Abstract
Free full text

Chromosome Structure and Human Immunodeficiency Virus Type 1 cDNA Integration: Centromeric Alphoid Repeats Are a Disfavored Target
Abstract
Integration of retroviral cDNA into host chromosomal DNA is an essential and distinctive step in viral replication. Despite considerable study, the host determinants of sites for integration have not been fully clarified. To investigate integration site selection in vivo, we used two approaches. (i) We have analyzed the host sequences flanking 61 human immunodeficiency virus type 1 (HIV-1) integration sites made by experimental infection and compared them to a library of 104 control sequences. (ii) We have also analyzed HIV-1 integration frequencies near several human repeated-sequence DNA families, using a repeat-specific PCR-based assay. At odds with previous reports from smaller-scale studies, we found no strong biases either for or against integration near repetitive sequences such as Alu or LINE-1 elements. We also did not find a clear bias for integration in transcription units as proposed previously, although transcription units were found somewhat more frequently near integration sites than near controls. However, we did find that centromeric alphoid repeats were selectively absent at integration sites. The repeat-specific PCR-based assay also indicated that alphoid repeats were disfavored for integration in vivo but not as naked DNA in vitro. Evidently the distinctive DNA organization at centromeres disfavors cDNA integration. We also found a weak consensus sequence for host DNA at integration sites, and assays of integration in vitro indicated that this sequence is favored as naked DNA, revealing in addition an influence of target primary sequence.
To replicate, a retrovirus must integrate a cDNA copy of its RNA genome into a chromosome of the host. The host integration acceptor sites are not expected to be present as naked DNA but rather associated with histones and other DNA-binding proteins in chromatin. DNA packaging in vivo is expected to influence integration site selection, and the choice of integration site may have profound effects on both the virus and the host (13, 57). The determinants of integration efficiency in vivo remain incompletely defined, despite their importance.
Previous surveys of in vivo integration sites have led to several proposals for factors influencing site selection. Studies of Moloney murine leukemia virus have supported a model in which open chromatin regions at transcription units were favored, since associated features such as DNase I-hypersensitive sites (45, 58) or CpG islands (47) were apparently enriched near integration sites. Another study proposed that unusual host DNA structures were common near integration sites (34). A recent study of avian leukosis virus integration frequencies at several chromosomal sites failed to show any major differences among the regions studied (62), contrary to an earlier report (50). For human immunodeficiency virus type 1 (HIV-1), it has been proposed that integration may be favored near repetitive elements (including LINE-1 elements [54] or Alu islands [55]) or topoisomerase cleavage sites (24).
Assays of integration in vitro have revealed several effects of proteins bound to target DNA. Simple DNA-binding proteins can block access of integration complexes to target DNA, creating regions refractory for integration (3, 9, 44). In contrast, wrapping DNA on nucleosomes can create hot spots for integration at sites of probable DNA distortion (40–42, 44). Distortion of DNA in several other protein-DNA complexes can also favor integration (3, 35), consistent with the possibility that DNA distortion is involved in the integrase mechanism (11, 48).
Here we present two experiments designed to address some of the questions surrounding integration site selection in vivo. We have (i) sequenced 61 integration junctions made after experimental infection of cultured human T cells and compared them with 104 control DNA fragments from uninfected human cells and (ii) used a region-specific PCR assay to assess the frequency of integration near several repeated-sequence families. In addition, we have identified a weakly conserved sequence at in vivo integration sites and determined that it is favored for integration when tested in vitro.
MATERIALS AND METHODS
DNA manipulation.
Plasmids containing synthetic integration target sites were prepared by annealing pairs of oligonucleotides (CH10-1–CH10-2, CH11-1–CH11-2, and CH13-1–CH13-2) (Table (Table1)1) and ligating them with pUC19 DNA that had been cleaved with EcoRI and HindIII. The standard cloning methods used were as described previously (46). Integration target DNAs were prepared by cleaving the plasmids mentioned above with PvuII, which releases the oligonucleotide insert together with flanking plasmid DNA.
TABLE 1
Oligonucleotides used in this study
study
| Oligonucleotide | Sequence | Comments |
|---|---|---|
| HUA | 5′-CTTTTTGCCTGTACTGGGTCTC-3′ | HIV U3 primer for inverse PCR |
| HUB | 5′-GATCAAGGATATCTTGTCTTCGT-3′ | HIV U3 primer for inverse PCR |
| IP3 | 5′-TCTTGTCTTCGTTGGGAGTGA | HIV U3 primer for inverse PCR |
| det3b | 5′-GAACCCACTGCTTAAGCCTC-3′ | HIV U3 primer for inverse PCR |
| det3a | 5′-CTTCGTTGGGAGTGAATTAG-3′ | Primer for detection of circle junctions |
| sc8 | 5′-CTTCAAGTAGTGTGTGCCCG-3′ | Primer for detection of circle junctions |
| sc10 | 5′-GGGTTTTCCAGTCACACCTCAGG-3′ | Primer for detection of the HIV internal fragment |
| TA6 | 5′-CATCAAGCTTGGTACCGAGC-3′ | Primer for sequencing from pTA vector |
| TA7 | 5′-TAATACGACTCACTATAGGG-3′ | Primer for sequencing from pTA vector |
| SC24 | 5′-TGGCGCAATCTCGGCTCAC-3′ | Primer for amplifying Alu1 sequences |
| CH12 | 5′-CTCCGCTTCCCGGGTTC-3′ | Primer for amplifying Alu1 sequences |
| CH5 | 5′-CTTCCAGTTTTTGCCCATTCAGT-3′ | Primer for amplifying LINE-1 sequences |
| CH6 | 5′-AGTATGATATTGGCTGTGGGTTTGTC-3′ | Primer for amplifying LINE-1 sequences |
| SC21 | 5′-GCAAGGGGATATGTGGACC-3′ | Primer for amplifying alphoid repeats |
