Abstract
Free full text

Succinoglycan Production by Rhizobium meliloti Is Regulated through the ExoS-ChvI Two-Component Regulatory System
Abstract
The Rhizobium meliloti exoS gene is involved in regulating the production of succinoglycan, which plays a crucial role in the establishment of the symbiosis between R. meliloti Rm1021 and its host plant, alfalfa. The exoS96::Tn5 mutation causes the upregulation of the succinoglycan biosynthetic genes, thereby resulting in the overproduction of succinoglycan. Through cloning and sequencing, we found that the exoS gene is a close homolog of the Agrobacterium tumefaciens chvG gene, which has been proposed to encode the sensor protein of the ChvG-ChvI two-component regulatory system, a member of the EnvZ-OmpR family. Further analyses revealed the existence of a newly discovered A. tumefaciens chvI homolog located just upstream of the R. meliloti exoS gene. R. meliloti ChvI may serve as the response regulator of ExoS in a two-component regulatory system. By using ExoS-specific antibodies, it was found that the ExoS protein cofractionated with membrane proteins, suggesting that it is located in the cytoplasmic membrane. By using the same antibodies, it was shown that the exoS96::Tn5 allele encodes an N-terminal truncated derivative of ExoS. The cytoplasmic histidine kinase domain of ExoS was expressed in Escherichia coli and purified, as was the R. meliloti ChvI protein. The ChvI protein autophosphorylated in the presence of acetylphosphate, and the ExoS cytoplasmic domain fragment autophosphorylated at a histidine residue in the presence of ATP. The ChvI protein was phosphorylated in the presence of ATP only when the histidine kinase domain of ExoS was also present. We propose a model for regulation of succinoglycan production by R. meliloti through the ExoS-ChvI two-component regulatory system.
Nodulation of the leguminous plant alfalfa by Rhizobium meliloti is a complex developmental process that requires a series of signal exchanges between the host and bacteria (4, 24, 43, 50). Plant flavonoids and bacterial Nod factors are among the best-characterized signals involved in this interorganismal communication process. These plant flavonoids are secreted from the roots and induce Nod factor production by the bacteria (34, 50). The Nod factor then induces root hair curling and initiates nodule development (9, 25). In order for R. meliloti cells to successfully invade the nodules they elicit on alfalfa roots, they must be able to synthesize at least one of the following polysaccharides: succinoglycan (13, 14, 40), exopolysaccharide II (15), or a particular 3-deoxy-d-manno-2-octulosonic acid-containing capsular polysaccharide (41, 42). Succinoglycan was the first such exopolysaccharide to be recognized as being crucial for bacterial invasion of nodules (12, 23). Succinoglycan is a polymer composed of repeating octasaccharide subunits containing seven glucose molecules and one galactose molecule with three modifications: succinyl, acetyl, and pyruvyl (1, 19, 38). Succinoglycan can be detected in R. meliloti culture supernatants in two size classes: high and low molecular weights (3, 22, 49). A low-molecular-weight oligosaccharide, thought to be a tetramer of the succinoglycan subunits, was reported to partially restore nodule invasion by exo mutants when added with the exo mutant to the roots of alfalfa plants (3). These observations have led to the suggestion that succinoglycan molecules function as bacterially derived signaling molecules that ensure the successful entry of the bacteria into developing nodules.
Succinoglycan synthesis has been shown to be regulated both in the free-living stage and during symbiosis. In the free-living stage, succinoglycan production by R. meliloti Rm1021 is sensitive to the concentration of ammonia, phosphate, and sulfate present in the growth media (23). For the symbiotic stage, gene fusions have been used to show that the exo genes are expressed only in the invasion zone inside the nodule, not in the more mature regions of the nodule (39). This suggests that succinoglycan production is turned off after the bacteria invade the plant cells but before they differentiate into bacteroids.
The first insights into the nature of the regulation of succinoglycan biosynthesis came from the isolation of two succinoglycan-overproducing mutants, those carrying exoR95::Tn5 and exoS96::Tn5 (10). In both mutants (36) the transcription of succinoglycan biosynthesis genes is increased, which leads to the overproduction of succinoglycan. This succinoglycan overproduction can be suppressed by transposon insertions in the promoter region of the exoY gene (37), which encodes the glycosyl transferase which acts first during succinoglycan biosynthesis (40). Interestingly, the exoS96 mutant induces nitrogen-fixing nodules on alfalfa roots but the exoR95 mutant does not. The exoS mutant, but not the exoR mutant, synthesizes less succinoglycan in the presence of ammonia. The exoR gene was sequenced (36), but no homolog has yet been found among genes of known function, which might suggest the mechanism of action of the exoR gene product. The exoS and the exoR genes are located on the R. meliloti chromosome and are therefore not linked to the exo genes, which are located on the second symbiotic megaplasmid (10).
In this report, we describe the cloning and sequencing of the R. meliloti exoS gene and our efforts to understand its role in regulating succinoglycan production. We present evidence suggesting that the exoS gene encodes a sensor histidine kinase, which acts together with the product of the previously identified R. meliloti chvI gene to form a two-component regulatory system involved in regulating succinoglycan biosynthesis.
MATERIALS AND METHODS
Bacterial strains and growth media.
