Abstract
Free full text

Phosphotyrosine (p-Tyr)-Dependent and -Independent Mechanisms of p190 RhoGAP-p120 RasGAP Interaction: Tyr 1105 of p190, a Substrate for c-Src, Is the Sole p-Tyr Mediator of Complex Formation
Abstract
p190 RhoGAP is a 190-kDa protein that stably associates with p120 RasGAP and regulates actin dynamics through members of the Rho family of small GTPases. Previous studies have indicated a direct relationship between levels of p190 tyrosine phosphorylation, the extent and kinetics of epidermal growth factor (EGF)-induced actin rearrangements, and EGF-induced cell cycle progression, suggesting that p190 links Ras-mediated mitogenic signaling with signaling through the actin cytoskeleton. Determining which tyrosine residues in p190 are phosphorylated, what factors regulate phosphorylation of these sites, and what effect tyrosine phosphorylation has on p190 function is key to understanding the role(s) that p190 may play in these processes. To begin investigating these questions, we used biochemical approaches to characterize the number and relative levels of in vivo-phosphorylated tyrosine residues on endogenous p190 from C3H10T1/2 murine fibroblasts. Only two tryptic phosphopeptides containing phosphotyrosine (p-Tyr), a major site, identified as Y1105, and a minor, unidentified site, were detected. Phosphorylation of Y1105, but not the minor site, was modulated in vivo to a greater extent by overexpression of c-Src than by the EGF receptor and was efficiently catalyzed by c-Src in vitro, indicating that Y1105 is a selective and preferential target of c-Src both in vitro and in vivo. In vitro and in vivo coprecipitation analysis using glutathione S-transferase (GST) fusion proteins containing wild-type and Y1105F variants of the p190 middle domain, variants of full-length p190 ectopically expressed in COS-7 cells, and endogenous p190 and p120 in C3H10T1/2 cells revealed that p190 could bind to p120 in the presence and absence of p190 tyrosine phosphorylation. p-Tyr-independent complexes comprised 10 to 20% of the complexes formed in the presence of p-Tyr. Mutation of Y1105 from Tyr to Phe resulted in complete loss of p-Tyr-dependent complex formation, indicating that p-Y1105 was the sole p-Tyr residue mediating binding to p120. These studies describe a specific mechanism by which c-Src can regulate p190-p120 association and also document a significant role for p-Tyr-independent means of p190-p120 binding.
p190 RhoGAP was initially identified as a tyrosine-phosphorylated protein from v-Src-transformed fibroblasts that coimmunoprecipitated with the GTPase-activating protein (GAP) of p21 Ras, p120 RasGAP (1, 8). Cloning and sequencing of the cDNA encoding p190 led to the identification of two functional domains, an N-terminal GTP binding domain and a C-terminal GAP domain which has been shown to activate members of the Rho subfamily of small GTPases (27, 28). This subfamily regulates actin stress fiber and focal adhesion formation (Rho), lamellipodium and membrane ruffling (Rac), and filopodium (Cdc42) formation (11, 19, 23, 24). In vivo, the GAP activity of p190 is specific for Rho, as microinjection of the RhoGAP domain into Swiss 3T3 fibroblasts inhibits actin stress fiber formation but not membrane ruffling (25).
Previous studies have suggested that tyrosine phosphorylation of p190 may influence its function (6). Specifically, it was demonstrated that the level of p190 tyrosine phosphorylation was enhanced when wild-type c-Src (K+ c-Src) was overexpressed in C3H10T1/2 murine fibroblasts and decreased below normal levels in cells overexpressing kinase-defective c-Src (K− c-Src) (4, 6). Treatment of these cells with epidermal growth factor (EGF) had no effect on the level of p190 tyrosine phosphorylation, indicating that p190 is a preferred substrate for c-Src compared to the EGF receptor. Upon EGF addition, however, p190 and p120 underwent simultaneous and striking but transient redistributions into perinuclear concentric arcs that coincided both temporally and spatially with EGF-induced actin stress fiber disassembly and reassembly (6). The magnitude and rate of actin cytoskeleton disassembly and p190/p120 redistribution correlated directly with the extent of p190 tyrosine phosphorylation in the various C3H10T1/2 cell lines and with the numbers of cells undergoing DNA synthesis in response to EGF (6, 12, 34). These results are consistent with the hypothesis that tyrosine phosphorylation of p190 plays a role in regulating EGF-dependent actin cytoskeletal reorganization in a linkage of the Ras and Rho signaling pathways. That p190 may play a significant role in regulation of cell cycle progression is also supported by the findings of Wang et al. (33), who demonstrated the ability of p190 to cause reversion of Ras-induced transformation. Together these results suggest that p190 may function as a negative regulator of mitogenesis through control of actin cytoskeleton dynamics.
The amino terminus of p120 contains a tandem arrangement of SH2-SH3-SH2 domains that have been proposed to mediate binding to p190. Structure-function analyses of p120 have demonstrated that the two SH2 domains in the N-terminal half of p120 synergistically bind tyrosine-phosphorylated p190 (3, 10, 15). These findings suggest that the interaction of p190 with p120 may be mediated by dual phosphotyrosine (p-Tyr) sites on p190. Indeed, mutagenesis and transient overexpression studies of p190 (9) have led to the hypothesis that complex formation between p190 and p120 requires phosphorylation of Y1087 and Y1105 in p190 and that the dual p-Tyr–SH2 interaction results in a conformational change in p120 that exposes the SH3 domain of p120 to additional binding proteins (9). However, coimmunoprecipitation of p190 with p120 has been observed when p190 is tyrosyl phosphorylated at low or even undetectable levels (3, 4, 9). These observations suggest that other mechanisms may also regulate the interaction of p190 with p120.
To gain further insights into the role of tyrosine phosphorylation in p190 function and particularly into the nature of the association between p190 and p120, we sought to identify by biochemical approaches the p-Tyr residues in endogenous p190 and to characterize their roles in mediating binding to p120. In contrast to previous reports, results demonstrate the presence of only one major p-Tyr residue (Y1105) in endogenous p190 and indicate that this residue serves as an in vitro and in vivo target for phosphorylation by c-Src. Furthermore, phosphorylation of Y1105 is shown to be sufficient for mediating p-Tyr-dependent binding of p190 to p120, but a surprising and significant portion of p120 could be associated with p190 in a p-Tyr-independent manner. Together, our results support a model whereby association between p190 and p120 is regulated by inputs from several signaling pathways, one of which involves tyrosine phosphorylation of p190 by c-Src.
MATERIALS AND METHODS
Cell lines.
The clonal cell lines used in this study were generated in our laboratory and were derived from the murine fibroblast cell line C3H10T1/2. They include Neo (control), K− c-Src (kinase-defective c-Src overexpressers that express an enzymatically inactive chicken c-Src which contains an A430V mutation in the Ala-Pro-Glu [APE] motif of the kinase domain), K+ c-Src (wild-type chicken c-Src overexpressers), EGFR (human EGF receptor overexpressers), EGFR/K+ c-Src (EGF receptor–c-Src double overexpressers), and IV5 (v-Src transformants). Details of their derivation and characterization have been described previously (12, 13, 34). K+ c-Src, K− c-Src, and EGFR/K+ c-Src cell lines express equal levels of c-Src (~25-fold over the endogenous level), and EGFR and EGFR/K+ c-Src express similar levels of cell surface EGF receptors (~2 × 105 receptors/cell, or ~40-fold over the endogenous level). Cells were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal calf serum, antibiotics, and G418 (400 μg/ml) in a humidified 37°C, 5% CO2 environment.
Antibodies.
The p190-specific mouse monoclonal antibody (MAb) 8C10 was derived and characterized in our laboratory (5, 6). 8C10 recognizes an epitope in the N-terminal region of p190 (unpublished data) and quantitatively immunoprecipitates endogenous p190 from cell extracts. The p120 RasGAP-specific mouse MAb 6F2 was also derived and characterized in our laboratory (5, 6). The anti-p-Tyr MAb 4G10 was purchased from Upstate Biotechnology, Inc. (UBI), Lake Placid, N.Y. ChromPure mouse immunoglobulin G (IgG), purchased from Jackson ImmunoResearch Laboratories, Inc., Bar Harbor, Maine, was used as the negative control antibody.
Metabolic labeling.
Cells were grown to ~75% confluence in 15-cm tissue culture dishes, equilibrated for 30 min in 15 ml of phosphate-free DMEM containing 10% dialyzed fetal bovine serum (Gibco BRL, Life Technologies, Gaithersburg, Md.), and radiolabeled for 18 to 36 h in the same medium with 0.5 mCi of [32P]orthophosphoric acid (carrier free; ICN, Irvine, Calif.) per ml. In cases where cells were serum starved (18 h), they were labeled overnight in the above phosphate-free medium without dialyzed serum and subsequently stimulated for 30 min with 100 ng of murine EGF (Sigma, St. Louis, Mo.) per ml dissolved in phosphate-free DMEM.