| SC23 | 5′-ACCACCGTAGGCCTGAAAGCAGTC-3′ | Primer for amplifying alphoid repeats |
| CH15 | 5′-CCTGAGGCCTCCCTCAGCCAT-3′ | Primer for amplifying THE 1 repeats |
| CH16 | 5′-GCCATGATTGTAAGTTTCCTGAGG-3′ | Primer for amplifying THE 1 repeats |
| NEB-40 | 5′-GTTTTCCCAGTCACGAC-3′ | Primer for amplifying integration products in pUC19 |
| FB652 | 5′-TGTGGAAAATCTCTAGCA-3′ | Primer for amplifying HIV U5 sequences |
| CH 11 | 5′-CTCCGCTTCCCGGGTTC-3′ | Primer for amplifying integration products in pUC19 |
| FB66 | 5′-GCCTAGATCCGTGTGGAAAATC-3′ | Primer for amplifying products made with purified integrase |
| FB64 | 5′-ACTGCTAGAGATTTTCCACACGGATCCTAGGC-3′ | Substrate for purified integrase (annealed to FB65-2) |
| FB65-2 | 5′-GCCTAGGATCCGTGTGGAAAATCTCTCTCTAGCA-3′ | Substrate for purified integrase (annealed to FB64) |
| AP1 | 5′-CCATCCTAATACGACTCACTATAGGGC-3′ | Adaptor primer 1 |
| AP2 | 5′-ACTCACTATAGGCTCGAGCGGC-3′ | Adaptor primer 2 |
| ADAPT1 | 5′-CTAATACGACTCACTATAGGGCTCGAGCGGCCGCCCGGGCAGGT-3′ | Vectorette adaptor primer (top strand) |
| ADAPT2 | 5′-ACCTGCCC-NH2-3′ | Vectorette adaptor primer (bottom strand) |
| CH10-1 | 5′-AATTCTTCTCGAGTAGGTTACCTATGATCAA-3′ | Insert for pCH10 (top strand) |
| CH10-2 | 5′-AGCTTTGATCATAGGTAACCTACTCGAGAAG-3′ | Insert for pCH10 (bottom strand) |
| CH11-1 | 5′-AATTCTTCTCGAGTAGTTTAACTATGATCAA-3′ | Insert for pCH11 (top strand) |
| CH11-2 | 5′-AGCTTTGATCATAGTTAAACTACTCGAGAAG-3′ | Insert for pCH11 (bottom strand) |
| CH13-1 | 5′-AATTCGTGTTAACTCGGTGACCGAAGGCCTA-3′ | Insert for pCH12 (top strand) |
| CH13-2 | 5′-AGCTTAGGCCTTCGGTCACCGAGTTAACACG-3′ | Insert for pCH12 (bottom strand) |
The oligonucleotides used in this study are shown in Table Table11.
Construction of DNA libraries.
To generate a large pool of independent integration events, SupT1 cells (2 × 107 cells) were infected with the HXB2 or R9 (56) (referred to as R8 in reference 22) HIV-1 strain. Viral stocks were assayed by measuring the concentration of p24, and the infectivity was scored by the MAGI assay (28). Cells were infected at a multiplicity of 1 to 10 and harvested 12 to 14 h later. The cellular genomic DNA was depleted of low-molecular-weight DNA prior to cloning as described previously (39).
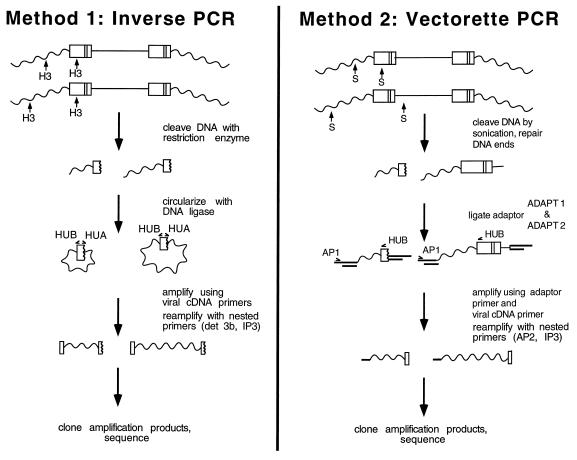
For construction of library 1 (Fig. (Fig.1,1, method 1), DNA from infected cells was cleaved with HindIII and circularized by ligation (31). Sixty-six nanograms of DNA was used as the template for PCR. HUA and HUB, divergently oriented primers complementary to the HIV long terminal repeats (LTRs), were used for the first amplification. Amplification was carried out for 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 3 min. The products were purified by using the Qiaquick PCR purification kit (Qiagen, Santa Clarita, Calif.). One microliter from the 50-μl column eluate was used as the template for the second-round PCR (20 cycles; program as described above) with nested primers det3b and IP3.
For construction of library 2 (Fig. (Fig.1,1, method 2) DNA fragments sheared by sonication (average length, about 1.5 kb) were made blunt-ended by treatment with Bal 31 followed by T4 DNA polymerase and deoxynucleoside triphosphates. Ligation of adapters, amplification, and cloning were carried out as described previously (51), except that primers HUB and IP3 were used as viral end primers for the first and second amplifications, respectively. PCR products were cloned by using the pCR II TA cloning vector from Invitrogen (San Diego, Calif.).
The products of PCRs contained two contaminants in addition to the desired integration junctions, one derived from a circular form of the viral DNA (2-LTR circle) and the second from the 3′ internal part of the viral DNA (for a discussion, see reference 31). Colonies containing host-virus junctions were distinguished from colonies containing contaminating sequences by PCR. Bacterial colonies containing plasmids were resuspended in PCR buffer and amplified with Taq polymerase for 20 cycles of 1 min at 94°C, 30 s at 60°C, and 1 min at 72°C. The circle junctions were detected using primers det3a and sc8. The internal fragment was detected using primers sc10 and IP3. The inserts were sequenced by using primers TA6 and TA7, which are complementary to the vector (pCR II; Invitrogen). Sequences of integration junctions and controls were determined by the dideoxy sequencing method.
Each sequence was determined at least twice. For each integration site clone, the sequence of 34 bases of viral DNA at the LTR tip was determined, in addition to the flanking host DNA. For most integration site clones (59 of 61), all of the cloned human DNA adjacent to the proviral DNA was sequenced.
A control experiment was carried out to exclude a possible artifact. Since DNA samples were treated with DNA ligase, free HIV genomes might have become joined to host DNA fragments by DNA ligase instead of integration. This is unlikely in the case of library 1, however, since the blunt-ended or 3′ cleaved forms of the HIV cDNA would not be expected to become ligated to the protruding 5′ ends generated by cleavage with HindIII. However, to document this expectation, a control experiment was performed in which purified unintegrated HIV cDNA was incubated in the presence of DNA ligase with HindIII-cleaved sequences and possible ligation was assayed by PCR across the ligation junction (one primer complementary to the HIV DNA and the other complementary to the HindIII-cleaved test DNA). No ligation was detected (data not shown). In the case of library 2, hypothetical ligation of unintegrated HIV cDNA should have yielded predominantly the vectorette linker joined directly to HIV cDNA, since DNA ends from the linkers were present in vast excess over ends from viral or human DNA. However, no such forms were detected (data not shown). Internal evidence also argues against this class of artifacts. For example, the 5-bp consensus host sequence flanking integration sites identified here closely resembles that found in a previous study employing conventional cloning and sequencing (55), an observation that helps validate each study.
