzees who have been infected by natural routes.
CONCLUSIONS
Although SIVcpz was first identified in chimpanzees in 1990 (36) and subsequently shown to be the source of human infection by HIV-1 (24), it is only very recently that investigators have begun to appreciate the complexities of the evolutionary origins, geographic distribution, prevalence, natural history, and pathogenesis of this virus. The preceding sections summarize what is currently known concerning the origins of HIV-1 groups M, N, and O; the evolutionary history of SIVcpz; the molecular epidemiology of distinct SIVcpz lineages in P. t. schweinfurthii in East Africa and P. t. troglodytes in west central Africa; and the most basic biological properties of SIVcpz in vivo and ex vivo. What is evident from this discussion is how remarkably limited our current understanding is in each of these areas. We would argue that one of the most important yet underdeveloped areas of scientific investigation that could contribute importantly to human AIDS research—including vaccine and therapeutic drug development—is a comprehensive analysis of SIVcpz infection in its natural host.
Important questions remain unanswered regarding the precise geographical origins of HIV-1 groups M, N, and O and the natural SIVcpz reservoirs that were their source. Virtually nothing is known about the biology of SIVcpz infection in chimpanzees and, even more importantly, the circumstances under which SIVcpz was transmitted to humans and became an epidemically spreading pathogen. It has been argued that adaptive changes were necessary for SIVcpz to become pathogenic and to spread within the human population (45), but there is no direct evidence to support either contention. If indeed SIVcpz is nonpathogenic in chimpanzees, then elucidating the molecular basis for this difference between SIVcpz infection of apes and HIV-1 infection of humans demands attention. And if adaptive changes in SIVcpz were required, either for epidemic spread or to confer pathogenicity in humans, then the identification of such changes at a molecular level could well lead to a deeper understanding of HIV-1 transmission and pathogenesis as well as to novel strategies for preventive or therapeutic intervention. Just as human cancer therapy has shifted to include strategies to achieve long-term control (as opposed to cure), so too could HIV/AIDS therapy encompass strategies to modify HIV-1 infection from a pathogenic illness to a nonpathogenic chronic infection. Given the genetic similarities between HIV-1 and SIVcpz and between humans and chimpanzees, this goal could well be within striking distance.
Acknowledgments
We thank John Mitani for the photograph of a wild chimpanzee from Kibale National Park and Wendy J. Abbott for artwork and manuscript preparation.
This work was supported in part by grants from the National Institutes of Health (R01 AI50529, R01 AI44596, R01 AI58715, and NO1 AI85338 to B.H.H. and P.M.S.) and the Howard Hughes Medical Institute (to G.M.S).
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.79.7.3891-3902.2005
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/79/7/3891.full.pdf
Citations & impact
Impact metrics
Article citations
Can ferritin be a surrogate marker for CD4 cells in human immunodeficiency virus patients? A cross-sectional study of association of serum ferritin levels with immunological staging of human immunodeficiency virus patients.
Indian J Sex Transm Dis AIDS, 45(1):31-33, 01 Jan 2024
Cited by: 0 articles | PMID: 38989078
Genetic adaptations to SIV across chimpanzee populations.
PLoS Genet, 18(8):e1010337, 25 Aug 2022
Cited by: 1 article | PMID: 36007015 | PMCID: PMC9467346
Analysis of evolutionary and genetic patterns in structural genes of primate lentiviruses.
Genes Genomics, 44(7):773-791, 05 May 2022
Cited by: 1 article | PMID: 35511321 | PMCID: PMC9068864
HIV and FIV glycoproteins increase cellular tau pathology via cGMP-dependent kinase II activation.
J Cell Sci, 135(12):jcs259764, 21 Jun 2022
Cited by: 5 articles | PMID: 35638570 | PMCID: PMC9270957
Close-up: HIV/SIV intasome structures shed new light on integrase inhibitor binding and viral escape mechanisms.
FEBS J, 288(2):427-433, 22 Jun 2020
Cited by: 6 articles | PMID: 32506843 | PMCID: PMC7719574
Review Free full text in Europe PMC
Go to all (92) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
AIDS virus traced to chimp subspecies.
Science, 283(5403):772-773, 01 Feb 1999
Cited by: 1 article | PMID: 10049113
Molecular screening for HIV-1 group N and simian immunodeficiency virus cpz-like virus infections in Cameroon.
AIDS, 14(6):750-752, 01 Apr 2000
Cited by: 5 articles | PMID: 10807202
Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes.
Nature, 397(6718):436-441, 01 Feb 1999
Cited by: 767 articles | PMID: 9989410
AIDS as a zoonosis: scientific and public health implications.
Science, 287(5453):607-614, 01 Jan 2000
Cited by: 584 articles | PMID: 10649986
Review
Funding
Funders who supported this work.
NIAID NIH HHS (8)
Grant ID: N01 AI85338
Grant ID: R01 AI44596
Grant ID: R01 AI050529
Grant ID: R01 AI058715
Grant ID: R01 AI58715
Grant ID: R01 AI044596
Grant ID: R01 AI50529
Grant ID: N01AI85338