Immunoprecipitation and Western blotting.
Cells were washed either in phosphate-free DMEM (after radiolabeling) or with Tris-buffered saline (TBS; 50 mM TrisHCl, 150 mM NaCl [pH 7.2]) and lysed in RIPA–p-Tyr lysis buffer (150 mM NaCl, 0.25% deoxycholic acid, 1% Nonidet P-40, 50 mM Tris [pH 7.2], 0.5% aprotinin, 12.5 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate). Extracts were clarified by centrifugation at 16,000 × g for 5 min at 4°C, and protein concentrations of the supernatants were determined by the bicinchoninic acid assay (Pierce Chemical Co., Rockford, Ill.). For tryptic mapping, 5 μg of purified MAb 8C10 IgG was used to quantitatively immunoprecipitate 32P-labeled p190 from the lysate of each 15-cm dish (2 to 5 mg of protein). In the mass spectrometry (MS) experiments, 50 μg of 8C10 IgG was used to immunoprecipitate unlabeled p190 from 100 mg of lysate.
For analysis of endogenous p190-p120 complexes, 5 μg of purified MAb 6F2 was used to quantitatively immunoprecipitate p120 from 1 mg of lysate. Primary immune complexes were captured on protein A-Sepharose, and pellets were washed three times in RIPA–p-Tyr lysis buffer. Immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 7% acrylamide gels and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore Corp., Bedford, Mass.) by using a graphite electroblotting apparatus (Millipore) at a current of 0.8 mA per cm2 of membrane for 3 h to ensure complete transfer of p190. The membrane was then incubated in blocking buffer (4% bovine serum albumin in TBS with 0.1% Tween 20) for 1 h at room temperature and subsequently in blocking buffer containing primary antibody 8C10, 6F2, or 4G10 at 1 to 5 μg/ml for 1 h at room temperature. Binding of primary antibody was detected with 125I-labeled goat anti-mouse IgG (NEN, Boston, Mass.) incubated at a concentration of 1 μCi/ml in blocking buffer for 30 min at room temperature. The membrane was then washed three times for 5 min each in TBS with 0.1% Tween 20, air dried, and subjected to autoradiography. Bound radioactivity was quantitated by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, Calif.) after 5 to 30 days of exposure and expressed as PhosphorImager units (PIU).
Two-dimensional phosphopeptide analysis and phosphoamino acid analysis.
Phosphopeptide mapping of metabolically 32P-labeled p190 was done by the method of Boyle et al. (2), with an approximately 80% efficiency of elution of labeled p190 from the gel slice. Trypsinized, oxidized samples (representing 70% of the p190 in the gel) were analyzed on 10- by 10-cm thin-layer cellulose (TLC) plates (E. Merck, Darmstadt, Germany) by electrophoresis in the first dimension at 1,500 V for 22 min in pH 1.9 buffer (2.2% formic acid, 7.8% glacial acetic acid), using a Hunter thin-layer electrophoresis unit (HTLE 7000; CBS Scientific, Del Mar, Calif.), and by chromatography in the second dimension in isobutyric acid buffer (isobutyric acid–n-butanol–pyridine–glacial acetic acid–H2O, 62.5:1.9:4.8:2.9:27.9 by volume). Labeled peptides were detected by autoradiography using Biomax film (Eastman Kodak, Rochester, N.Y.).
For phosphoamino acid analysis, individual phosphopeptides were scraped from TLC plates and subjected to acid hydrolysis and two-dimensional electrophoresis as described by Boyle et al. (2). The first dimension was carried out in pH 1.9 buffer at 1,500 V for 37 min, and the second was carried out in pH 3.5 buffer (5% glacial acetic acid, 0.5% pyridine) at 1,300 V for 29 min. Levels of p-Tyr, phosphoserine (p-Ser), and phosphothreonine (p-Thr) were quantitated by PhosphorImager scanning. Relative p-Tyr levels in p190 from Neo, K− c-Src, and K+ c-Src cell lines as determined by phosphoamino acid analysis of metabolically labeled p190 were similar to those obtained from p-Tyr immunoblots of p190.
Sequential Edman analysis.
Peptide 3 was isolated and eluted from multiple phosphotryptic maps of p190 from IV5 cells that had been metabolically labeled with 32Pi. Labeling with 60 mCi of 32P yielded 2,300 cpm of peptide 3. Peptide 3 was subjected to Edman degradation at the University of Virginia Biomolecular Research Facility according to the method of Shannon and Fox (29). Briefly, the peptide was covalently coupled to a modified Sequelon membrane and washed with acetonitrile-trifluoroacetic acid and methanol. Approximately 900 cpm of membrane-coupled peptide 3 was then subjected to analysis in an Applied Biosystems 470A Sequenator. The amount of radioactivity in each fraction was measured by Cerenkov counting on a Beckman model LS 5801 liquid scintillation counter.
MS analysis.
Unlabeled p190 was immunoprecipitated from ~75 mg of clarified K+ c-Src cellular lysate by using MAb 8C10. Precipitated proteins were separated by SDS-PAGE and localized by Coomassie blue staining. Judging from the intensity of the stain, ~1 μg (5 pmol) of p190 was present in the gel. p190 was then excised from the gel, and gel pieces were minced, washed and soaked in 50% methanol overnight, dehydrated in acetonitrile, reduced in 10 mM dithiothreitol in 0.1 M NH4HCO3 for 1 h at 55°C, and alkylated in 50 mM iodoacetamide–0.1 M NH4HCO3 for 1 h at room temperature. After two cycles of washing with 0.1 M NH4HCO3 and dehydrating for 5 min in acetonitrile, the gel pieces were rehydrated in a solution containing trypsin (12.5 ng/μl) in 50 mM NH4HCO3 and incubated on ice for 45 min. Excess trypsin was removed, and samples were digested overnight at 37°C in the presence of 50 mM NH4HCO3. The resulting peptides were extracted from the polyacrylamide in two 200-μl aliquots of 50% acetonitrile–5% formic acid. These extracts were combined and evaporated to ~20 μl for liquid chromatography (LC)-MS analysis. Approximately 1 pmol of trypsinized p190 (50 fmol/μl) was extracted from the gel. LC-MS analysis was accomplished with a Finnigan-MAT TSQ7000 system equipped with an electrospray ion source interfaced to a 10-cm by 75-μm-internal-diameter POROS 10 R2 reversed-phase capillary column. For each analysis, 50 fmol of digest was injected, and peptides were eluted by an acetonitrile–0.1 M acetic acid gradient at a flow rate of 0.6 μl/min. Molecular weights of the peptides were determined by capillary LC-MS, and peptide sequences were determined by collision-activated dissociation (CAD) using LC-electrospray-tandem MS with argon as the collision gas. To verify the sequence, Tyr and Ser phosphorylated peptides were synthesized at the University of Virginia Biomolecular Research Facility. The MS work was performed at the W. M. Keck Biomedical Mass Spectrometry Laboratory at the University of Virginia.
Construction of p190 RhoGAP expression plasmids.
A bacterial expression plasmid encoding glutathione S-transferase (GST) fused to the N terminus of the middle domain of p190 RhoGAP (residues 380 to 1180) was generated by PCR-based amplification of the middle domain and incorporation of an N-terminal BamHI restriction site and a C-terminal EcoRI restriction site, which allowed subsequent insertion of the PCR product into the pGEX-2TK vector (Pharmacia, Uppsala, Sweden). Two point mutants (Y1105F and Y1087F) of this construct were generated by using a Quickchange mutagenesis kit (Stratagene). Fusion proteins were purified from extracts of transformed Escherichia coli INVαF′ One Shot competent cells (Invitrogen, San Diego, Calif.) by glutathione-Sepharose chromatography (Pharmacia).
The mammalian expression vector pRc/CMV (Invitrogen), encoding full-length hemagglutinin (HA)-tagged wild-type (wt) p190 RhoGAP, was provided by J. Settleman (27). Three variants of p190 (Y1087F, Y1105F, and Y1087/Y1105F) were generated from this vector, using a Quickchange mutagenesis kit (Stratagene). All constructs were sequenced and verified to be correct.
Tryptic phosphopeptide analysis of in vitro-phosphorylated p190 middle domain.
Six micrograms of purified GST-p190 middle domain was incubated in the presence of 150 μCi of [γ-32P]ATP (6,000 Ci/mmol, 150 mCi/ml; Dupont, NEN, Boston, Mass.) and 9 U of purified baculovirus c-Src (UBI) for 30 min at 30°C in a final volume of 68 μl of kinase buffer (50 mM Tris, 10 mM MnCl2, 5 mM glutathione [pH 7.2]). Reactions were stopped by the addition of 2× SDS sample buffer and boiled for 5 min. Proteins were separated by SDS-PAGE and stained with Coomassie blue. The gel was dried, and the 32P-labeled GST-p190 middle domain was identified by autoradiography, excised, and subjected to tryptic phosphopeptide analysis by the method of Boyle et al. (2). As a control, an equivalent molar amount of GST alone was phosphorylated and analyzed under identical conditions.