R. meliloti Rm7096 carrying the exoS96::Tn5 mutant allele was derived from wild-type strain Rm1021 (10). The following media were used for the growth of bacteria. Luria-Bertani LB medium was used for Escherichia coli, and LB medium with 2.5 mM MgSO4 and 2.5 mM CaCl2 was used for R. meliloti (13). Mannitol-glutamate-salt (MGS) medium was used in experiments in which succinoglycan production was measured (15). Agarose was used at 1.5% as a solid medium. The following antibiotics were also used at the concentrations indicated: ampicillin, 100 μg/μl; gentamicin, 15 μg/μl; kanamycin, 25 μg/μl; neomycin, 200 μg/μl; and streptomycin, 100 μg/μl.
Cloning and sequencing.
The Tn5-derived kanamycin resistance marker and IS50L along with adjacent R. meliloti DNA was cloned into plasmid pNEB193 (New England Biolabs, Beverly, Mass.) from SalI-digested genomic DNA of the exoS96::Tn5 mutant and sequenced. The exoS gene carrying the 5.3-kb BglII fragment of pM13 (10) was cloned into pMCS5 (17) and named pHC50. The 5.3-kb region was then subcloned into the same vector in three fragments, and nested deletions from opposite directions were made from the subclones. Both strands of DNA of the exoS gene and part of the downstream region have been sequenced from these nested deletion constructs manually with a sequencing kit from U.S. Biochemicals (Arlington Heights, Ill.). Parts of the region were also sequenced from pHC50 with an automated sequencer from Perkin-Elmer ABI (Branchburg, N.J.).
Protein purifications and analyses.
All the proteins that were used in this study were cloned into the pET16b vector from Novagen (Madison, Wis.) so that they were expressed with an N-terminal 10-histidine tag attached. The cloned genes were sequenced for confirmation. The overexpressed protein was purified over a nickel column with the HisBind buffer kit from Novagen in accordance with the protocol for cell lysis and subsequent chromatography provided by the manufacturer. Protein products were resolved in polyacrylamide gel with the Hoffer minigel system (San Francisco, Calif.). The concentrations of polyacrylamide gel are indicated in the figure legends.
In the case of the incomplete and complete predicted cytoplasmic domains of ExoS, ExoScyto67, and ExoScyto72, respectively, both proteins had to be solubilized from inclusion bodies in 6 M urea and purified on a nickel column in 6 M urea. Both purified proteins were renatured as described by Jin et al. (20) by being dialyzed, without stirring for 18 h and then with stirring for 18 h, against the renaturing buffer (20 mM HEPES [pH 7.4], 50 mM KCl, 5 mM dithiothreitol, 50% glycerol).
A membrane protein fraction was prepared in accordance with the protocol described by Chervitz et al. (6) with some modification. Cells from 400 ml of saturated culture were collected and resuspended in 5 ml of lysis buffer. Cells were lysed with a microtip powered by a Sonicator from Heat Systems (Farmingdale N.Y.) at power level 3 with 2.5 s on and 2.5 s off. The total on time was 7 min for E. coli cells and 10 min for R. meliloti cells. The suspension was then subjected to centrifugation at 7,600 × g for 20 min to remove intact cells; this was followed by centrifugation at 27,000 × g for 20 min to collect membrane protein embedded in the cytoplasmic membrane.
Antibody generation and immunoblot analysis.
To obtain polyclonal antibodies against ExoS, the purified cytoplasmic domain of ExoS was resolved on a polyacrylamide gel. The gel was stained in 0.05% Coomassie brilliant blue R-250 prepared in water for 10 min at room temperature with shaking. The lightly stained protein band was then cut out. The gel slices containing protein were used by HRP Inc. (Denver, Pa.) to raise antibodies against ExoS in rabbits. Western blot analyses with ExoS-specific antibodies were conducted by use of the Western Light chemiluminescent detection system from Tropix, Inc. (Bedford, Mass.) according to the manufacturer’s instructions.
Autophosphorylation and kinase assay of ExoScyto72.
The purified ExoScyto72 protein was mixed with [γ-32P]ATP in the reaction buffer (30 mM HEPES [pH 7.4], 10 mM MgCl2, 5 mM CaCl2, 50 mM KCl) along with enough unlabeled ATP to give a final concentration of 0.4 μM. The reaction mixture was incubated at room temperature for 10 min. The volumes of the reaction mixtures were reduced under centrifugation in Microcon-10 units (Amicon, Beverly, Mass.). The concentrated, labeled protein was mixed with or without the purified ChvI protein for an additional 5 min before being mixed with an equal volume of 2× loading buffer to stop the reactions. The samples containing ExoScyto72 were boiled before being resolved on gels. The samples containing labeled ChvI protein were not boiled because boiling causes dephosphorylation of phosphorylated ChvI (data not shown). To test the pH stability of phosphorylated ExoScyto72, the labeled protein was further incubated in 0.1 N HCl or 0.1 N NaOH for an additional 5 min before the addition of 2× loading buffer.
Autophosphorylation assays of ChvI.