DNA sequence analysis.
Sequences were analyzed by comparison to the nonredundant human sequence (nr) database, the human cDNA (dbEST) database, and the MONTH (November 1997) database by using BLASTN with Search Launcher and Repeat Masker. Default parameters were used. For comparisons between integration sites and control libraries, only a subset of the available sequence was considered (see Table Table2),2), with either an average length of 144 bp or a length of exactly 50 bp (see Table Table3).3). A total of 8,809 bp of human DNA flanking 61 integration sites was sequenced and analyzed for the integration site libraries (see Tables Tables22 and and3).3). The lengths of flanking human DNA sequences analyzed ranged from 37 to 430 bp. For the control human DNA fragments, a total of 14,989 bp in a total of 104 DNA clones were sequenced. Lengths of sequences analyzed ranged from 51 to 264 bp. Links to integration site and control sequences can be found at http://www.salk.edu/faculty/bushman.html.
TABLE 2
Integration sites analyzed and their similarities to known sequences
sequences
| Sequence namea | Length (bp)b | Dup seqc | Identified similaritiesd | Identified similarities truncated to 50 bpe |
|---|---|---|---|---|
| MolH 1 | 106 | ATGTC | *f | * |
| MolH 2 | 60 | CAAGC | * | * |
| SupH 1 | 156 | TCTTC | LINE-1 [2–153, SW = 508] | * |
| SupH 2 | 132 | GCTAC | * | * |
| SupH 3 | 91 | GGAAA | * | * |
| SupH 4 | 139 | GTGGT | * | * |
| SupH 5 | 140 | TATAT | * | * |
| SupH 6 | 114 | ATCCC | * | * |
| SupH 7 | 230 | GCATG | * | * |
| SupH 9 | 82 | CTATA | * | * |
| SupH 10 | 212 | TACAC | LINE-1 [2–107, SW = 251] | * |
| SupH 11 | 166 | CATGC | Alu [15–110, SW = 716] | Alu [SW = 304] |
| SupH 12 | 89 | GTTGG | * | * |
| SupH 13 | 63 | CTCAC | Transcription unit (cDNA) [5–62, P = 1.6 × 10−16] | Transcription unit (cDNA) [P = 1.9 × 10−12] |
| SupH 14 | 111 | GTCAC | * | * |
| SupH 15 | 164 | TATGG | LINE-1 [2–107, SW = 400] | * |
| SupH 16 | 66 | AACAG | * | * |
| SupH 17 | 54 | CTCAC | * | * |
| SupH 18 | 159 | GTTGT | * | * |
| SupH 20 | 342 | GTTTC | Alu [3–125, SW = 956] | Alu [SW = 373] |
| SupH 21 | 173 | CATAT | * | * |
| SupH 22 | 38 | CACAC | * | Excluded |
| SupH 23 | 258 | CATTC | * | * |
| SupH 24 | 110 | GTAAT | * | * |
| SupH 25 | 37 | CTTTT | * | Excluded |
| SupH 27 | 160 | CCATT | * | * |
| SupH 28 | 93 | AATAC | Transcription unit (cDNA) [1–93, P = 3.7 × 10−33] | Transcription unit (cDNA) [P = 1.5 × 10−13] |
| SupH 29 | 143 | GCCCA | * | * |
| SupH 31 | 188 | ATATT | * | * |
| SupH 32 | 157 | GTTGA | Transcription unit (cDNA) [59–157, P = 5.9 × 10−34] | * |
| SupH 33 | 50 | CTTCA | Transcription unit (VACH1 gene) [1–50, P = 6 × 10−13] | Transcription unit (VACH1 gene) [P = 6 × 10−13] |
| SupH 34 | 50 | AGTTG | * | * |
| SupH 35 | 420 | TTAAC | Transcription unit (cDNA) [52–143, P = 2.8 × 10−25]; LINE-2 [223–274, SW = 252] | * |
| SupH 36 | 237 | CTTGT | * | * |
| SupH 37 | 69 | CACAC | Alu [1–69, SW = 471] | Alu [SW = 371] |
| SupH 38 | 68 | GTTAT | * | * |
| SupH 39 | 89 | CAAAA | * | * |
| SupH 41 | 41 | ATGGC | * | Excluded |
| SupH 42 | 437 | AAAAC | LINE-1 [1–437, SW = 2684] | LINE-1 [SW = 264] |
| SupH 43 | 179 | ATAGT | Transcription unit (cDNA) [1–179, P = 9.4 × 10−65]; other repeat (LTR element) [98–152, SW = 198] | Transcription unit (cDNA) [P = 3.8 × 10−13] |
| SupH 44 | 337 | GAAAC | Other repeat (MIR, SINE) [191–315, SW = 493] | * |
| SupH 46 | 81 | GGGAG | Transcription unit (cDNA) [1–33, P = 3.9 × 10−6] | Transcription unit (cDNA) [P = 4.6 × 10−6] |
| SupH 47 | 111 | AAAAC | Transcription unit (cDNA) [1–57, P = 2.1 × 10−13] | Transcription unit (cDNA) [P = 2.2 × 10−9] |
| SupH 48 | 125 | CTGTG | Other repeat (MIR, SINE) [1–123, SW = 474] | Other repeat (MIR, SINE) [SW = 245] |
| SupH 49 | 260 | TTTTG | Alu [1–128, SW = 698] | Alu [SW = 300] |
| SupS 1 | 176 | GCAGG | Transcription unit (CD27 gene) [1–176, P = 2.7 × 1062] | Transcription unit (cDNA) [P = 5.4 × 10−13] |
| SupS 2 | 113 | GTTCT | * | * |
| SupS 3 | 125 | ATACC | Alu [4–115, SW = 540] | Alu [SW = 195] |
| SupS 4 | 215 | CCCTC | Other repeat (MER74, LTR element) [1–213, SW = 599] | Other repeat (MER74, LTR element) [SW = 277] |
| SupS 5 | 147 | CAGCA | * | * |
| SupS 7 | 171 | GAGTC | * | * |
| SupS 8 | 85 | TGAGT | Transcription unit (cDNA) [1–81, 3.2 × 10−26] | Transcription unit (cDNA) [P = 3.6 × 10−13] |
| SupS 9 | 86 | GTACC | * | * |
| SupS 10 | 52 | AAAGC | Alu [2–59, SW = 356] | Alu [SW = 310] |
| SupS 11 | 147 | CTAAC | * | * |
| SupS 12 | 131 | GTTTC | * | * |
| SupS 13 | 94 | ATGTG | Transcription unit (cDNA) [1–94, P = 5.1 × 10−28] | Transcription unit (cDNA) [P = 3.4 × 10−12] |
| SupS 14 | 184 | GAGAC | * | * |
| SupS 15 | 120 | AAATG | * | * |
| SupS 16 | 161 | CTCTG | * | * |
| SupS 17 | 215 | GTATG | * | * |
| Total bp | 8,809 | 2,900 | ||
| Avg | 144 | 50 |
TABLE 3
Sequence composition of libraries of integration sites and control DNA fragments
fragments
| Sequence class | Analysis of 144-bp sequences (avg length)a
| Reanalysis of 50-bp sequencesb
| ||
|---|---|---|---|---|
| Integration sites (%) | Genomic DNA (%) | Integration sites (%) | Genomic DNA (%) | |
| Anonymous | 61 | 43 | 69 | 71 |
| Alu element | 10 | 9 | 10 | 6 |
| LINE element | 8 | 13 | 2 | 6 |
| Alphoid repeat | 0 | 6 | 0 | 3 |
| Other repeats | 7 | 22 | 3 | 10 |
| Transcription unit | 18 | 8 | 16 | 4 |