GST fusion protein coassociation experiments.
Purified GST fusion proteins attached to glutathione-Sepharose beads were phosphorylated by c-Src as described above (except that [γ-32P]ATP was replaced by 100 μM unlabeled ATP) or subjected to mock phosphorylation by incubation in kinase buffer without c-Src. In a separate experiment that used both unlabeled and radiolabeled ATP, the stoichiometry of phosphorylation was determined to be approximately 0.7 mol of PO4/mol of fusion protein. After phosphorylation, the beads were washed, and 30 μg of bound GST fusion protein was incubated with 1 mg of clarified RIPA–p-Tyr cellular lysate protein for 1, 3, 6, or 24 h at 4°C with constant rotation. An equal molar amount of GST alone was incubated with whole-cell lysates to control for nonspecific protein interactions. Samples were then washed three times with RIPA–p-Tyr lysis buffer, subjected to SDS-PAGE, transferred to Immobilon-P, and immunoblotted for p120 or p-Tyr, using 125I-labeled goat anti-mouse Ig as the developing reagent. Radioisotope binding was visualized by autoradiography and quantitated by PhosphorImager analysis after 5 days of exposure.
Transient overexpression studies.
COS-7 cells were transfected with 4 μg of the pRc/CMV vector (lacking the p190 insert), or the same vector containing HA-tagged, wt full-length p190, the Y1105F mutant of p190, the Y1087F mutant, or the Y1087F/Y1105F mutant with or without 8 μg of plasmid pcDNAc-Src, using SuperFect transfection reagent (Qiagen Inc., Valencia, Calif.) according to the manufacturer’s directions; 48 h later, cells were lysed with RIPA–p-Tyr buffer, and ectopically expressed p190 was immunoprecipitated with the anti-HA MAb 12CA5 (Babco, Richmond, Calif.) from equal amounts of lysate protein. Precipitated proteins were separated by SDS-PAGE, transferred to Immobilon-P, and analyzed for p190, p120, or levels of p-Tyr on p190 by Western immunoblotting. 125I-labeled goat anti-mouse Ig binding to primary antibodies was visualized by autoradiography and quantitated by PhosphorImager analysis.
RESULTS
Characterization of p190 phosphorylation by tryptic phosphopeptide mapping and phosphoamino acid analysis.
Characterization of p190 tyrosine phosphorylation was first approached by determining the relative levels of phosphoamino acids in the total pool of endogenous, 32P-labeled p190 isolated from our panel of cell lines. v-Src-transformed (IV5) cells were included in this analysis, because p190 was first observed in v-Src-transformed fibroblasts and because p190 is heavily tyrosine phosphorylated in the presence of v-Src (8). Quantitation by PhosphorImager analysis showed that p190 from IV5 cells contained ~10% p-Tyr, 90% p-Ser, and <0.1% p-Thr, while p190 from K+ c-Src cells contained 2 to 5% p-Tyr, 95 to 98% p-Ser, and <0.1% p-Thr and that from Neo cells contained <1% p-Tyr ~99% p-Ser, and <0.1% p-Thr. No p-Tyr was detected by this method in p190 from K− c-Src cells, indicating that the protein was almost exclusively phosphorylated on serine residues. To determine if different p190 peptides were tyrosine phosphorylated in IV5 cells compared to K+ c-Src cells, tryptic phosphopeptide maps of p190 from each cell line were generated (Fig. (Fig.1A1A and B). The tryptic map of p190 from IV5 cells revealed a complex pattern of approximately 27 phosphopeptides (Fig. (Fig.1A).1A). Phosphoamino acid analysis of each peptide in the tryptic map showed that five peptides contained p-Tyr (peptides 3, 8, 24, 25, and 27) (Table (Table1)1) and that peptide 3 contained the greatest amount of p-Tyr, representing 80% of the total p-Tyr in p190 (Table (Table2).2). Peptide 8, the next most abundant p-Tyr-containing peptide, contained only 15% of the total p-Tyr (Table (Table2).2).
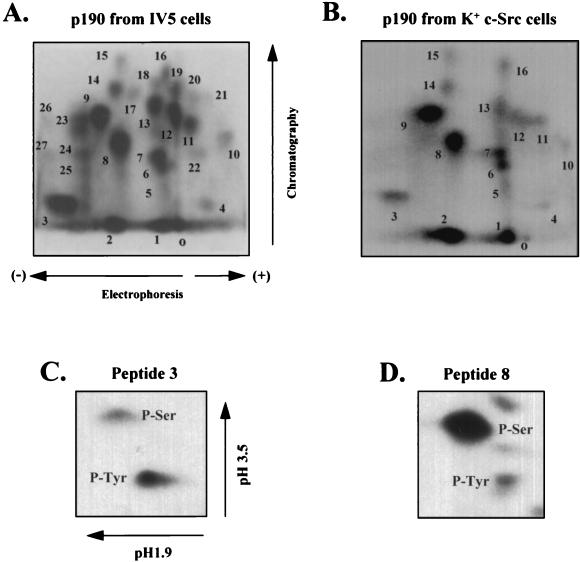
Two-dimensional tryptic phosphopeptide analysis of metabolically 32P-labeled p190 and phosphoamino acid analysis of p-Tyr-containing peptides. (A and B) Two-dimensional tryptic phosphopeptide maps were generated from metabolically 32P-labeled p190, immunoprecipitated with MAb 8C10 from v-Src-transformed IV5 cells (A) or K+ c-Src cells (B) and processed as described in Materials and Methods. Peptides were spotted onto TLC plates at the origin (marked “o”), resolved by electrophoresis in the horizontal dimension and chromatography in the vertical dimension, and visualized by autoradiography. Peptides displaying the same migration pattern in both maps are numbered identically. In panel A, 1,150 sample cpm was applied, and plates were exposed for 5 days on Kodak BioMax film. In panel B, 2,200 cpm was applied, and plates were exposed for 48 h on the same film. (C and D) Two-dimensional phosphoamino acid analysis. Phosphopeptides 3 (C) and 8 (D) were isolated from peptide maps of p190 from K+ c-Src cells and hydrolyzed as described in Materials and Methods. Unlabeled p-Ser, p-Thr, and p-Tyr standards were added, and the samples were spotted onto cellulose plates and separated by electrophoresis in pH 1.9 buffer in the horizontal dimension and in pH 3.5 buffer in the vertical dimension. Standards were visualized with 0.25% ninhydrin; 400 cpm of peptide 3 and 1,500 cpm of peptide 8 were applied, and plates were exposed for 54 h on BioMax film.
TABLE 1
Phosphoamino acid content of p190 phosphopeptides from 32P-labeled IV5 cells
cells
| Peptide no. | Content (%)a
| ||
|---|---|---|---|
| p-Tyr | p-Ser | p-Thr | |
| 1 | 0 | 100 | 0 |
| 2 | 0 | 100 | 0 |
| 3 | 70–100 |  0–30 0–30 | 0 |
| 4 | 0 | 100 | 0 |
| 5 | ND | ND | ND |
| 6 | 0 | 100 | 0 |
| 7 | 0 | 100 | 0 |
| 8 |  5–14 5–14 | 86–95 | <0.1 |
| 9–20 | 0 | 100 | 0 |
| 21 | ND | ND | ND |
| 22 | 0 | 100 | 0 |
| 23 | 0 | 100 | 0 |
| 24 | 40 | 60 | 0 |
| 25 | 60 | 40 | 0 |
| 26 | 0 | 100 | 0 |
| 27 | 35 | 50 | 15 |
TABLE 2
Percentage of total p-Tyr contained in individual p190 phosphopeptides from 32P-labeled IV5 cells
cells
| Peptide no. | p-Tyr contenta
| |
|---|---|---|
| PIU | % of total p-Tyr in p190 | |
| 3 | 27,100 | 79.8 |
| 8 | 5,072 | 14.9 |
| 24 | 595 | 1.8 |
| 25 | 1,087 | 3.2 |
| 27 | 115 | 0.3 |
 Total Total | 33,969 | 100 |
The phosphopeptide map of p190 from K+ c-Src cells was significantly less complex than that from IV5 cells, as only 16 phosphopeptides were reproducibly generated (Fig. (Fig.1B)1B) and only 2 (peptides 3 and 8 [Fig. 1C and D]) contained p-Tyr. The phosphoamino acid content of peptides 3 and 8 was nearly identical to that of p190 from IV5 cells (Table (Table1),1), but the p-Tyr content of peptide 3 represented more of the total p-Tyr in p190 from K+ c-Src cells (95%) than the same peptide from IV5 cells (80%), while peptide 8 contained only 5% of the total pTyr in K+ c-Src cells, versus 15% in IV5 cells. The remaining 14 peptides from K+ c-Src cells were identical in phosphoamino acid content to those from IV5 cells (only p-Ser was detected). The finding that peptide 3 accounted for such a high percentage of the total p-Tyr content of p190 suggested that it contained either multiple p-Tyr sites or a single tyrosine residue that was highly phosphorylated. To distinguish between these possibilities, we identified the site(s) of tyrosine phosphorylation in peptide 3.