The autophosphorylation of ChvI protein was assayed as previously described (35), with some modifications. [32P]acetylphosphate was synthesized with 0.3 U of E. coli acetate kinase (Sigma) in a buffer containing 25 mM Tris-HCl (pH 7.4), 60 mM KOAc, 10 mM MgCl2, and 10 μCi of [γ-32P]ATP. The reaction was conducted at room temperature for 20 min; acetylphosphate was separated from the enzyme with a Microcon-10 microconcentrator (10,000-molecular-weight cut-off; Amicon), 3 μg of purified His-ChvI protein was added, and the sample was further incubated for 10 min. The phosphorylated ChvI was further incubated with 0.1 N HCl or 0.1 N NaOH for 5 min and neutralized by the addition of 0.1 N NaOH or 0.1 HCl, respectively. All reactions were stopped by the addition of equal volumes of 2× sodium dodecyl sulfate loading buffer.
Nucleotide sequence accession number.
The GenBank accession number for the DNA sequence of the exoS region is AF027298.
RESULTS
Cloning and sequencing the exoS gene.
To identify the exoS gene, we took advantage of the transposon-generated exoS96::Tn5 allele and the exoS+ cosmid, pM13 (10). The exoS+ gene had been localized to a 5.3-kb BglII fragment of genomic DNA. We first cloned a SalI DNA fragment containing IS50L and the kanamycin resistance gene portion of the Tn5 DNA as well as the flanking R. meliloti DNA. Although sequencing this R. meliloti DNA did not reveal any homologies to previously sequenced genes, primers derived from this DNA allowed us to obtain a sequence from a subclone carrying the 5.3-kb BglII fragment that represented the DNA flanking the opposite side of Tn5 in the mutant strain. Analyses of the DNA sequence revealed a 1,737-bp open reading frame which, in the exoS96 mutant, is interrupted by the insertion of Tn5 57 bp downstream of the start of the open reading frame. The predicted ExoS protein from this open reading frame was found to have an overall identity of 68% and a similarity of 78% to the Agrobacterium tumefaciens ChvG protein (Fig. (Fig.1).1). The sharp drop in the homology between ExoS and ChvG in the region of amino acids 135 to 206 of ExoS could be the result of sequencing artifacts. Compared with our repeatedly sequenced exoS DNA sequence, the DNA sequences in the region differ by two +1 frameshifts and two −1 frameshifts. Eliminating the difference would boost the homology between ExoS and ChvG to 78% overall identity. Based on the homology of its predicted kinase domain to that of the E. coli PhoR protein, the A. tumefaciens ChvG protein has been proposed to be the sensor of the ChvG-ChvI two-component regulatory system (5). The R. meliloti ExoS protein has an even higher level of identity with the putative histidine kinase domain of A. tumefaciens ChvG protein (Fig. (Fig.1).1). Furthermore, the four most conserved signature sequence patterns, the H, N, D/F, and G boxes that are present in all of the sensor histidine kinases, are also found in these two proteins (Fig. (Fig.1).1). The homology of the R. meliloti ExoS protein to the A. tumefaciens ChvG protein and other known sensor histidine kinases strongly suggests that ExoS serves as the sensor of a two-component regulatory system that regulates succinoglycan synthesis in R. meliloti.
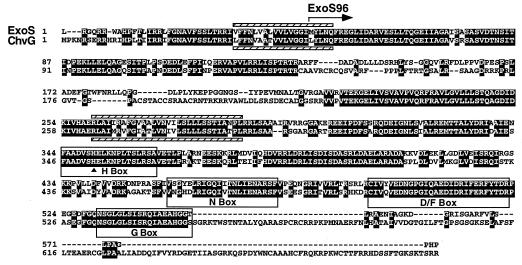
Alignment of the putative amino acid sequences of the R. meliloti ExoS and A. tumefaciens ChvG proteins. The two proteins have an overall identity of 68% and a similarity of 78%. Identical residues are shaded. A possible reason for the sharp drop in homology between residues 135 and 206 is discussed in the text. The putative N terminus of ExoS96 is indicated by an arrow. The four boxes highlight the four most important regions, the H, N, D/F, and G boxes, within the histidine kinase domains of sensor proteins in two-component regulatory systems. The site of phosphorylation is indicated by a triangle. The hatched bars indicate predicted transmembrane domains for ExoS (above the sequence) and ChvG (below the sequence).
Not only does it appear that ExoS is a sensor kinase, but also the characteristics of its predicted sequence suggest that it belongs to the subset of sensor kinases that are integral membrane proteins and that have substantial periplasmic and cytoplasmic domains. Analyses of the hydrophobicity plots of R. meliloti ExoS and A. tumefaciens ChvG and of their peptide sequences suggest that they have very similar membrane topologies (data not shown). The domain between the first and second transmembrane domains of the A. tumefaciens ChvG protein was determined to be a periplasmic domain based on the isolation of two TnphoA fusions (5). The domain between the second and third transmembrane domain contains the highly conserved histidine kinase domain which typically is part of a cytoplasmic domain (5). This N-terminal periplasmic domain and a C-terminal cytoplasmic domain arrangement have been found in many membrane histidine kinase sensor proteins (45). These observations suggest that ExoS similarly has an N-terminal periplasmic domain and a C-terminal cytoplasmic domain which also contains the highly conserved histidine kinase domain.