Similarities to repeated sequences were ranked in accordance with the Smith-Waterman parameter (SW) generated by Repeat Masker (see A. F. A. Smit and P. Green, RepeatMasker at http://ftp.genome.washington.edu/RM/RepeatMasker.html) or by the probability of matching by chance generated by BLASTN (1) (P value) (see http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-blast?Jform=0). Minimum similarities for each sequence class considered to be significant matches are as follows: cDNA, P = 4.6 × 10−6; LINE 1, SW = 217; Alu repeat, SW = 195; alphoid repeat, SW = 218; other repeats, SW = 190. Most regions of sequence similarity extended over at least 50 bp, although in the case of the lowest scoring cDNA, a 31-bp perfect match was judged to be significant.
Integration in vitro.
Preintegration complexes (PICs) were extracted from a 6-h coculture of SupT1 cells grown in RPMI 1640 medium containing 10% fetal calf serum and chronically infected MoltIIIB cells stimulated with phorbol 12-myristate 13-acetate as previously described by Farnet and Haseltine (19). In vitro integration was achieved by incubating 400 μl of PIC extract with 1.2 μg of DNA from uninfected SupT1 cells for 45 min. The integration product was recovered by incubating it with proteinase K in 0.5% sodium dodecyl sulfate followed by extraction with phenol-chloroform. The same procedure was followed for the inactive PICs after first incubating the concentrated PICs in 15 mM EDTA for 5 min prior to adding target DNA. Integration assays with recombinant HIV-1 integrase were carried out essentially as described previously (4, 10).
Region-specific analysis of integration acceptor sites.
Integration junctions were amplified essentially as described previously (9, 30, 44). Cellular DNA templates were prepared from infected and uninfected samples as described above. Integration products were visualized by nested PCR. Products were first amplified with viral primer HUB and a repeat primer. Products were then reamplified with the viral primer IP3 which had been end labeled by treatment with [γ-32P]ATP and kinase and a nested repeat primer. The primers for repeated sequences were designed by aligning multiple repeat copies and identifying conserved regions. Primers for amplifying repeated sequences were as follows (see Table Table11 for sequences; in each case, the second primer is the nested second primer). Alu1, SC24 and CH12 (27); LINE-1, CH5 and CH6 (64); alphoid repeat, SC21 and SC23 (61); and THE 1, CH15 and CH16 (52). The amounts of integration products generated in vivo and in vitro that were used as templates for PCR were adjusted to provide equal numbers of proviruses in each case. The first round of PCR was carried out for 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. For the second round of PCR, 2 μl from the initial PCR was added to a 25-μl reaction mixture and the mixture was amplified for 20 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. TaqStart antibody (Clontech, Palo Alto, Calif.) was used in both amplifications (hot-start PCR) in accordance with the manufacturer’s recommendations.
Assays of integration into cloned target DNAs were carried out as described previously (for PICs [4, 8] and for purified integrase [3, 33]). PICs were concentrated and partially purified by pelleting through 20% sucrose as described before (4). Integration targets were (i) a purified PvuII fragment containing the sequence of interest (PICs) or (ii) uncleaved plasmid DNA (purified integrase). Similar results were also obtained with PICs when uncleaved plasmid DNAs were used as the target. Primers for amplifying integration products were as follows: PIC reactions, top strand, NEB-40 and FB 652 (4); PIC reactions, bottom strand, CH 11 and FB 652; purified integrase reactions, top strand, FB 66 (4) and NEB-40; purified integrase reactions, bottom strand, FB 66 and CH 11.
RESULTS
Construction of integration site libraries.
DNA for library construction was obtained from a human T-cell line (SupT1) acutely infected with cell-free stocks of HIV-1. Cellular DNA was harvested 12 to 14 h after initiation of infection, allowing initial integration to be studied separately from selection during subsequent growth of cells.
Libraries were constructed by two different methods in an effort to control for possible biases introduced in the DNA cloning steps (Fig. (Fig.1).1). For library 1, genomic DNA from infected cells was digested with HindIII, which cleaved the population of proviruses near the viral DNA ends and at numerous positions in flanking host DNA. HindIII-cleaved DNA was then circularized by treatment with DNA ligase, and virus-host DNA junctions were amplified with divergent primers complementary to viral end sequences (inverse PCR) (31, 49). For library 2, DNA fragments were made blunt ended by treatment with Bal 31 nuclease and T4 DNA polymerase and ligated to short linkers. DNA fragments were amplified with primers complementary to the linker and the HIV cDNA end (vectorette PCR) (51). PCR fragments were then cloned and sequenced. Sixty-one integration sites were analyzed by this means.
To aid in interpretation of the data, control libraries were constructed from uninfected SupT1 cell DNA by methods parallel to those used for cloning integration sites. SupT1 DNA fragments were generated by cleavage with HindIII (control library 1) or sonication and end repair (control library 2), cloned into plasmid vectors, and sequenced. One hundred four control clones from uninfected human DNA were characterized by this means.
