| PMC full text: | Published online 2024 Feb 20. doi: 10.1186/s12864-024-10104-9
|
Fig. 3
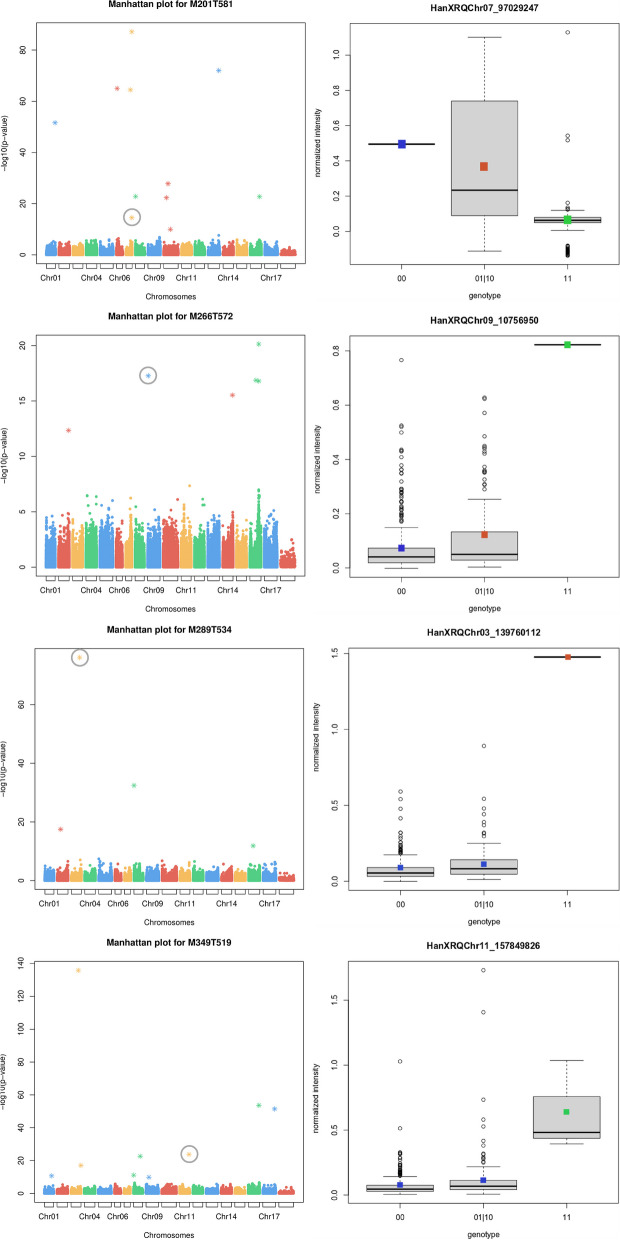
Manhattan plots and genotypic boxplots of the functionally characterized metabolites, here indicated using the codes of LC-MS features. For each metabolite, the corresponding Manhattan plot (left) and genotypic boxplot (right) are shown. Manhattan plots show the reference SNPs obtained from GWAS on the X-axis and the corresponding p-values on the Y-axis. SNPs filtered according to the eBIC criterion are shown as asterisks (Table S03). The reference SNP associated to a most likely candidate gene is highlighted by a grey circle. Genotypic boxplots show the normalized intensity values of the corresponding LC-MS feature (Y-axis) grouped according to the three possible allelic states (i.e. 00, 01|10, and 11). The classes identified with the Tukey’s test are indicated using colored squares. It is to note that the SNPs used to produce the genotypic boxplots are, by definition, reference SNPs, and therefore they are not the same ones found associated to the corresponding most likely candidate gene, which belong instead to co-inherited sets. The correspondences among the reference SNPs used for genotypic plots and the co-inherited used to identify most likely candidate genes are given in Table Table22