As we were beginning to sequence DNA in the vicinity of the exoS gene in the hopes of identifying a gene encoding the corresponding regulator of the two-component system, Østerås et al. (31) published data suggesting that R. meliloti Rm1021 contained a two-component regulatory system that is similar to the A. tumefaciens ChvG-ChvI two-component regulatory system. They reported the sequence of an R. meliloti gene homologous to the A. tumefaciens chvI gene and that of 360 bp homologous to the 5′ end of the A. tumefaciens chvG gene (27). As in A. tumefaciens, the chvI gene is located upstream of the pckA gene (phosphoenolpyruvate carboxykinase), which was the focus of their study (31). The DNA sequence of the R. meliloti chvI-chvG intergenic region and the first part of the chvG gene determined by Østerås et al. was identical to that of our exoS gene, which indicates that the exoS gene is the putative chvG gene postulated by Østerås et al. Taken together, the combined DNA sequence data indicate that R. meliloti encodes a putative two-component regulatory system, ExoS-ChvI, that is closely related to the ChvG-ChvI two-component regulatory system of A. tumefaciens (Fig. (Fig.2).2).
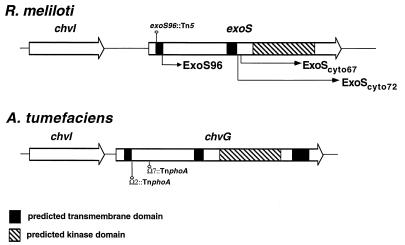
Schematic representation of the organization of the R. meliloti ExoS-ChvI and A. tumefaciens ChvG-ChvI two-component regulatory systems showing the similarity between the two systems. The predicted transmembrane domains and kinase domains are indicated, as are the two cytoplasmic domains of ExoS (ExoScyto67 and ExoScyto72) that were used in the study. The lengths of the two arrows indicate the sizes of the cytoplasmic domains. The sites of TnphoA insertions in the chvG open reading frame and Tn5 insertion in the exoS96 mutant are indicated.
R. meliloti exoS96 mutation is suppressed by the A. tumefaciens chvG gene.
In order to determine whether R. meliloti ExoS and A. tumefaciens ChvG are functionally related, as suggested by the high level of homology between the two, plasmids carrying either the A. tumefaciens chvG+ gene or the chvG gene interrupted by a transposon were introduced into the R. meliloti exoS96 mutant. The levels of succinoglycan production by the resulting strains were then examined on plates. The presence of the A. tumefaciens chvG+ gene caused the reduction of succinoglycan production by the R. meliloti exoS96 mutant, while the transposon-interrupted chvG gene did not cause a reduction. This indicates that the A. tumefaciens chvG gene can suppress the elevated, constitutive level of succinoglycan synthesis observed in an exoS96 mutant, suggesting that the R. meliloti ExoS and A. tumefaciens ChvG proteins are functionally interchangeable in R. meliloti cells.
Additional evidence that these two proteins are functionally interchangeable came from the work of Østerås et al. They interrupted the cloned R. meliloti chvG gene, which we have now shown is actually the exoS gene, and introduced it into A. tumefaciens chvG mutants. They found that only the uninterrupted R. meliloti exoS gene can suppress the mutant phenotype of the A. tumefaciens chvG mutant, which is sensitive to complex growth media (31). They have also shown that the R. meliloti chvI gene can suppress the A. tumefaciens chvI mutant. These results suggest that the heterologous pairs of sensors and response regulators from the two organisms can interact effectively.
ExoS cofractionates with the membrane.
In the hope of obtaining sufficient ExoS protein for raising antibodies and for biochemical characterization, we expressed N-terminal His-tagged ExoS in E. coli under the control of the T7 promoter. However, after induction no extra protein band corresponding to the predicted molecular weight of ExoS was detected when all the proteins from the induced cells were resolved on a polyacrylamide gel. For increased sensitivity, we separated the proteins from the induced cells into cytoplasmic and membrane fractions. One small extra band having the predicted molecular weight for ExoS could be seen when membrane proteins were separated on a gel. The very small amount of ExoS protein present in the induced cells would have made it difficult to purify enough protein for raising antibodies. Therefore, we instead constructed a plasmid that expressed an N-terminal His-tagged fragment of ExoS, ExoScyto67, consisting of amino acids 301 to 547, the region constituting most of the predicted histidine kinase domain. This ExoScyto67 protein was produced at high levels but was insoluble. We thus solubilized inclusion bodies in urea, purified the ExoScyto67 protein over a nickel column, resolved the purified protein on a polyacrylamide gel (Fig. (Fig.3),3), and raised antibodies against ExoS with the gel-purified protein. The removal of urea from the purified protein resulted in the precipitation of the protein.
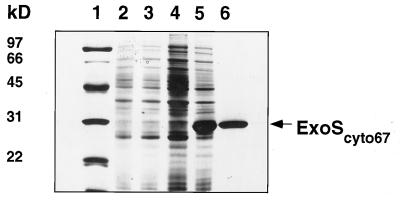
Comassie blue-stained sodium dodecyl sulfate-polyacrylamide gel of protein samples from the purification of the cytoplasmic domain of ExoS, ExoScyto67. Lane 1, protein size standards; lanes 2 and 3, total cellular proteins from cells carrying cloning vectors only that were grown under uninduced and induced conditions, respectively; lanes 4 and 5, total cellular proteins from cells carrying the plasmid encoding ExoScyto67 that were grown under uninduced and induced conditions, respectively; lane 6, ExoScyto67 purified over a nickel column.