Analysis of integration site libraries.
Analysis of the sequencing data presented several challenges. Our raw sequence data contained different numbers of base pairs determined for each DNA clone analyzed. To compare the integration site and control data sets in a meaningful fashion, it was necessary to compare matching numbers of base pairs in each DNA clone and then compare the frequencies of appearance of different types of sequences in each data set. The average length of host DNA flanking integration sites was 144 bp, so sequences in the control library, which were slightly longer, were each truncated to yield test sequences with an average length of 144 bp (further parameters describing the data sets are presented in Materials and Methods).
Some copies of the human repeated DNA sequences are quite divergent from the family consensus sequence, presenting a challenge for identification. Repeated sequences were identified here by a two-step process. The program Repeat Masker, which compares unknown sequences to a set of consensus sequences derived from human repeat sequences (52), was used first. In a second step, all sequences were compared to the nr, dbEST, and MONTH (November 1997) databases by using BLASTN with default settings. In some cases, highly repeated sequences missed by Repeat Masker were identified by BLASTN and further analysis allowed them to be grouped into known sequence classes. The minimum degrees of similarity scored as matches are given in Materials and Methods.
Analysis of cDNA matches presented another challenge. New sequences are being added to the dbEST database at a high rate, and even during the course of this work many anonymous sequences were found in later searches to match new cDNAs. The data presented here represent the number of matches to cDNAs as of November 1997, but new additions to the database will likely increase the number of matches in the future. For cDNAs, there was a natural partitioning of sequences into plausible and unlikely matches, since integration into a transcribed region should yield a near-perfect match over a discrete region.
Integration sites sequenced and the matches to known sequences are summarized in Table Table22 and and3.3. Sequences were classified as transcription units, Alu elements, LINE elements, alphoid repeats, other repeats, or anonymous. Transcription units were identified in database searches either as cDNAs or as sequences within the transcribed regions of known genes. Alu elements and LINE elements are the familiar interspersed nuclear repeats characteristic of human DNA. Alphoid repeats comprise the alpha satellite DNA, tandem arrays of 171-bp repeats associated with centromeric heterochromatin (38, 61). The “other repeat” class included several types, namely, SINE elements apart from Alu elements, low-complexity repeats, and retrovirus-related sequences such as THE 1 elements (36) and MLT1 sequences (14, 52) (for a recent summary of nomenclature, see reference 52). Anonymous sequences were defined as sequences contained in none of the classes.
For the control libraries, Alu sequences were identified in 10% of clones. Previous studies suggest that Alu elements comprise 8 to 15% of the human genome (53). LINE-1 elements comprised 13% of the control sequences; 5 to 18% was expected (16, 25, 53). Information available on transcription units, alphoid repeats, and the other repeats was insufficient to allow their abundance to be predicted with confidence. Analysis of the %GC of DNA in control library clones and in human DNA flanking integration sites revealed no obvious differences from that of bulk human DNA (data not shown). Thus, in those cases that could be checked, sequences in our control libraries had compositions close to those expected for randomly selected human genomic DNA fragments.
Comparison of the integration site and control libraries revealed that centromeric alphoid repeats were absent among integration sites but that six alphoid repeats were present in the control libraries (Tables (Tables22 and and3).3). Alphoid repeats were also absent among previously characterized HIV-1 integration sites (37, 59).
Other types of sequences were differentially distributed between integration site sequences and control sequences, although none showed the all-or-nothing partitioning characteristic of alphoid repeats. Transcription units were more abundant in the integration sites (18%) than in controls (8%). The other repeats were also differentially distributed (7%) in integration sites versus 23% in controls), although in this case many different sequence types contributed to the totals. Alu elements and LINE elements were not obviously differentially distributed.
As a test of the robustness of our conclusions, integration site sequences were reanalyzed after truncation so that only 50 bp of host DNA remained at the junction between viral and host sequences for all clones. The control data was similarly truncated to 50 bp in each sequence, arbitrarily starting from one junction with the DNA vector used for cloning. Sequence similarities were identified in the 50-bp data set by using the criteria described above (Table (Table3).3). Fewer matches were detected, as expected, since the sequences were shorter. However, in this case also, alphoid repeats were detected in the control library and not the integration site library.
A weak consensus sequence at integration sites.
Figure Figure22 presents an analysis of the 5 bp of host DNA at the junction between virus and host sequences expected to be duplicated upon integration. A weak consensus sequence can be derived from this data [5′ GT(A/T)AC 3′]. Only one end was sequenced for each integrant, so the duplicated nature of this sequence is inferred. The consensus sequence is rotationally symmetric, as expected, since each end of the HIV cDNA is joined to the 5′ end of each strand of this sequence (Fig. (Fig.2).2). A closely related sequence was derived from a previous study of HIV integration sites by Stevens and Griffith [5′ GTA(A/T)(T/C) 3′] (55). In this study, DNA from HIV-infected cells was cloned in lambda vectors, followed by isolation of provirus-containing clones by hybridization and sequencing of 29 proviral integration sites. The observation that our methods and that of Stevens and Griffith yielded similar integration site consensus sequences strongly validates each study.
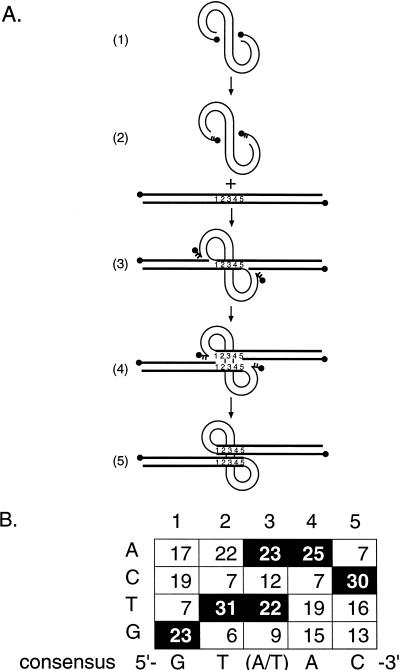
Consensus sequence at the junctions between HIV cDNA and host DNA and the mechanism of generation of the host sequence duplication. (A) Integration pathway. HIV cDNA is shown as the curved line in part 1. Two nucleotides are removed from each 3′ end of the cDNA (part 2). Host target DNA is shown as a straight line. The host DNA that becomes duplicated is indicated by the numbers 1 to 5. The recessed 3′ ends of the cDNA are then attached to protruding 5′ ends in the target DNA (part 3), and the integration intermediate melts to yield single-stranded gaps at each end (part 4). The in vitro integration reactions with PICs stop at this stage. Repair of the DNA gaps at each host-virus DNA junction results in the production of the 5-bp duplication of target DNA (part 5). (B) Tabulation of the host sequence inferred to be duplicated in our integration site collection. HIV cDNA is joined to target DNA just 5′ of position 1, as illustrated, and similarly on the other strand. Sixty-six duplications are included in this compilation, 61 from the sites listed in Table Table22 and 5 additional integration sites with the following duplication sequences: 5′-AGAGT-3′, 5′-GGTAC-3′, 5′-AACAT-3′, 5′-GTAAC-3′, 5′-AATGT-3′ (data not shown).
