Abstract
Free full text

Point mutation in AML1 disrupts subnuclear targeting, prevents myeloid differentiation, and effects a transformation-like phenotype
Abstract
The multifunctional C terminus of the hematopoietic AML1 transcription factor interacts with coregulatory proteins, supports the convergence and integration of physiological signals, and contains the nuclear matrix targeting signal, the protein motif that is necessary and sufficient to target AML1 to subnuclear sites. The (8;21) chromosomal translocation, which replaces the C terminus of AML1 with the ETO protein, modifies subnuclear targeting of AML1 in acute myeloid leukemia (AML) and results in defective myelopoiesis. We therefore addressed the relevance of AML1 subnuclear targeting and associated functions that reside in the C terminus to myeloid differentiation. A single amino acid substitution that abrogates intranuclear localization was introduced in the AML1 subnuclear targeting signal. Expression of the mutant AML1 protein blocks differentiation of myeloid progenitors to granulocytes in the presence of endogenous AML1 protein, as also occurs in the (8;21) chromosomal translocation, where only one allele of the AML1 gene is affected. The cells expressing the mutant AML1 protein continue to proliferate, maintain an immature blast-like morphology, and exhibit transformed properties that are hallmarks of leukemogenesis. These findings functionally link AML1 subnuclear targeting with competency for myeloid differentiation and expression of the transformed/leukemia phenotype.
Changes in nuclear morphology are diagnostic hallmarks of leukemic cells and have been traditionally linked to structural and biochemical properties of hematopoietic lineage cells (1-3). In addition, the mammalian nucleus is functionally organized into subnuclear domains that contain regulatory machinery for transcription, replication, and repair (4-6). Alterations in the composition and distribution of these regulatory domains are observed in leukemic phenotypes (5, 7-9). Thus, modifications in nuclear structure and subnuclear organization are associated with normal myeloid cell growth/differentiation and leukemic transformation.
Acute myeloid leukemia (AML) is a prevalent hematopoietic malignancy characterized by abnormal proliferation and abrogated differentiation of myeloid progenitor cells (10, 11). Numerous cytogenetic abnormalities associated with AML involve the gene encoding acute myelogenous leukemia transcription factor 1 (AML1; also designated as Runt-related transcription factor 1, Runx1) (12, 13). AML1 is a nuclear matrix-associated transcription factor essential for hematopoiesis, thereby providing means to investigate nuclear structure-function interrelationships that are linked to biological control and are altered during disease (5, 14). For example, the (8;21) translocation, a predominant mutation identified in ≈15% of adult AML patients, produces a chimeric protein (AML1-ETO) in which the C terminus of AML1 is replaced by the ETO protein (13). The AML1-ETO translocation-fusion protein is a dominant-negative inhibitor of the endogenous AML1 protein and exhibits intranuclear misrouting as well as modified gene-regulatory functions (15-17).
Intranuclear misrouting of the AML1-ETO fusion protein is caused by the absence of the multifunctional C-terminal region of AML1, which is relinquished in the (8;21) translocation (17). The AML1 C terminus supports the convergence and integration of physiological signals by interacting with several regulatory proteins that include end-point effectors of the TGFβ, bone morphogenetic protein (BMP) and Src signaling pathways, chromatin-modifying proteins, coactivators, and corepressors (14, 18-20). The C terminus contains the critical determinant of intranuclear routing [designated the nuclear matrix targeting signal (NMTS)]. The NMTS is necessary and sufficient to direct AML1 to nuclear matrix-associated subnuclear sites where the gene-regulatory machinery resides (21). The AML1-ETO fusion protein is aberrantly targeted to intranuclear sites that do not coincide with AML1 domains (17). Instead, subnuclear targeting of the fusion protein is controlled by ETO intranuclear targeting signals (17, 22). Therefore, the misrouting of regulatory factors as a consequence of gene rearrangement appears to be mechanistically linked to translocation-associated leukemias (5).
It is important to understand the involvement of specific acquired and lost properties of the AML1-ETO fusion protein in myeloid differentiation and leukemogenesis. Here, we directly addressed the contribution of AML1 intranuclear trafficking and associated functions to competency for myeloid differentiation by introducing a single amino acid mutation in the AML1 NMTS that alters intranuclear trafficking in myeloid progenitor cells. Granulocytic maturation is arrested in cells expressing the mutant AML1 protein, which is similar to the in vivo condition in AML patients where only one AML1 allele is mutated. These cells continue to proliferate, maintain an immature blast-like morphology, and exhibit transformed properties characteristic of leukemogenesis. Changes in myeloid cell growth and differentiation are accompanied by altered gene expression profiles. Thus, our results establish that a point mutation in the AML1 NMTS that impacts subnuclear targeting also blocks granulocyte differentiation and promotes transformation-like phenotype. These findings suggest that the AML1 subnuclear organization is linked with competency for myeloid differentiation and expression of the transformed/leukemia phenotype.