The ExoS-specific antibodies were used to determine the cellular location of the ExoS protein. The total cellular protein of the wild-type strain Rm1021 cells was fractionated into cytoplasmic and membrane proteins after sonication and centrifugation. Equivalent amounts of protein from both fractions were resolved on a polyacrylamide gel, blotted, and probed with ExoS-specific antibodies. As shown in Fig. Fig.4A,4A, one strong band could be seen in the lane containing membrane proteins while one band of much lower intensity migrated at the same position in the lane containing cytoplasmic proteins. This lower-intensity band disappeared when the cytoplasmic protein fraction was subjected to further centrifugation at higher speed, which suggests that the band represents a small amount of ExoS that was associated with the smaller vesicle of the cytoplasmic membrane. These results indicate that ExoS is a membrane protein. This conclusion is consistent with the computer analyses of its putative peptide sequence and with the fact that ExoS cofractionated with membrane proteins in E. coli. Thus, it appears that ExoS is a sensor protein located in the cytoplasmic membrane.
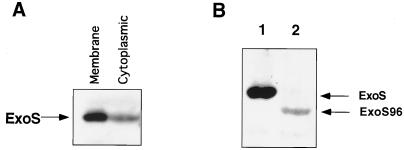
Cellular distribution of the ExoS protein examined in an immunoblot by using ExoS-specific antibodies. (A) Total cellular proteins from wild-type strain Rm1021 were separated into membrane and cytoplasmic fractions and examined in an immunoblot by using ExoS-specific antibodies. The ExoS protein was found to cofractionate predominantly in the membrane protein fraction. The cross-reacting material in the cytoplasmic protein fraction can be removed by additional high-speed centrifugation, suggesting that it represents ExoS protein which associated with smaller vesicles of cytoplasmic membrane. (B) Membrane proteins from Rm1021 and the exoS96 mutant were examined in an immunoblot by using ExoS-specific antibodies. Lane 1, wild-type strain proteins; lane 2, exoS96 mutant proteins. The ExoS band is missing from the lane containing ExoS96 membrane proteins. A faint new band, which is about 1/20 of the intensity of that of ExoS is seen; the band represents a protein that is 5 kDa smaller than ExoS.
exoS96 encodes an N-terminal truncated version of ExoS that cofractionates with the membrane.
We were interested in the properties of the exoS96::Tn5 allele for several reasons. We had initially assumed that it was a null allele because it was caused by a Tn5 insertion and because the exoS96::Tn5 allele was recessive to exoS+ carried on pM13 (10), an IncP-based cosmid vector. However, we were unsuccessful in homogenotizing a construct of the exoS gene in which part of the open reading frame had been replaced by a drug marker, despite several attempts employing minimal medium and the procedures that we have used routinely to generate hundreds of disruptions in R. meliloti. We were also unable to interupt the chvI gene by using the same approach. Furthermore, Østerås et al. (31) had reported that they were unsuccessful in homogenotizing chvG::Tn5 mutations into the genome. These observations raised the possibility that the R. meliloti chvI and exoS genes could not be deleted in a wild-type R. meliloti background and that there was something special about the exoS96 allele.
In the exoS96::Tn5 mutant, the site of insertion is within the exoS open reading frame, 57 bp downstream from the putative start site of the open reading frame. There is an in-frame ATG start codon 135 bp downstream from the beginning of the open reading frame. Since there is an E. coli outward-facing promoter within 69 bp of the end of IS50R (7), it seemed possible that the rest of the exoS open reading frame could still be transcribed in R. meliloti at a low level and then translated. This would result in the synthesis of a truncated ExoS missing the first 45 amino acids at the amino terminus, which would make the ExoS96 protein 5 kDa smaller than the ExoS protein. To examine this possibility, equal amounts of membrane proteins from the exoS96 mutant were resolved side by side with those from the wild-type strain and were probed with anti-ExoS antibodies (Fig. (Fig.4B).4B). For the exoS96 mutant, the wild-type ExoS band was missing. However there was a band which was about 5 kDa smaller. Its intensity was only 1/20 that of the ExoS band. These results suggest that in the exoS96 mutant there is a truncated ExoS being made at approximately 1/20 the level of ExoS in the wild-type strain. The size reduction detected in the experiment was consistent with our prediction, which suggests that the ATG located 136 bp inside the exoS open reading frame is being used to start the translation. The lower level of the ExoS96 protein compared to the ExoS protein could be due to a low level of transcription, a lack of a proper ribosomal binding site in front of the ATG start codon, or both.
The cytoplasmic domain of the ExoS protein can autophosphorylate at a histidine residue in vitro in the presence of ATP.