Sequential Edman analysis of the major tyrosine-containing phosphopeptide from p190.
To determine the identity of the phosphorylated residue(s) in peptide 3, multiple phosphotryptic maps of 32P-labeled p190 from IV5 cells were generated, and peptide 3 was scraped from the TLC plates and eluted from the cellulose. Eluates were pooled and subjected to Edman degradation. Radiolabel was released at cycle 7 (data not shown), indicating that the seventh residue was phosphorylated. Although peptide 3 was composed of as much as 30% p-Ser in some analyses (Table (Table1),1), the level of the signal seen at residue 7 for peptide 3 was consistent with it representing the p-Tyr moiety of this peptide. The four tryptic peptides in p190 that contain a Tyr residue at position 7 are listed in Table Table3.3. The relative mobilities of these peptides, based on mass/electrical charge ratios (m/z) and overall hydrophobicity, were calculated (2). Peptide 3 exhibited high mobility in the electrophoretic dimension and low mobility in the chromatographic dimension (Fig. (Fig.1A1A and B). The peptides STALQPY116IK and NEEENIY1105SVPHDSTQGK have the greatest mobilities in the electrophoretic dimension, since they have the lowest m/z ratios among the candidates (Table (Table3).3). The peptide NEEENIY1105SVPHDSTQGK has the lowest mobility in the chromatography dimension, as it is the most hydrophilic peptide in the group (Table (Table3).3). Based on both of these parameters, peptide NEEENIY1105SVPHDSTQGK most closely matched the migration pattern of peptide 3.
TABLE 3
Candidates for p190 peptide 3 and their physical characteristics
characteristics
| Peptide sequence | Molecular mass (M+H, Da)
| Expected charge | m/z
| Relative hydrophobicity | ||
|---|---|---|---|---|---|---|
| Unphosphorylated | Phosphorylated | Unphosphorylated | Phosphorylated | |||
| STALQPY116IK | 1,021.2 | 1,101.2 | +2 | 511.1 | 551.1 | 0.653 |
| MQASPEY305QDYVYLEGTQK | 2,151.4 | 2,231.4 | +2 | 1,076.2 | 1,116.2 | 0.603 |
| NEEENIY1105SVPHDSTQGK | 1,948.0 | 2,028.0 | +3 | 650.0 | 676.7 | 0.572 |
| ATWESNY1246FGVPLTTVVTPEK | 2,240.5 | 2,320.5 | +2 | 1,120.8 | 1,160.8 | 0.650 |
Identification by MS of Y1105 as an in vivo phosphorylation site in p190.
To determine which of the four p-Tyr candidate peptides was actually tyrosine phosphorylated in vivo, tandem MS analysis was carried out on unlabeled tryptic peptides of p190 from K+ c-Src cells as described in Materials and Methods. The predicted m/z ratio for each of the candidate peptides in the phosphorylated and unphosphorylated states is shown in Table Table3.3. It was estimated that over 1,000 different ions, distinguished by either m/z ratio or retention time, were present in the digest above a 10% relative abundance threshold. This data set of ions was then searched, using the predicted m/z ratios of the candidate peptides. LC-MS analysis detected peptide candidate NEEENIY1105SVPHDSTQGK in the unphosphorylated state (m/z = 650 for +3 peptide and 974 for +2 peptide) and the phosphorylated state (m/z = 676.7 for +3 peptide and 1,014.5 for +2 peptide). In the mass spectrum obtained at this retention time, the triply charged unphosphorylated and phosphorylated ions of this peptide were present at ~20% relative abundance and therefore were readily detectable in a complex spectrum containing many other ions (data not shown). Moreover, it appeared that approximately 50% of this peptide was in the phosphorylated state, as both the phosphorylated and unphosphorylated ions were present at the same relative intensity.
Tandem MS coupled with CAD analysis was then used to confirm the sequence of the triply charged, phosphorylated form of this peptide ion and to determine whether the phosphorylation was on Y1105, S1106, S1111, or T1112. Figure Figure2A2A shows the detection of ion y5 (representing peptide fragment S1111TQGK) with an m/z ratio of 520.4, consistent with the unphosphorylated state of this peptide fragment. This ruled out both S1111 and T1112 as the phosphorylated residue in this peptide. The y11 ion (representing peptide fragment Y1105SVPHDSTQGK) was detected with a m/z of 650.1, consistent with the phosphorylated state of this peptide. However, the y10 ion (m/z = 528) was not detected, and thus it could not be determined from this spectrum alone whether the phosphorylation in the y11 ion was on Y1105 or S1106. Therefore, peptides phosphorylated on either site were synthesized and analyzed by MS. Figures Figures2B2B and C show that the CAD spectrum for the synthetic tyrosine-phosphorylated peptide matches the spectrum for the endogenous peptide. The similarity of the spectra for the synthetic tyrosine-phosphorylated peptide and the endogenous peptide is particularly apparent in the ions at m/z 650, 764, and 893, which correspond to the y11, y13, and y15 ions in the doubly charged species. These ions are not seen in the serine-phosphorylated synthetic peptide due to the loss of 50 units (loss of the 98-Da H3PO4, m/z = 49 in the +2 y11 ion), a characteristic of serine-phosphorylated peptides. Taken together, these data demonstrate that Y1105 is phosphorylated in endogenous p190. Using a similar strategy, we found no evidence for phosphorylation of Y1087.
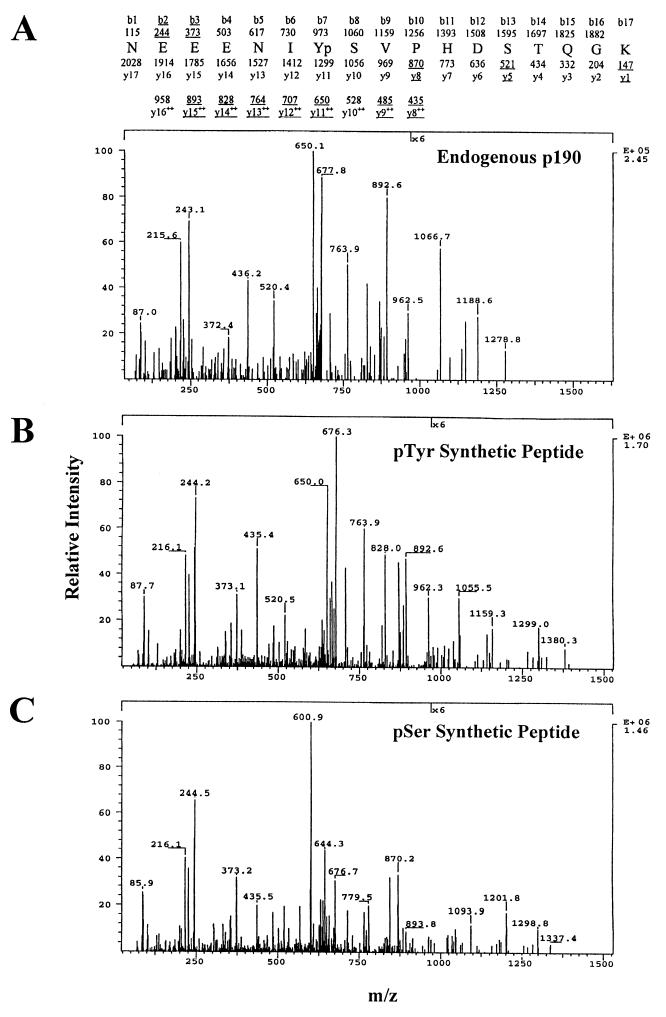
CAD mass spectra of unlabeled phosphorylated p190 peptides. (A) The sequence of the p190 tryptic peptide spanning amino acids 1099 to 1115 and the predicted m/z ratios of ion fragments of this peptide are shown above the CAD spectrum of this tryptic peptide from endogenous p190 from K+ c-Src overexpressers. Ion fragments originating from the amino terminus are listed as b1 to b17. Ions originating from the carboxy terminus are listed as y1 to y17. y8 to y16 are listed in both the singly and doubly charged forms due to the contribution of a positive charge by the His1109 residue. Underlined ions and m/z ratios represent values detected in the spectrum. (B) CAD spectrum of the synthetic peptide 1099-1115 phosphorylated at Tyr1105. (C) CAD spectrum of the synthetic peptide 1099-1115 phosphorylated at Ser1106.
Direct phosphorylation of Y1105 by c-Src in vitro.