Region-specific assays of integration target sites.
Several features of the sequencing data complicated interpretation. (i) The number of matching sequences detected was determined in part by the choice of parameters in the similarity search. (ii) In some clones the integration junctions were within the identified cDNA or repeated sequence, while in others the junctions were near but not within the identified sequence. In Tables Tables22 and and3,3, these were considered together. (iii) Although this study of HIV-1 integration site sequences is the largest yet reported, the differences between integration sites and controls were generally not clearly significant, as evaluated by the chi-square or Fisher’s exact test. No finding was clearly significant in the analysis of both the 144-bp flanking sequences and the 50-bp sequence data. For these reasons, it was important to test some of the hypotheses generated by the sequence analysis by an independent method.
To this end, integration near repeated sequences was studied by using an assay based on PCR amplification of host-virus DNA junctions. In each reaction, one primer was complementary to an HIV-1 LTR end and the second primer was complementary to a repeated sequence (alphoid, Alu, LINE-1, or THE 1 repeats) (Fig. (Fig.3)3) (30, 44, 62). The first PCR amplification was followed by a second PCR with nested primers. The LTR primer in the second amplification was labeled at the 5′ end with 32P. Amplification products were separated on DNA sequencing-type gels and analyzed by autoradiography. An integration event in or near the repeated sequence studied gave rise to a labeled band by amplification. Amplification of many such integration events gave rise to a ladder of labeled bands on the final autoradiogram.
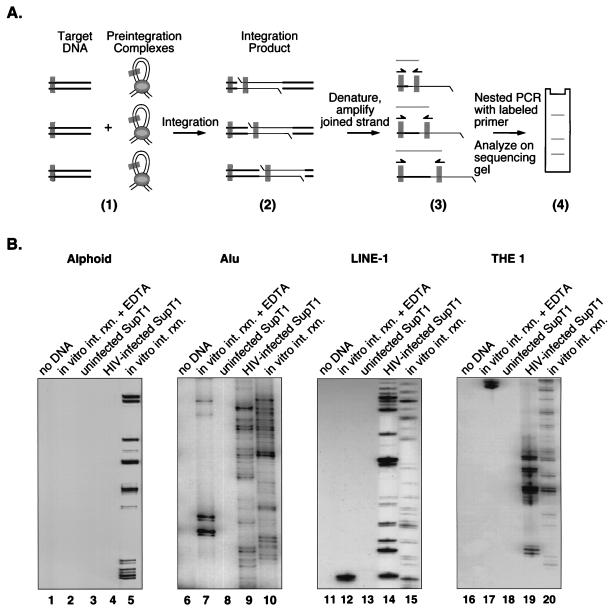
Analysis of integration sites near several repeat families using a PCR-based assay. (A) Diagram of the PCR method used to analyze integration sites. Primer binding sites are shown as gray rectangles. Part 1 illustrates either integration in vivo into cellular chromosomes or integration in vitro into deproteinized DNA. Products of integration reactions in vitro differ from products made in vivo in that only the former has the DNA breaks indicated in part 2 (the gapped integration intermediate is quickly repaired in vivo). In part 4, the three bands on the sequencing gel arose from three different integration events. (B) Results of PCR assays using primers complementary to alphoid repeats (lanes 1 to 5), Alu elements (lanes 6 to 10), LINE-1 elements (lanes 11 to 14), and THE 1 elements (lanes 16 to 20). The presence of a ladder of bands indicates that the template DNA contained HIV cDNA integrated near the repeat family specified. Lanes: 1, 6, 11, and 16, control amplification reactions with no added template; 2, 7, 12, and 17, amplification of inactive PICs and SupT1 DNA; 3, 8, 13, and 18, amplification from uninfected SupT1 DNA; 4, 9, 14, and 19, amplification of DNA from HIV-1 infected SupT1 cells; 5, 10, 15, and 20, amplification of deproteinized DNA that had been incubated with active PICs in vitro. Cellular DNA was detectable as a contaminant of the PIC preparations (data not shown); cellular DNA might have served as an integration target during PIC preparation or participated in recombination during PCR, possibly giving rise to the artifactual bands in lanes 7 and 12.
The importance of the in vivo setting was assessed by comparing integration sites from infected cells with sites made in vitro by integration into deproteinized chromosomal DNA. The in vitro reactions were carried out by using PICs purified from infected cells as a source of integration activity (5, 15, 19). PICs contain the viral cDNA in association with the virus-encoded integrase protein and other viral and cellular proteins (7, 17, 20, 22, 32). Previous studies have demonstrated that incubation of PICs with naked DNA targets results in the covalent integration of some of the HIV cDNA into target (for reviews, see references 13 and 18). The DNA samples from in vivo infections or in vitro integration reactions used for PCR contained similar numbers of proviruses (data not shown).
Amplification of DNA from in vitro integration reactions with the alphoid primer yielded a ladder of labeled bands indicative of integration (Fig. (Fig.3B,3B, lane 5). However, amplification of DNA from infected cells with the alphoid primer did not yield a ladder of labeled bands (Fig. (Fig.3B,3B, lane 4), indicating that integration did not take place in or near these sequences in vivo. Similar assays using primers complementary to Alu1 elements (Fig. (Fig.3B,3B, compare lanes 9 and 10), LINE-1 elements (Fig. (Fig.3B,3B, compare lanes 14 and 15), and THE 1 repeats (Fig. (Fig.3B,3B, compare lanes 19 and 20) yielded integration bands in both in vivo- and in vitro-integrated samples. This finding bolsters the idea that alphoid sequences are competent for integration in naked DNA but masked in vivo. Alu, LINE-1, and THE 1 elements, in contrast, are competent in both cases.