The high level of homology of ExoS to the histidine kinase domains of E. coli PhoR and EnvZ strongly suggests that ExoS is a histidine kinase which would autophosphorylate at the conserved histidine residue in the presence of ATP (Fig. (Fig.1).1). To confirm biochemically that ExoS has histidine kinase activities, plasmid pHC72 was constructed to express the entire cytoplasmic domain of ExoS, ExoScyto72, with an N-terminal histidine tag (Fig. (Fig.2).2). The ExoScyto72 protein consists of amino acids 281 to 578 from ExoS and forms inclusion bodies when it is overexpressed. It was purified under denaturing conditions and renatured as described in Materials and Methods. When the purified and renatured ExoScyto72 protein was incubated with [γ-32P]ATP and then resolved on a polyacrylamide gel, a band appeared on the autoradiograph at the position corresponding to that of ExoScyto72 (Fig. (Fig.5).5). This indicated that the cytoplasmic domain of ExoS can autophosphorylate in the presence of [γ-32P]ATP, a property which is a characteristic of all tested sensor proteins of two-component regulatory systems.
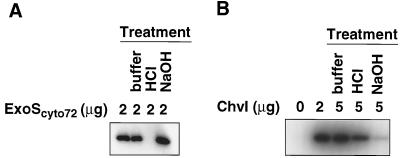
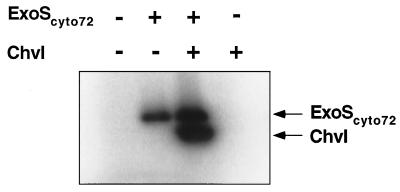
Autoradiograph demonstrating autophosphorylation of the ExoS and ChvI proteins. (A) ExoScyto72 protein autophosphorylates in the presence of [γ-32P]ATP. The phosphorylated ExoScyto72 loses its phosphate at acidic pH (0.1 N HCl), but not at neutral (buffer; pH 7.4), and alkaline (0.1 N NaOH) pHs. (B) ChvI protein autophosphorylates in the presence of [32P]acetylphosphate. The phosphorylated ChvI protein loses its phosphate at alkaline pH (0.1 NaOH), but not at neutral (pH 7.4) and acidic (0.1 N HCl) pHs.
To test whether a histidine residue is the site of phosphorylation, we examined the stability of phosphorylated ExoScyto72 protein at different pHs (28). Histidyl-phosphate is stable at neutral and alkaline pH but is not stable at acidic pH. The phosphorylated ExoScyto72 protein was further incubated for 5 min at neutral (no additional buffer), acidic (0.1 N hydrochloric acid), or alkaline (0.1 N sodium hydroxide) pH (Fig. (Fig.5A).5A). The ExoScyto72 only lost the phosphate label after the acid treatment, an observation consistent with a histidine residue being the site of phosphorylation.
The ChvI protein can autophosphorylate at an aspartate residue in vitro in the presence of acetylphosphate.
On the basis of its sequence homology, and because of the genetic analysis of its A. tumefaciens homolog, the R. meliloti ChvI protein has been said to function as a response regulator in a two-component regulatory system (31). Because many response regulators have been shown to autophosphorylate in the presence of acetylphosphate (26, 45), we decided to test whether the R. meliloti ChvI protein shared this distinctive property.
We therefore cloned the chvI open reading frame and expressed a derivative carrying an N-terminal oligohistidine tag in E. coli. The resulting soluble protein was purified through a nickel column. The purified ChvI protein was then incubated with 32P-acetylphosphate. As shown in Fig. Fig.5B,5B, the ChvI protein became radiolabeled when [32P]acetylphosphate was present, suggesting that ChvI can autophosphorylate by using acetylphosphate as the phosphate donor. The phosphorylated ChvI is stable under acidic but not alkaline conditions; this is consistent with the phosphorylation occurring at an acidic residue (35), most likely Asp-52 by analogy to other response regulators. The ChvI protein was not phosphorylated when it was incubated with [γ-32P]ATP (Fig. (Fig.6).6). All of these results are consistent with ChvI being a response regulator of a two-component regulatory system.
The cytoplasmic domain of ExoS phosphorylates ChvI in vitro.
To examine whether the cytoplasmic domain of ExoS has kinase activity, purified ExoScyto72 was tested for its ability to phosphorylate the R. meliloti ChvI protein. If the ExoScyto72 protein was first allowed to autophosphorylate by incubation with [γ-32P]ATP and then was mixed with purified ChvI protein, the ChvI protein became radiolabeled (Fig. (Fig.6).6). The ChvI protein was not labeled if ExoScyto72 was omitted from the reaction. This indicates that the cytoplasmic domain of ExoS phosphorylates ChvI, which in turn suggests that the ExoS protein is a protein kinase. These results suggest that ExoS is a membrane-bound histidine kinase sensor protein of the ExoS-ChvI two-component regulatory system. As had already been noted (31), the ChvI protein is homologous to response regulators that are phosphorylated on an aspartate residue by the histidine kinase activity of the corresponding sensor protein.
DISCUSSION
Taken together, the data presented in this paper suggest that the exoS and chvI genes constitute a two-component regulatory system that plays a role in controlling succinoglycan production by R. meliloti. The organization of the exoS and chvI genes is also typical of that of a two-component regulatory system. ExoS contains all four highly conserved protein sequence signatures that have been found in all of the histidine kinase sensor proteins, suggesting that ExoS is also a histidine kinase sensor protein. Our biochemical data support the respective roles for these two proteins that are suggested by analyses of their sequences.