While MS analysis identified Y1105 as a phosphorylated residue in K+ c-Src cells, it did not directly designate peptide 3 from the phosphopeptide map of in vivo-labeled p190 as the p-Y1105-containing peptide. To investigate the ability of c-Src to directly phosphorylate p190 on Y1105 and to determine if peptide 3 contained Y1105, in vitro kinase assays were performed with purified, baculovirus-expressed c-Src and a GST fusion protein spanning amino acids 380 to 1180 of p190. As shown in the phosphotryptic map in Fig. Fig.3A,3A, several peptides were phosphorylated by c-Src in vitro, but one peptide, labeled A, was phosphorylated to a much greater extent than the others. Phosphorylation of GST alone yielded one minor peptide which migrated significantly differently from peptide A (not shown). Peptide A was scraped from multiple phosphopeptide maps, and the isolated peptide was analyzed for purity by two-dimensional phosphopeptide mapping (Fig. (Fig.3B).3B). Metabolically labeled p190 from K+ c-Src cells was also analyzed by tryptic phosphopeptide mapping, either alone (Fig. (Fig.3C)3C) or in combination with the isolated in vitro-phosphorylated peptide A. Figure Figure3D3D shows that peptide A comigrated with in vivo-labeled peptide 3. Phosphoamino acid analysis of peptide A showed that it contained, as expected, only p-Tyr, and Edman degradation analysis revealed that peptide A was phosphorylated at position 7. Of the four tryptic peptide candidates that contain a Tyr at position 7, only the peptide NEEENIY1105SVPHDSTQGK is contained within the p190 GST fusion protein used in this in vitro kinase reaction. These results, together with the MS data, demonstrate that peptide 3 from the in vivo phosphotryptic map contains p-Y1105. The results also show that purified c-Src can directly phosphorylate p190 at Y1105. Further support for the identification of peptide 3 as the p-Y1105-containing peptide is provided by the finding that the synthetic peptide NEEENIpY1105SVPHDSTQGK comigrated with the in vivo-labeled peptide 3 (data not shown).
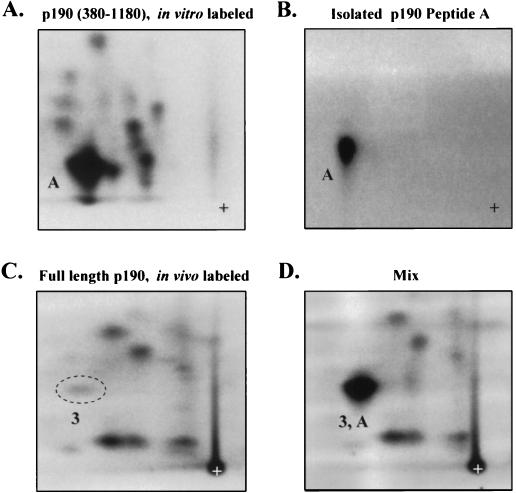
Phosphorylation of Y1105 by c-Src in vitro. (A) Phosphotryptic peptide map of the GST-middle domain (p190 residues 380 to 1180) phosphorylated in vitro with [γ-32P]ATP by purified c-Src (UBI); ~1,000 cpm was loaded. (B) Rechromatography of the isolated peptide A depicted in panel A. (C) Phosphotryptic peptide map of full-length p190, metabolically labeled with 32P; ~1,000 cpm was loaded. (D) Mix of samples depicted in panels B (~600 cpm) and C (~1,000 cpm). The comigrating phosphopeptides are labeled A and 3, respectively. Maps represent one experiment of three, all giving identical results.
Y1105 is a preferred in vivo target of c-Src.
Previous studies in our laboratory indicated that the overall level of p190 tyrosine phosphorylation (as determined by p-Tyr Western immunoblotting) was affected by the levels and catalytic activity of c-Src in cells (4, 6). However, other investigators have described increases in p190 p-Tyr content upon relatively long-term (30-min) EGF stimulation of cells overexpressing the EGF receptor (1, 8), suggesting that the EGF receptor may also phosphorylate p190. To determine if c-Src or EGF receptor overexpression or EGF stimulation resulted in the appearance of novel phosphorylation sites (compared to control cells) or increased the stoichiometry of basally phosphorylated residues, we examined the complexity of phosphotryptic peptide maps and the p-Tyr content of peptides 3 and 8 of p190 from 32P-labeled Neo control, K− c-Src, K+ c-Src, EGFR, and EGFR/K+ c-Src 10T1/2 cells that had been mock stimulated or stimulated with EGF for 30 min. Figure Figure44 shows that the maps from all cell lines, under serum-starved or EGF-stimulated conditions, were nearly identical in overall pattern, indicating that none of the manipulations resulted in phosphorylations that generated novel phosphopeptides. To determine if changes in phosphoamino acid content occurred in either peptide 3 or 8, these peptides were isolated from each of the maps and subjected to phosphoamino acid analysis. p-Tyr levels were determined relative to the total phosphoamino acid content of p190 on each cellulose plate (Table (Table4).4). Figure Figure44 and Table Table44 show that peptides 3 and 8 were tyrosine phosphorylated in Neo control cells and that the p-Tyr content of these peptides did not change appreciably upon EGF stimulation. With overexpression of either wt or kinase-defective c-Src and to a lesser extent with overexpression of EGF receptors, however, peptide 3 (Y1105) exhibited striking changes in p-Tyr level. Overexpression of K+ c-Src resulted in an ~4-fold increase in the level of Y1105 phosphorylation, whereas overexpression of K− c-Src led to a 3- to 4-fold decrease, compared to the Neo control cells. As with the Neo control cells, treatment of the K− c-Src and K+ c-Src cells with EGF for 30 min did not result in a significant change in the tyrosine phosphorylation of Y1105. Overexpression of the EGF receptor, in the presence or absence of K+ c-Src overexpression, resulted in a modest (~1.5-fold) increase in phosphorylation of Y1105 in the absence of EGF stimulation and an ~2-fold increase upon treatment with EGF. This increase was not as great as that seen with c-Src overexpression. In contrast to peptide 3, the p-Tyr level of peptide 8 remained constant throughout the analysis. Interestingly, differences in levels of Y1105 phosphorylation in the different cell lines paralleled the differences seen by Western immunoblotting (4, 6), suggesting that Y1105 represents the major residue whose phosphorylation is regulated. Taken together, these results indicate that Y1105 is a preferred in vivo target for c-Src compared to the EGF receptor and that peptide 8 is not a target for either tyrosine kinase. Furthermore, peptide 8 contained less p-Tyr than peptide 3 in all cell lines except K− c-Src (ranging from ratios of 1:4 in Neo control cells to 1:60 in EGFR/K+ c-Src cells stimulated with EGF). The potential significance of this difference is discussed below.
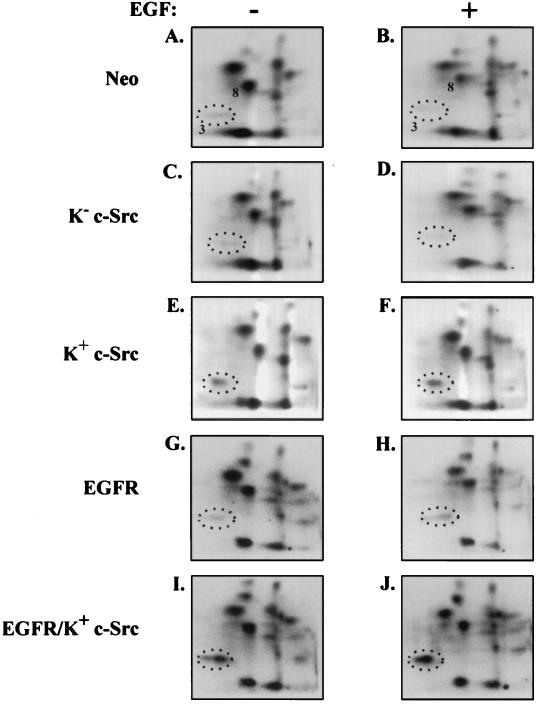
Y1105 is a preferred in vivo c-Src phosphorylation site. Indicated cell lines were brought to near confluence, serum starved and labeled with 32P for 18 h, and either left unstimulated or stimulated for 30 min with EGF (100 ng/ml). P190 was immunoprecipitated and processed to generate phosphotryptic peptides. Phosphopeptides were analyzed by electrophoresis in the first dimension and chromatography in the second dimension on TLC plates. Amounts (in counts per minute) of sample loaded: (A) 2,025; (B) 2,450; (C) 2,360; (D) 1,796; (E) 2,212; (F) 2,254; (G) 2,117; (H) 1,834; (I) 1,389; (J) 760. Times of exposure on BioMax film were 20 (A to F), 24 (G and H), and 72 (I and J) h.