Materials and Methods
Cell Culture and Differentiation Assay. The murine myeloid progenitor 32D.cl3 cell line (32D cells) was grown in RPMI medium 1640 supplemented with 10% FBS, antibiotics, l-glutamine, and 20 units/ml murine interleukin-3 (IL-3) (a gift from Gerard Zambetti, St. Jude Children's Research Hospital, Memphis, TN). 32D cells were induced to differentiate to granulocytes by removing IL-3, washing twice in PBS, and adding granulocyte colony-stimulating factor (G-CSF) (R & D Systems) to a final concentration of 25 ng/ml. Cells were cytocentrifuged (Cytospin 4, Thermo Shandon, Pittsburgh), and cell morphologies were evaluated at 12 days after G-CSF addition by using a Hema-3 staining set (Fisher).
Retroviral Transduction and Generation of Stable Cell Lines. The Xpress-tagged wild-type and mutant AML1 (AML1 Y380A) cDNAs were cloned in pMSCV (mouse stem cell virus) retroviral construct. The final constructs (pMSCV AML1 or pMSCV AML1 Y380A) were obtained through a three-way ligation reaction by using HindIII (blunted)-EcoRI-digested Xpress tag [from pcDNA3.1/His A (Invitrogen)], EcoRI-ClaI (blunted) AML1 cDNA (from pCMV5-AML1), and HpaI-digested pMSCV Puro vector. The GP2-293 packaging cell line was cotransfected with pMSCV vector alone, pMSCV-AML1 or pMSCV-AML1 Y380A and pVSVG (containing the viral envelope gene) by the calcium phosphate method. The medium containing viral particles was used to infect 32D cells in the presence of IL-3 and Polybrene (8 μg/ml). After 48 h, puromycin was added to a final concentration of 1 μg/ml. Cells were selected for 2 weeks in puromycin to generate stable cell lines expressing pMSCV (EV), pMSCV-AML1 (WT), or pMSCV-AML1 Y380A (MT).
Biochemical Fractionation and Western Blotting. Cells were biochemically fractionated as described in ref. 23. Total cell lysates (2 × 106 cells per lane) or various cellular fractions were subjected to Western blot analysis. Antibodies used were monoclonal mouse anti-lamin B1 (Zymed), AML-1/Runx1 rabbit polyclonal antibody (Active Motif, Carlsbad, CA), and anti-Xpress antibody (Invitrogen).
In Situ Immunofluorescence Analysis. Cells (8 × 104 per slide) were cytocentrifuged for 5 min at 400 rpm in a Cytospin, fixed by using formaldehyde (3.7%), and permeabilized with 0.5% Triton X-100 for whole-cell preparations. Nuclear matrix intermediate filament preparations were obtained as described in ref. 24. A monoclonal mouse Xpress antibody (Invitrogen; 1:500 dilution) and/or a polyclonal rabbit AML1/Runx1 antibody (Active Motif) was used to detect AML1/Runx1 proteins followed by appropriate fluorochrome-conjugated Alexa secondary antibodies (Molecular Probes; 1:800 dilution). Cells were mounted in Prolong Gold antifade mounting medium (Molecular Probes). Fluorescence and transmitted light images were captured by using a Zeiss Axioplan 2 microscope equipped with a digital charged-coupled device (CCD) camera (Hamamatsu Photonic, Bridgewater, NJ) interfaced with the MetaMorph Imaging System (Universal Imaging, Downingtown, PA).
Cytochemical Analysis of Myeloperoxidase (MPO) Activity. The stable cell lines were cytocentrifuged and stained for MPO according to the supplier's protocol (leukocyte peroxidase, Sigma-Aldrich). Briefly, 1 × 105 cells on slides were fixed for 30 sec in the fixative solution, washed, and placed in the peroxidase indicator reagent solution in a 37°C water bath for 30 min in the dark. Slides were then washed, dried, counterstained in acid hematoxylin solution for 30 sec, and examined microscopically for the presence of brown-black insoluble granules.
Cell Surface Marker Analysis by Flow Cytometry. One million cells of each of the EV, WT, and MT cell lines were washed twice in PBS/1% BSA (PBSA) and resuspended in 50 μl of diluent [0.01 M PBS (pH 7.4), containing 1% BSA and 0.1% NaN3] and incubated with saturating amounts of mouse fluorochrome-conjugated granzyme 1 (Gr-1) and CD11b antibodies (BD Biosciences Pharmingen) at 4°C for 45 min in the dark. The cells were then washed twice in PBSA, resuspended in 500 μl of PBSA containing 0.5% paraformaldehyde, and stored at 4°C. Data were collected by using a FACSCalibur (Becton Dickinson) flow cytometer for fluorescence at 488 nm as compared to a FITC and phycoerythrin-conjugated isotype controls (BD Pharmingen) and analyzed by using flowjo software (Tree Star, Ashland, OR). Annexin V staining was carried out according to manufacturer's instructions (Calbiochem).