From the computer analyses of its amino acid sequence, ExoS was predicted to be a membrane protein with two transmembrane domains, one close to the N terminus and the other in the middle of the protein. The proposed membrane location of ExoS is consistent with our observations that intact ExoS proteins cofractionated with membrane proteins in both R. meliloti and E. coli. Since ExoS and A. tumefaciens ChvG are functionally exchangeable and have similar hydrophobicity profiles, they most likely have similar membrane topologies. It has been shown that the A. tumefaciens ChvG protein has an N-terminal periplasmic domain and a C-terminal cytoplasmic domain (5). The ChvG protein was also proposed to possibly have a third transmembrane domain at the C terminus (5) which ExoS does not have. We have shown that the putative cytoplasmic domain of ExoS has kinase activity. Thus, we propose that ExoS is a histidine kinase sensor protein located in the cytoplasmic membrane that has an N-terminal periplasmic domain and a C-terminal cytoplasmic kinase domain. This is the typical membrane topology for a membrane-bound sensor protein of a two-component regulatory system (45).
Our discoveries concerning R. meliloti exoS and chvI, together with a consideration of other two-component regulatory systems (52), suggest a possible model for the regulation of succinoglycan production by the ExoS-ChvI two-component regulatory system. According to our model, ExoS forms homodimers in the cytoplasmic membrane, and the periplasmic sensor domain of ExoS can switch between activated and nonactivated forms. In the presence of an as yet unknown environmental signal, the sensor domain stays in the activated form. The sensing of the signal is passed through the membrane by means of a minor structural change in the sensing domain and activates the cytoplasmic kinase. The response regulator, ChvI, is then phosphorylated and activates the transcription of the exo genes, thereby increasing production of succinoglycan.
We were initially surprised to find that the exoS96::Tn5 mutant expresses a smaller ExoS derivative with an N-terminal truncation because the exoS96::Tn5 allele is recessive to an exoS+ gene carried on an IncP cosmid (10). Since most of the sensors have both kinase and phosphatase activity (2, 29, 45), our data are most easily explained by postulating that it is this truncation that makes ExoS96 into a constitutively active kinase or a defective phosphatase and that results in the consequent overexpression of the exo genes and the overproduction of succinoglycan. A consideration of the findings for other two-component regulatory systems, which are described below, provides a possible explanation for the phenotype of the exoS96 mutant and helped us refine our model.
It has been shown for E. coli CheA (45), EnvZ (53), and NtrB (30) and for A. tumefaciens VirA (32) that homodimers are the active forms of the two-component sensor proteins. The autophosphorylation of NtrB and EnvZ has been shown to involve trans-phosphorylation between subunits of a dimer based on the in vivo complementation between wild-type proteins and various mutant forms of the proteins (30, 53). The dimerized form of phosphorylated CheA serves as a phosphate donor, and monomers of phosphorylated CheA have to dimerize before they can serve as phosphate donors (46). The A. tumefaciens VirA protein has been shown to form dimers in the cytoplasmic membrane that can be cross-linked on the surfaces of live cells (32). Coexpression of mutant forms of the VirA protein in the wild-type strain inhibits the function of the VirA protein, which suggests the formation of an inactive heterodimer of active and inactive forms of the VirA protein (32).
The amino-terminal peptide leading to the first transmembrane domain and the two transmembrane domains around the periplasmic domain have been found to have a large impact on the sensing ability of the periplasmic domain in sensors of several other two-component regulatory systems. Point mutations in the first transmembrane domain of the A. tumefaciens VirA protein (11, 21) can turn VirA into a constitutively active or inactive sensor kinase (11, 33, 48). Replacing either of the two transmembrane domains of the VirA protein with that of the E. coli aspartate sensor, Tar, reduces the ability of the VirA protein to respond to its inducer (48). In the nitrate- and nitrite-sensing dual two-component regulatory system, a single substitution of one of the amino acids that precedes the first transmembrane domain made one of the sensors, NarX, stay constitutively active, resulting in upregulation of the relevant genes (51). In the E. coli EnvZ-OmpR system, sensor EnvZ has both kinase and phosphatase activities. One particular amino acid substitution at the end of the first transmembrane domain causes EnvZ to lose its phosphatase activity but not its kinase activity (47). The effect of this mutation can be suppressed by a single mutation in the first or the second transmembrane domain of EnvZ (47). These findings suggest that alterations of either of the transmembrane domains that flank the periplasmic domain in this class of sensors can perturb the constitutive state of the sensor and/or its ability to sense the relevant environmental signals.