TABLE 4
Percentages of p-Tyr in peptide 3 (Y1105) and peptide 8 relative to total p190 phosphoamino acid content in various c-Src/EGFR overexpressers
overexpressers
| Cell line | EGF | p-Tyr content (%)a
| |
|---|---|---|---|
| Peptide 3 | Peptide 8 | ||
| Neo | − | 0.29 ± ± 0.07 0.07 | 0.07 ± ± 0.04 0.04 |
| + | 0.19 ± ± 0.06 0.06 | 0.06 ± ± 0.03 0.03 | |
| K− c-Src | − | 0.09 ± ± 0.04 0.04 | 0.05 ± ± 0.05 0.05 |
| + | 0.04 ± ± 0.01 0.01 | 0.08 | |
| K+ c-Src | − | 1.12 ± ± 0.11 0.11 | 0.05 |
| + | 1.01 ± ± 0.41 0.41 | 0.01 | |
| EGFR | − | 0.38 | 0.02 |
| + | 0.74 | 0.01 | |
| K+ c-Src/EGFR | − | 1.54 | 0.03 ± ± 0.02 0.02 |
| + | 3.05 | 0.03 | |
p-Tyr-dependent and -independent mechanisms of p190-p120 association.
Previous studies demonstrated that the two SH2 domains of p120 RasGAP synergistically bind tyrosine phosphorylated p190 (3, 10). These findings suggest that the interaction of p190 with p120 is mediated by p-Tyr–SH2 interactions, possibly involving Y1105 and another phosphorylated Tyr in p190. Indeed, Hu and Settleman (9) reported that mutagenesis of both Y1105 and Y1087 in p190 disrupts the p190-p120 interaction in cells overexpressing both proteins. However, as yet we have not detected in vivo phosphorylation of Y1087, and although we have observed two p-Tyr-containing peptides in p190 (one of which is Y1105), the two peptides are disparately phosphorylated. These findings suggest that if tandem p-Tyr–SH2 interactions are required for complex formation, only a minority of the p190 molecules in a cell qualify for interactions via this mechanism. Furthermore, previous studies using the 10T1/2 cell system demonstrated that all tyrosine-phosphorylated p190 is complexed with p120 (4); in the present study, immunoprecipitation of p120 from 32P-labeled cells, followed by p190 immunoprecipitation and tryptic phosphopeptide analysis of complexed and free p190, confirmed that all p190 phosphorylated on Tyr1105 was associated with p120 (not shown). These considerations raise the possibility that mechanisms other than paired p-Tyr–SH2 interactions regulate binding of p190 to p120, including single p-Tyr–SH2 interactions.
To examine this issue further, we carried out p120 immunoprecipitation studies in the various c-Src overexpressor cell lines that exhibited different levels of p190 tyrosyl phosphorylation and compared both the amounts of p190 and the levels of p-Tyr in p190 that associated with p120. We reasoned that if the level of p-Tyr in p190 were the sole regulator of the ability of p190 to complex with p120, the amount of p190 associated with p120 would vary proportionately with the p-Tyr level in p190 associated with p120. Figure Figure5A5A shows a general correlation between the p-Tyr content and the amount of p190 protein that associates with p120 in the three cell lines, i.e., that overexpression of K+ c-Src resulted in an increase in both p-Tyr content and amount of p190 protein associated with p120, while overexpression of K− c-Src resulted in a decrease in both, relative to Neo controls. Furthermore, in a parallel experiment we observed no effect of EGF stimulation on either the p-Tyr content or the amount of p190 associated with p120 in any of the cell lines (data not shown). Figure Figure5B5B shows that upon quantitation of these and additional experiments, the levels of 190 protein associated with p120 were ~3.5-fold lower in K+ c-Src cells (Fig. (Fig.5B,5B, inset) and ~2-fold higher in K− c-Src cells than would be expected from the levels of p190 tyrosine phosphorylation. These data strongly suggest that p-Tyr-independent as well as -dependent mechanisms regulate the interaction between these two proteins and that phosphorylation of Y1105 may have functions other than regulating association with p120.
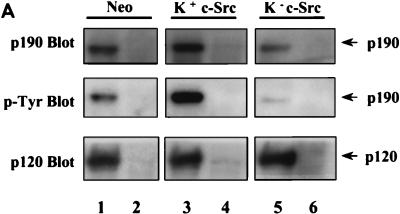
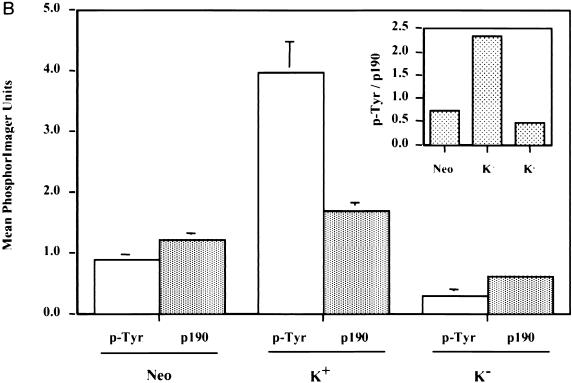
p-Tyr-dependent and -independent mechanisms of p190-p120 association. (A) p120 RasGAP was quantitatively immunoprecipitated from 500 μg of lysate derived from serum-deprived, nonstimulated Neo cells (lane 1), K+ c-Src cells (lane 3), or K− c-Src cells (lane 5), using MAb 6F2. In lanes 2, 4, and 6, mouse IgG (Jackson ImmunoResearch Laboratories) was used as a negative antibody control. Precipitated proteins were resolved by SDS-PAGE, transferred to an Immobilon membrane, and immunoblotted with anti-p120 MAb 6F2 (bottom lanes) and anti-p190 MAb 8C10 (top lanes). The membrane slice containing p190 was stripped of 8C10 and reprobed with anti-p-Tyr MAb 4G10 (middle lanes). Localization of primary antibodies was revealed by binding of 125I-labeled rabbit anti-mouse IgG and autoradiography. (B) The amount of radioactivity in each lane of panel A was quantitated by PhosphorImager analysis, and the PIU for p190 protein and p-Tyr p190 were adjusted to the amount of p120 in each immunoprecipitate. Adjusted PIU for each antibody (anti-p-Tyr or anti-p190) were then compared across the cell lines, arbitrarily setting the normalized value for Neo control cells to 1.00 in each experiment for each antibody. Results from two independent experiments, one done in triplicate and one done in quadruplicate for each cell line and antibody, were pooled and expressed as the mean ± standard error. Results from both nonstimulated and EGF-stimulated (not shown in panel A) cells were included in the analysis. Differences in mean p-Tyr levels of the three cell lines are significantly different by Student’s paired t test (P < 0.005). Differences in mean p190 protein associated with p120 between the three cell lines are significant to a P value of <0.02. To assess the relative differences in p190 p-Tyr content versus p190 protein associated with p120 RasGAP between the different cell lines, the mean p-Tyr/p190 protein ratio was calculated for each cell line and is shown in the inset.
To test directly the requirement for tyrosine phosphorylation of p190 in general and Y1105 specifically for association of p190 with p120, two experiments were performed. In the first, GST fusion proteins containing the middle domain of p190, either wt or a variant harboring a Y1105F mutation, were examined for the ability to bind and coprecipitate p120 from extracts of Neo cells. Before incubation with the extract, fusion proteins were either phosphorylated in vitro by c-Src or subjected to mock phosphorylation as described in Materials and Methods. A fusion protein containing a Y1087F mutation was also included in the analysis. Figure Figure6A6A shows that p120 associated with both the wt and Y1087F mutant forms of the middle domain in both c-Src phosphorylation-dependent and -independent manners, while no association was detected with the Y1105F mutant. In the wt and Y1087F variants, binding of p120 to the phosphorylated forms was significantly greater than binding to the unphosphorylated forms, a finding that is consistent with the analysis of endogenous p190 depicted in Fig. Fig.5.5. Figure Figure6B6B shows that mutation of Y1105 to F ablated the ability of c-Src to phosphorylate the middle domain, while the Y1087F mutation had no effect. No phosphotyrosine could be detected on the fusion protein containing wt p190 middle domain that had been mock phosphorylated, indicating that binding of p120 under these conditions occurred via a p-Tyr-independent mechanism. To control for nonspecific association, GST alone was included in the analysis, and the p120 membrane was also reprobed with anti-mitogen-activated protein kinase antibody. No nonspecific binding could be detected in either case. The same pattern of p120 binding was observed when extracts from K+ c-Src or K− c-Src cells were used and when incubations were conducted for various lengths of time from 1 to 24 h. We conclude from these results that p190 and p120 bind one another through both p-Tyr-dependent and p-Tyr-independent mechanisms and that phosphorylation of Y1105 by c-Src is sufficient for mediating the p-Tyr-dependent association of p190 with p120.