Control amplification reactions with no added template DNA (Fig. (Fig.3B,3B, lanes 1, 6, 11, and 16) or with DNA from uninfected human T cells did not yield labeled bands (Fig. (Fig.3B,3B, lanes 3, 8, 13, and 18). A further control containing integration reactions in vitro carried out in the presence of EDTA to chelate the required metal was mainly negative, although occasional artifactual bands of unknown origin were seen (Fig. (Fig.3B,3B, lanes 7 and 12).
Primary DNA sequences favored for integration.
Alignment of human DNA sequences at integration junctions yielded a consensus sequence (Fig. (Fig.22 and and4).4). A related sequence has been reported by Stevens and Griffith (55). To determine whether this sequence was favored for integration as naked DNA, several model sequences were synthesized and tested using integration in vitro. Target 1 contained the favored motif embedded in an arbitrary DNA sequence (Fig. (Fig.4A,4A, target 1). Target 2 is identical to target 1 except for changes at the two most conserved positions (Fig. (Fig.4A,4A, nucleotide positions 1 and 5) from the most favored nucleotide to the least favored. Target 3, like target 1, contained the favored target sequence but embedded in different arbitrary flanking DNA.
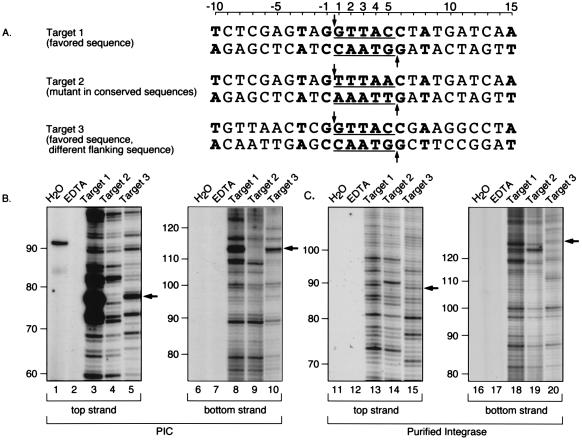
A conserved sequence at integration sites and analysis of integration at such sites in vitro. (A) Integration target sites tested. The host sequences duplicated upon integration are underlined; the points at which covalent strand transfer takes place on each strand are indicated by arrows; bases favored at integration sites are in boldface type. (B) Integration into targets 1 to 3 directed by PICs. Lanes: 1 and 6, H2O instead of template; 2 and 7, EDTA added to integration reactions. 3 and 8, target 1; 4 and 9, target 2; 5 and 10, target 3. Arrows indicate the location of the expected integration hotspots (5′ of position 1 on the top strand and 5′ of position 5 on the bottom strand). (C) Integration into targets 1 to 3 directed by purified HIV-1 integrase. Lanes 11 to 20 correspond to lanes 1 to 10, respectively, in panel B. Sizes were assigned by coelectrophoresis adjacent to several DNA sequencing ladders generated by the Sanger method.
Integration assays were carried out to examine favored sites in each sequence. Since previous work indicated that target site selection in naked DNA differed between PICs and the simpler integration complexes formed with recombinant HIV integrase protein (4), the two sources of integration activity were compared. As for the experiment illustrated in Fig. Fig.3,3, integration products were analyzed by amplification using one primer complementary to the viral DNA end and a second primer complementary to target sequences flanking the region of interest. Thus, each band on the final autoradiogram represents integration at a single target phosphodiester, and the intensity of the band represents the relative number of integration events.
Assays of PICs revealed the presence of a strong integration band at the position expected for the hot spot in target 1 (Fig. (Fig.4B,4B, lanes 3 and 8). Altering the two most favored bases (target 2) greatly reduced the signal at this position (Fig. (Fig.4B,4B, lanes 4 and 9). Assays of target 3, in which the flanking DNA was changed but the favored sequence was preserved, displayed favored integration at the expected hot spot sequence (Fig. (Fig.4B,4B, lanes 5 and 10). PCR assays to which no template was added (Fig. (Fig.4B,4B, lanes 1 and 6), or which contained mock integration reactions carried out in the presence of EDTA instead of the required divalent metal (Fig. (Fig.4B,4B, lanes 2 and 7), revealed no reproducible amplification products. Taken together, these data indicate that the favored target sequence identified from studies in vivo is sufficient to act as a hot spot for PICs in vitro.
Figure Figure4C4C presents an analysis of integration directed by purified HIV integrase into targets 1 to 3. The arrows mark the expected location of integration at the hot spot. A band is visible for targets 1 and 3 on the top strand (Fig. (Fig.4C,4C, lanes 13 and 15) and bottom strand (Fig. (Fig.4C,4C, lanes 18 and 20), although integration by purified integrase at the hot spot for PIC integration is much less prominent. This difference in target site selection highlights the differences between the two sources of integration activity, paralleling previous studies (for review and references, see reference 18).
DISCUSSION
We have used two methods to characterize chromosomal sites used by HIV-1 for integration in human SupT1 cells. We have sequenced a collection of integration sites and a collection of control sites and also analyzed integration near various repetitive sequences by using a PCR-based assay. DNA to be analyzed was prepared only 12 h after initiation of infection in an effort to obtain a population of sites unbiased by subsequent outgrowth of infected cells. In addition, the importance of a conserved host sequence at integration sites was tested by using integration in vitro. These studies clarify several factors influencing the selection of chromosomal sites for integration.
Comparison with integration site selection by yeast retrotransposons.
Previous studies of Ty retrotransposons in yeast reveal that retroelement integration can be highly site specific. The yeast Ty retrotransposons replicate by transcription, reverse transcription, and integration by using reverse transcriptase and integrase enzymes similar in function and sequence to their retroviral counterparts (2). Ty elements differ from retroviruses in that all steps in replication take place in a single cell. For this reason, Ty retrotransposons must be fastidious in their selection of integration sites, since integration into a required cellular gene would be lethal for the host and suicidal for the transposon.
Ty elements integrate selectively in benign locations in host DNA. Ty1 integrates in a window of several hundred base pairs upstream of host polymerase III (Pol III)-transcribed genes (26). Ty3 is the most selective, integrating at the start site of transcription of Pol III-transcribed genes (12, 29). Ty5 shows a different specificity, integrating in telomeres and in the silent mating cassette DNA (65, 66).