In the wild-type strain, ExoS has a 30-amino-acid N-terminal segment which precedes the first putative transmembrane domain, which consists of 20 amino acids. Our data suggest that ExoS96 is missing the first 45 amino acids, thereby leaving it with 5 amino acids of the first transmembrane domain. This shortened transmembrane domain may not be able to anchor the periplasmic domain to the membrane as strongly as the intact one and thus might allow the periplasmic domain of ExoS96 to stay in an active form continuously. The cytoplasmic domain would then also stay active and increase the phosphorylation of ChvI either because its kinase activity is constitutive or because its putative phosphatase activity is diminished. This would lead to the overexpression of the exo genes and the overproduction of succinoglycan. We suggest that, when the exoS96 mutant is complemented with the wild-type exoS+ gene, heterodimers of ExoS and ExoS96 are formed. The interactions between the periplasmic domain of ExoS and ExoS96 restore the ability of the periplasmic domain of ExoS96 to switch between activated and nonactivated forms and suppress the overproduction of succinoglycan observed for the exoS96 mutant. This hypothesis can be further tested by increasing the amount of ExoS96 in an exoS+ background to see if the production of succinoglycan will be increased or decreased. We are in the process of developing different approaches to test the hypothesis.
Succinoglycan production by R. meliloti is also negatively regulated at the transcriptional level by the exoR gene, which is involved in sensing ammonia in the media (10). Although, at present, there is no evidence to link ExoR to the ExoS-ChvI two-component regulatory system, extra factors have been found to work with several two-component regulatory systems. For example, a third factor, RcsA, is required for the regulation of E. coli capsular polysaccharide production by the rcsC-rcsB two-component regulatory system (16). A. tumefaciens ChvE, a periplasmic sugar-binding protein, is required for the virA-virG two-component regulatory system to be able to sense an inducer when the inducer is at low concentrations (11). The expression of eps and other virulence genes of Pseudomonas solanacearum is regulated by a combination of two-component regulatory systems (18).
We are also interested in finding the environmental signal that ExoS senses. Osmotic shock and the disruption of outer membrane proteins have been suggested as being environmental signals that E. coli RcsC senses in regulating capsule production in E. coli (44) and possibly the expression of a set of outer membrane protein genes, tolQRA (8). The amount of ammonia, phosphate, and sulfur in the media can change the rate of succinoglycan production (23). Since the exoS96 mutant responds to ammonia in the media in the same way as the wild type while the exoR95 mutant does not respond at all, it is unlikely that ExoS is directly involved in sensing ammonia (10). Identifying the environmental signal(s) sensed by ExoS should help us to gain a better understanding of the regulation of succinoglycan production both in the free-living state and during nodulation and to gain insights into the role(s) of succinoglycan in nodule invasion.
ACKNOWLEDGMENTS
We thank John A. Leigh, Stephen C. Winans, Turlough M. Finan, and Tevor C. Charles for strains, unpublished results, and helpful suggestions. We thank members of this laboratory for many helpful suggestions and critical discussions.
This work is supported by Public Health Service grant GM31030 to G.C.W. from the National Institutes of Health.
REFERENCES
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.180.1.20-26.1998
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/jb/180/1/20.full.pdf
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/content/full/180/1/20
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/180/1/20
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/180/1/20
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jb.180.1.20-26.1998
Article citations
A sensor histidine kinase from a plant-endosymbiont bacterium restores the virulence of a mammalian intracellular pathogen.
Microb Pathog, 185:106442, 08 Nov 2023
Cited by: 0 articles | PMID: 37944675
A protease and a lipoprotein jointly modulate the conserved ExoR-ExoS-ChvI signaling pathway critical in Sinorhizobium meliloti for symbiosis with legume hosts.
PLoS Genet, 19(10):e1010776, 23 Oct 2023
Cited by: 1 article | PMID: 37871041 | PMCID: PMC10659215
A bvrR/bvrS Non-Polar Brucella abortus Mutant Confirms the Role of the Two-Component System BvrR/BvrS in Virulence and Membrane Integrity.
Microorganisms, 11(8):2014, 05 Aug 2023
Cited by: 0 articles | PMID: 37630574 | PMCID: PMC10459465
The Brucella abortus two-component system response regulator BvrR binds to three DNA regulatory boxes in the upstream region of omp25.
Front Microbiol, 14:1241143, 14 Sep 2023
Cited by: 0 articles | PMID: 37779712 | PMCID: PMC10538546
Comparative Proteomic Analysis Revealing ActJ-Regulated Proteins in Sinorhizobium meliloti.
J Proteome Res, 22(6):1682-1694, 05 Apr 2023
Cited by: 1 article | PMID: 37017314
Go to all (105) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - AF027298
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Null mutations in Sinorhizobium meliloti exoS and chvI demonstrate the importance of this two-component regulatory system for symbiosis.
Mol Microbiol, 74(5):1223-1237, 19 Oct 2009
Cited by: 42 articles | PMID: 19843226
ExoR is genetically coupled to the ExoS-ChvI two-component system and located in the periplasm of Sinorhizobium meliloti.
Mol Microbiol, 64(3):647-664, 01 May 2007
Cited by: 64 articles | PMID: 17462014
Identification of direct transcriptional target genes of ExoS/ChvI two-component signaling in Sinorhizobium meliloti.
J Bacteriol, 191(22):6833-6842, 11 Sep 2009
Cited by: 34 articles | PMID: 19749054 | PMCID: PMC2772461
Rhizobium meliloti exopolysaccharides: genetic analyses and symbiotic importance.
Biochem Soc Trans, 19(3):636-641, 01 Aug 1991
Cited by: 10 articles | PMID: 1783190
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: GM31030
Grant ID: R01 GM031030