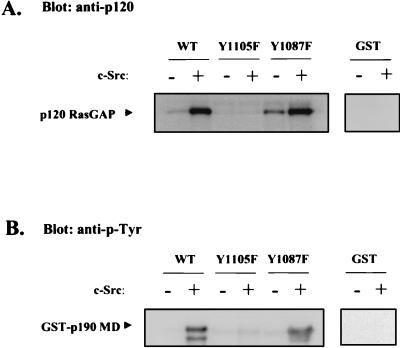
Binding of p120 RasGAP to GST fusion proteins containing p190 middle domain variants. GST fusion proteins containing a variant (wt, Y1105F, or Y1087F) of the p190 middle domain (MD; residues 380 to 1180) or GST alone attached to glutathione-agarose beads were either mock phosphorylated (−) or phosphorylated (+) by c-Src in vitro, washed, and incubated with detergent lysates of Neo cells for 3 h at room temperature. Beads were pelleted and washed, and precipitated proteins were subjected to SDS-PAGE and Western immunoblotting with 6F2 anti-p120 RasGAP (A) or 4G10 anti-p-Tyr (B) MAb and 125I-labeled goat anti-mouse Ig.
In the second experiment, cytomegalovirus-based mammalian expression vectors encoding full-length, HA-tagged wt, Y1105F, Y1087F, or Y1105F/Y1087F p190 were transiently transfected with or without a vector encoding wt c-Src into COS-7 cells, and exogenously expressed p190 variants were examined for the ability to bind endogenous p120 in a coprecipitation assay. The results of one such experiment are depicted in Fig. Fig.7.7. Mutation of Y1105 (lane 5), but not of Y1087 (lane 6), ablated the ability of p190 to associate with p120 (center blot). This was the case whether Y1105F was present as a single mutation (lane 5) or as one of a pair with Y1087 (lane 7). Interestingly, in vivo, all forms of p190 became tyrosine phosphorylated (to various extents in multiple experiments) when expressed with c-Src (lower blot), but only when Y1105 was present was binding to p120 detected (lanes 4 and 6). These results provide additional evidence for the critical role of phosphorylated Y1105 in regulating p120 binding. They further suggest that when c-Src is overexpressed together with p190 in COS-7 cells, aberrant phosphorylations that are not seen with the endogenous protein appear to occur. In the absence of ectopically expressed c-Src, ectopically expressed wt p190 was also observed to associate with endogenous p120 (lane 2), although the p-Tyr immunoblot of p190 revealed no detectable tyrosine phosphorylation in the absence of c-Src overexpression. This result indicated that the association occurred via p-Tyr-independent mechanisms. Similar results were observed for each of the p190 variants. Quantification of three independent experiments revealed that p-Tyr-independent complexes represented 10 to 20% of the p-Tyr-dependent interactions. Overall, the results of this experiment are consistent with those depicted in Fig. Fig.55 and and6,6, i.e., that both p-Tyr-dependent and p-Tyr-independent interactions mediate association between p190 and p120, with p-Y1105 being the major regulator.
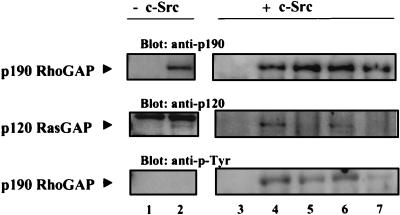
In vivo binding of p120 RasGAP to ectopically expressed p190 variants. The mammalian expression plasmid pRc/CMV, encoding full-length, HA-tagged wt p190 (lanes 2 and 4) or variant Y1105F (lane 5), Y1087F (lane 6), or Y1105F/Y1087F (lane 7), was transiently transfected into COS-7 cells with (lanes 3 to 7) or without (lanes 1 and 2) a pcDNA3 plasmid encoding wt c-Src. pRc/CMV plasmid lacking the p190 coding sequences was transfected to control for nonspecific protein interactions (lanes 1 and 3). Ectopically expressed p190 was immunoprecipitated with anti-HA MAb 12CA5, immunoprecipitates were divided into two equal parts, and each was subjected to SDS-PAGE. Western immunoblotting was then carried out, probing one membrane first with anti-p190 MAb 8C10 (top panel, lanes 3 to 7) or anti-HA MAb 12CA5 (lanes 1 and 2) and then with anti-p120 MAb 6F2 (middle panel). The second membrane was probed with anti-p-Tyr MAb 4G10 (bottom panel). Primary antibodies were visualized with 125I-labeled goat anti-mouse Ig. Results are representative of three independent experiments.
DISCUSSION
Tyrosine phosphorylation of p190 RhoGAP has been reported to play a role in mediating both the disassembly of actin stress fibers induced by EGF (6) and the interaction of p190 with p120 RasGAP (3, 4, 9). A first step in understanding the potential relationship between these two events is to determine the complexity and identity of p-Tyr residues on p190 and to characterize the nature of the association between p190 and p120. Given the size of the p190 molecule, the large number of potential p-Tyr sites, and the suggestion from previous mutagenesis studies that at least two sites would be phosphorylated to the same extent (9), it was surprising to find only one major site of tyrosine phosphorylation (Y1105) on p190 (Fig. (Fig.22 and Table Table3).3). However, this result was reproduced whether p190 was derived from v-Src or EGF receptor-transformed cells, from wt c-Src overexpressers, or from normal control fibroblasts, stimulated or not with EGF (Fig. (Fig.4).4). Furthermore, both MS analysis (Fig. (Fig.2)2) and mutagenesis (Fig. (Fig.66 and and7)7) failed to support a role for phosphorylation of Y1087 (the second site proposed by Hu and Settleman [9]). Together, these results confirm that only one major site of tyrosine phosphorylation (Y1105) exists on p190.
Y1105 was further shown to be a preferred substrate of c-Src both in vitro and in vivo (Fig. (Fig.33 and and4)4) and to be the sole mediator of the p-Tyr-dependent association of p190 with p120 (Fig. (Fig.66 and and7),7), thus directly linking c-Src to the regulation of p190-p120 complex formation. Indeed, in 10T1/2 fibroblasts that overexpressed K+ or K− c-Src, the level of p190 complexed with p120 generally correlated with the overall level of p190 tyrosine phosphorylation (Fig. (Fig.5).5). Further analysis also revealed that the level of p-Y1105 was a close measure of total p190 tyrosine phosphorylation (p-Y1105 represented 95% of the total p-Tyr in p190 from K+ c-Src cells and 80% in p190 from Neo control cells, while the minor site represented 5 and 20%, respectively [Fig. 4 and Table Table4]).4]). In addition, all of the tyrosine-phosphorylated p190 was found in complex with p120. Together these data are consistent with a model in which formation of the p190-p120 complex is regulated by a single p-Tyr–SH2 interaction.
However, further investigation revealed that such a model may be an oversimplification. Results from three different experiments (Fig. (Fig.55 to to7)7) provided evidence for a p-Tyr-independent mechanism of association as well as a p-Tyr-dependent mechanism. While p-Tyr-independent complexes were less abundant than the p-Tyr-dependent complexes, they still represented a significant proportion (10 to 20% of the p-Tyr-dependent complexes). Furthermore, quantitative analysis indicated that under circumstances of relatively high stoichiometry of p190 tyrosine phosphorylation (such as in K+ c-Src overexpressers [Fig. 5]), the ratio of tyrosine-phosphorylated p190 to the amount of p190 protein complexed with p120 was greater than 1 (i.e., 3.5), suggesting that some p-Y1105 was not bound by the SH2 domain of p120 and was free to carry out other functions. Since all of the tyrosine-phosphorylated p190 is complexed with p120, we reasoned that under such circumstances, p190 also must be binding to p120 in a p-Tyr-independent manner. Thus, a more complex picture emerges regarding the mechanisms regulating p190-p120 interaction, as well as the possibility that p-Y1105 has more than one role to play in p190 function.
Although the nature of the p-Tyr-independent interaction is unclear, several models could explain our data while not contradicting the published report that both SH2 domains of p120 are required for association (3). One possibility is that p190 and p120 are bridged by an as yet unidentified tyrosine-phosphorylated protein that binds one of the SH2 domains of p120, while p-Y1105 of p190 binds the other SH2 domain. In support of this model, we have observed several cellular proteins that coprecipitate with p190 and become phosphorylated on tyrosine when the immune complexes are subjected to an in vitro kinase assay. SH3-polyproline interactions could play a role in the association between p190 and its coprecipitating proteins, because close examination of the amino acid sequence of p190 reveals 13 PXXP motifs scattered throughout the molecule but localized predominantly to the middle domain and the extreme carboxyl terminus. Lastly, because p190 is so heavily phosphorylated on serine, it has been suggested that p-Ser residues contribute to the binding of p190 to p120 (10, 17). Indeed, there are numerous examples of non-p-Tyr (p-Ser) interactions with SH2 domains in signaling proteins (7, 14, 18, 20–22, 32).
We therefore carried out an experiment to test the possibility that p-Ser mediates p190-p120 interactions. In this experiment, transiently expressed, metabolically 32P-labeled wt p190 was immunoprecipitated from COS-7 cell extracts, subjected to treatment with alkaline phosphatase or protein phosphatase 2a, and subsequently examined for levels of coprecipitating p120. Our data indicate that under these conditions, p190 was labeled almost exclusively on p-Ser (<0.5% p-Tyr) and underwent >90% dephosphorylation upon incubation with phosphatase. Compared to p190 that was subjected to mock dephosphorylation, binding of p120 to p190 was completely unaffected by removal of the p-Ser (data not shown). While these experiments are preliminary, they suggest that p-Ser alone or p-Ser–SH2 interactions are not major participants in the p-Tyr-independent mechanism of association between p120 and p190.