The potential for extreme integration site bias revealed in the Ty studies formed part of the motive for carrying out a large-scale investigation of integration site selection by HIV-1. In humans, integration in Pol III transcription units or telomeric repeats should have been detectable but no strong bias in favor of such sequences was found here or in previous studies with HIV or other retroviruses (23, 45, 47, 55, 58, 62). Evidently, HIV and Ty elements differ in this respect.
Favored integration near active genes?
Our data neither strengthen nor exclude the model that integration is favored in open chromatin near active genes (23, 45, 47, 58). Identifiable transcription units were present more frequently in the integration site libraries than in the control libraries. However, the difference was not statistically significant for the 144-bp sequence comparison, although it was significant for the 50-bp sequence comparison (Table (Table33).
Conclusions concerning integration site location will need to be reevaluated as new information becomes available. It will be particularly interesting to compile and analyze all the known integration site sequences (references 55, 59, and 60 and present study) when the sequence of the human genome is completed and cDNAs and regulatory regions are mapped onto the genomic DNA.
Lack of evidence for favored integration near Alu or LINE elements.
The data did not indicate that integration was favored near LINE elements or Alu elements as previously proposed (54, 55). Both the sequencing study and the region-specific PCR study failed to show any clear biases. One previous proposal was not directly tested. Stevens and Griffith proposed that integration might be favored near Alu islands, chromosomal regions containing clustered Alu repeats (55). Because our sequencing study examined relatively short flanking sequences (average length, 144 bp), clustering of Alu repeats near integration sites could not be assessed.
An effect of primary sequence.
The data presented here also reveal a modest favoring of integration at a particular host DNA sequence. Previous studies of integration site sequences have revealed weakly conserved motifs for several retroviruses, including HIV (21, 43, 55). Two mechanisms might account for the observed sequence bias: the integration machinery might interact favorably with a factor bound at the conserved site, or the PIC itself might interact favorably with the conserved sequence as naked DNA. We found that the conserved sequence was favored in vitro as naked DNA, supporting the idea that the conserved sequence is favored in vivo due to interaction with the PIC itself.
Disfavored integration at centromeric alphoid repeats.
The most striking feature of our data is the absence of integration in vivo into centromeric alphoid repeats. Alphoid repeats were absent in integration site sequences but present in controls, and alphoid sequences were selectively disfavored in the repeat-specific PCR integration assay. Several lines of evidence indicate that centromeric heterochromatin is organized differently than euchromatin. (i) Heterochromatic centromeres are seen to be more compact than euchromatin in fixed chromosome spreads (6). (ii) Alphoid sequences are more resistant to digestion with DNase I in isolated nuclei than are most DNAs (38, 63). (iii) Alphoid repeats are associated with the centromere-specific proteins CENP-A, CENP-B, and CENP-C (38, 63). On the basis of the data reported here, we propose that HIV-1 cDNA integration is obstructed by packaging DNA in centromeric heterochromatin. These data provide an unexpected demonstration of the long-standing possibility that certain types of chromatin may obstruct cDNA integration.
The mechanism of the integration block is unclear. The wrapping of DNA in heterochromatin may itself provide a steric block to integration, a possibility supported by the observation of condensed structures at centromeres. Other models are also possible. Since gene activity is probably reduced in heterochromatin, HIV may have evolved to avoid integration in heterochromatin to optimize gene expression. Alternatively, centromeric DNA might be sequestered at a nuclear location inaccessible to incoming PICs.
ACKNOWLEDGMENTS
S.C. and C.H. contributed equally to this work.
We thank Gary Karpen, Leslie Orgel, and members of the Bushman laboratory for suggestions and comments on the manuscript, Arian Smit for advice on identifying repeated sequences, and Leslie Barden and Allison Bocksruker for artwork and help in preparing the manuscript.
This work was supported by grants AI 34786 and AI 37489. S.C. was supported in part by the Rau Foundation. F.B. is a Scholar of the Leukemia Society of America.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.72.5.4005-4014.1998
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/72/5/4005.full.pdf
Free after 4 months at jvi.asm.org
http://jvi.asm.org/cgi/content/full/72/5/4005
Free after 4 months at jvi.asm.org
http://jvi.asm.org/cgi/reprint/72/5/4005
Free to read at jvi.asm.org
http://jvi.asm.org/cgi/content/abstract/72/5/4005
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.72.5.4005-4014.1998
Article citations
Unraveling the palindromic and nonpalindromic motifs of retroviral integration site sequences by statistical mixture models.
Genome Res, 33(8):1395-1408, 18 Jul 2023
Cited by: 1 article | PMID: 37463751 | PMCID: PMC10547254
Progressive transformation of the HIV-1 reservoir cell profile over two decades of antiviral therapy.
Cell Host Microbe, 31(1):83-96.e5, 02 Jan 2023
Cited by: 31 articles | PMID: 36596305 | PMCID: PMC9839361
G-Quadruplex DNA and Other Non-Canonical B-Form DNA Motifs Influence Productive and Latent HIV-1 Integration and Reactivation Potential.
Viruses, 14(11):2494, 11 Nov 2022
Cited by: 2 articles | PMID: 36423103 | PMCID: PMC9692945
A point mutation in HIV-1 integrase redirects proviral integration into centromeric repeats.
Nat Commun, 13(1):1474, 18 Mar 2022
Cited by: 7 articles | PMID: 35304442 | PMCID: PMC8933506
Spatial and Genomic Correlates of HIV-1 Integration Site Targeting.
Cells, 11(4):655, 14 Feb 2022
Cited by: 11 articles | PMID: 35203306 | PMCID: PMC8869898
Go to all (100) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Functional cloning of centromere protein B (CENP-B) box-enriched alphoid DNA repeats utilizing the sequence-specific DNA binding activity of human CENP-B in vitro.
Chromosome Res, 2(6):453-459, 01 Nov 1994
Cited by: 7 articles | PMID: 7834222
Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration.
J Virol, 70(9):6459-6462, 01 Sep 1996
Cited by: 70 articles | PMID: 8709282 | PMCID: PMC190680
SATB1-binding sequences and Alu-like motifs define a unique chromatin context in the vicinity of human immunodeficiency virus type 1 integration sites.
J Virol, 81(11):5617-5627, 21 Mar 2007
Cited by: 11 articles | PMID: 17376900 | PMCID: PMC1900249
Funding
Funders who supported this work.
NIAID NIH HHS (3)
Grant ID: AI 34786
Grant ID: AI 37489
Grant ID: R01 AI034786