What then might the nature of the interaction be, and what purpose might the p-Tyr-independent pool of p190/p120 serve? Does the p-Tyr-independent pool constitute a signaling entity separate from and parallel to the p-Tyr-dependent pool, or does it exist in dynamic equilibrium with the p-Tyr-dependent pool? For example, does the p-Tyr-independent pool represent a low-affinity interaction between p190 and p120 that becomes high affinity upon phosphorylation of Y1105? These questions are subjects for future investigation, but given the current evidence, we favor the hypothesis that the p-Tyr-independent interaction involves a third protein (possibly binding to p190 through an SH3-polyproline interaction and to p120 through a p-Tyr– or p-Ser–SH2 mechanism) and that this interaction is involved in establishing a low-affinity complex between p190 and p120, which acquires high-affinity status upon phosphorylation of Y1105. Indeed, if the p-Tyr-independent complex represents a low-affinity interaction, then the 10 to 20% figure may be an underestimation of its abundance, due to the difficulty in maintaining association during isolation.
The quantitative analysis depicted in Fig. Fig.55 is, however, in part supportive of the dual, direct p-Tyr–SH2 model. It shows that overall, the amount of p120 found in association with p190 correlates with the level of p190 tyrosine phosphorylation. In addition, the phosphopeptide analysis revealed the presence of two p-Tyr-containing peptides, peptides 3 and 8. However, the differences in the phosphotyrosine levels of these two peptides (4:1, respectively, in Neo control cells and 20:1 in K+ c-Src cells) indicate that only a minority of p190 molecules are capable of interacting by this mechanism under steady-state conditions. Under certain transient circumstances (such as growth factor activation or adhesion/motility) when the two sites may be differential targets for phosphatases, with different turnover rates, the amounts of complex formed by this mechanism could vary.
Quantitation of the level of p190 associated with p120 and the level of p-Tyr in p190 in K+ c-Src cells suggested additional roles for Y1105 other than mediating the interaction with p120. Previous experiments (6, 12, 34) showed that the level of p190 tyrosine phosphorylation correlates with the extent of EGF-induced (i) subcellular redistribution of p190, (ii) actin stress fiber disassembly, and (iii) mitogenesis, suggesting possible roles for tyrosine phosphorylation of p190 in regulating actin cytoskeleton contributions to mitogenesis. These contributions would most likely be mediated through Rho, a small GTPase known to regulate actin stress fiber assembly (25). In this context, tyrosine phosphorylation of p190 may modulate its own enzymatic (GTP binding/hydrolysis or RhoGAP) activities through phosphorylation-induced conformational changes. Alternatively, p-Y1105 could serve as a docking site for SH2-containing signaling proteins other than p120. This raises the possibility of competition between p120 and other proteins for binding to p-Y1105, which could offer dynamic linkages to other signaling pathways, including those that regulate the actin cytoskeleton. Overall, the identification and characterization of the p-Tyr sites on endogenous p190 provides us the framework to model previous results into a more complete picture of the potential mechanisms that are involved in regulating p190 function.
The increase in p190 tyrosine phosphorylation upon overexpression of wt c-Src, coupled with the dominant negative effect of kinase deficient c-Src overexpression on p190 tyrosine phosphorylation, strongly suggests that p190 is a direct substrate of c-Src in vivo (4). It remains a formal possibility, however, that c-Src affects p190 tyrosine phosphorylation indirectly, through a downstream tyrosine kinase. The data presented here, which demonstrate that c-Src selectively phosphorylates Y1105 in vitro (Fig. (Fig.3),3), argue for a direct phosphorylation of p190 at Y1105 by c-Src in vivo. Direct phosphorylation is also supported by the work of Songyang et al. (30), who report that c-Src preferentially phosphorylates synthetic peptides containing the sequence EEIY, with isoleucine in the −1 position being the most critical. Y1105 is located in a peptide with the sequence EENIY1105, similar to the preferred synthetic sequence and with the isoleucine appropriately positioned.
Several investigators have reported increases in p190 tyrosine phosphorylation in response to polypeptide growth factors, suggesting that p190 may also be a substrate of tyrosine kinase receptors. Ellis et al. (8) showed that in Rat1 cells overexpressing the human EGF receptor, p190 tyrosine phosphorylation gradually increased over a 60-min time course of EGF stimulation, with only a small increase detected after 2 min. Tyrosine phosphorylation of p190 has also been shown to increase in response to mast cell growth factor and granulocyte-macrophage colony-stimulating factor stimulation of the myeloid cell line M07e (16) and in v-Fms (oncogenic form of the receptor for colony-stimulating factor 1)-transformed fibroblasts (31). In contrast to these reports, we have not detected an EGF-induced increase in the tyrosine phosphorylation of p190 in cells expressing endogenous levels of the EGF receptor. However, we have seen a modest increase in the overall tyrosine phosphorylation of p190 and specifically phosphorylation of Tyr 1105 (Fig. (Fig.4)4) in cells overexpressing the receptor after 30 min of EGF treatment. Whether this increase in phosphorylation is mediated by the receptor directly or indirectly is not known. Several studies indicate that Src family members play an integral role in EGF and colony-stimulating factor 1 signaling (12, 26, 34), suggesting that p190 may be phosphorylated by c-Src following EGF stimulation. Indeed, the approximately twofold increase in Y1105 phosphorylation in EGFR and EGFR/K+ c-Src cells following EGF stimulation (Fig. (Fig.4)4) is consistent with phosphorylation by endogenous and overexpressed c-Src, respectively.
ACKNOWLEDGMENTS
We thank J. Shannon for Edman analysis; J. Settleman for the pRc/CMVp190 expression plasmid, helpful discussions, and communicating results prior to publication; and members of the S. Parsons, M. Weber, and J. T. Parsons laboratories for advice throughout the course of this study and for critical analyses of the data.
This work was supported by PHS grant CA39438 from NCI.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.18.12.7052
Read article for free, from open access legal sources, via Unpaywall:
https://mcb.asm.org/content/mcb/18/12/7052.full.pdf
Free to read at mcb.asm.org
http://mcb.asm.org/cgi/content/abstract/18/12/7052
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/reprint/18/12/7052
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/content/full/18/12/7052
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.18.12.7052
Article citations
Diverse p120RasGAP interactions with doubly phosphorylated partners EphB4, p190RhoGAP, and Dok1.
J Biol Chem, 299(9):105098, 27 Jul 2023
Cited by: 2 articles | PMID: 37507023 | PMCID: PMC10470053
Tandem engagement of phosphotyrosines by the dual SH2 domains of p120RasGAP.
Structure, 30(12):1603-1614.e5, 22 Nov 2022
Cited by: 3 articles | PMID: 36417908 | PMCID: PMC9722645
Structure Determination of SH2-Phosphopeptide Complexes by X-Ray Crystallography: The Example of p120RasGAP.
Methods Mol Biol, 2705:77-89, 01 Jan 2023
Cited by: 0 articles | PMID: 37668970
Identification of an inhibitory domain in GTPase-activating protein p190RhoGAP responsible for masking its functional GAP domain.
J Biol Chem, 299(1):102792, 11 Dec 2022
Cited by: 1 article | PMID: 36516886 | PMCID: PMC9840978
Regulatory Roles of the N-Terminal Intrinsically Disordered Region of Modular Src.
Int J Mol Sci, 23(4):2241, 17 Feb 2022
Cited by: 5 articles | PMID: 35216357 | PMCID: PMC8874404
Review Free full text in Europe PMC
Go to all (91) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Two SH2 domains of p120 Ras GTPase-activating protein bind synergistically to tyrosine phosphorylated p190 Rho GTPase-activating protein.
J Biol Chem, 270(30):17947-17952, 01 Jul 1995
Cited by: 28 articles | PMID: 7629101
Increased levels of p21ras-GTP and enhanced DNA synthesis accompany elevated tyrosyl phosphorylation of GAP-associated proteins, p190 and p62, in c-src overexpressors.
Oncogene, 8(4):959-967, 01 Apr 1993
Cited by: 29 articles | PMID: 7681161
Aberrant Ras regulation and reduced p190 tyrosine phosphorylation in cells lacking p120-Gap.
Mol Cell Biol, 17(4):1840-1847, 01 Apr 1997
Cited by: 39 articles | PMID: 9121432 | PMCID: PMC232031
c-Src regulates the simultaneous rearrangement of actin cytoskeleton, p190RhoGAP, and p120RasGAP following epidermal growth factor stimulation.
J Cell Biol, 130(2):355-368, 01 Jul 1995
Cited by: 166 articles | PMID: 7542246 | PMCID: PMC2199934
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: CA39438
Grant ID: R01 CA039438