Colony-Forming Assay. Each of the stable cell lines (2 × 104 cells) was plated in duplicate in 35-mm dishes containing Methocult methyl cellulose medium (StemCell Technologies, Vancouver), supplemented with mouse stem cell factor and mouse IL-3, and incubated for 10-14 days at 37°C with 5% CO2/95% air and 95% relative humidity.
Results and Discussion
Molecular Determinants for Intranuclear Compartmentalization of AML1 in Hematopoietic Cells. To determine the functional relationship between NMTS-dependent localization of AML1 to subnuclear domains and regulation of hematopoietic cell growth and differentiation, we introduced a point mutation in the NMTS region of the AML1 protein that abrogates its subnuclear targeting. The mutant AML1 protein enters the nucleus, binds to DNA, and retains interaction with several coregulatory proteins that include SMADs and TLE (unpublished results). Wild-type or mutated AML1 (AML1 Y380A) was cloned in murine stem cell retrovirus-based pMSCV vectors to facilitate stable expression of the proteins in hematopoietic cells. We used the 32D.cl3 myeloid progenitor cell line for our studies. This cell line expresses Runx1 (Fig. 7, which is published as supporting information on the PNAS web site) and can be made to differentiate to either granulocytes or monocytes, thus providing a model system to study the effects of chromosomal translocation on myeloid lineage. The 32D cell line was transduced with the vector alone or with AML1 wild type or mutant retroviruses. Cells were selected for puromycin resistance to enrich cell populations with stably integrated transgenes. Three independent cell lines were generated: EV (with stably integrated vector alone), WT (stably expressing wild-type AML1), and MT (stably expressing AML1 Y380A mutant). Western blot analysis shows that the exogenous proteins are expressed and are of the expected sizes (Fig. 1A).
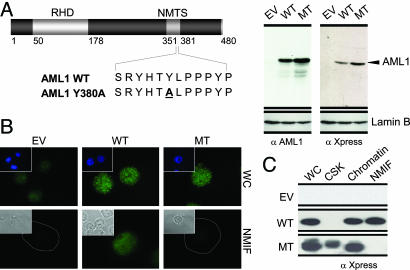
Generation of myeloid progenitor stable cell lines expressing AML1 with abrogated subnuclear targeting. (A) Stable expression of the wild-type and mutant AML1 proteins in myeloid progenitors. (Left) The AML1 proteins (wild type or mutant) used in this study are diagrammatically depicted; the runt homology domain (RHD) and the nuclear matrix targeting signal (NMTS) are shown. Numerals indicate the amino acid position within the protein. (Right) Expression of exogenous AML1 proteins in 32D cells infected with retrovirus carrying pMSCV empty vector (EV), wild-type AML1 (WT), or mutant AML1 (MT). Western blot analysis was carried out with antibodies raised against either AML1 or Xpress tag. Blots were also incubated with lamin B antibody as a control for protein loading. (B) A single amino acid substitution in the AML1 NMTS results in loss of nuclear matrix association. All three 32D stable cell lines were cytocentrifuged on precoated slides and were subjected to cellular fractionation and in situ immunofluorescence microscopy. A mouse monoclonal antibody against Xpress tag was used to assess the expression and localization of exogenous proteins. WC, whole cells; NMIF, nuclear matrix intermediate filaments. Insets in Upper show nuclei as visualized by DAPI staining. Insets in Lower show phase-contrast images. The dotted line represents the boundary of the nucleus. (C) The mutant AML1 protein is present in the soluble chromatin fraction. A total of 10 million cells for each cell line were used for biochemical fractionation, and an equivalent of 2 million cells per fraction was resolved by SDS/PAGE. An antibody against the Xpress tag was used to detect the stably expressed AML1 proteins. CSK, cytosolic fraction.
We determined the subnuclear distribution of wild-type and the NMTS mutant proteins in 32D stable cell lines by in situ immunofluorescence microscopy and biochemical approaches. Both wild-type and mutant AML1 proteins exhibit a punctate subnuclear distribution and are excluded from the nucleolar interior. As reported in ref. 21, the wild-type AML1 protein is associated with the nuclear matrix. However, the mutant AML1 protein is absent from nuclear matrix-intermediate filament preparations (Fig. 1B). These results were confirmed by biochemically assessing the partitioning of AML1 proteins among cytosolic, chromatin, and nuclear matrix compartments. As shown in Fig. 1C, the mutant protein is extracted into the cytosolic and chromatin fractions. Thus, mutation of the AML1 NMTS (Y380A) results in loss of nuclear matrix association in myeloid progenitor cells. The mutant protein enters the nucleus, binds to DNA, and retains interaction with coregulatory proteins that interact with the C terminus of AML1 transcription factor. However, it is important to note that there may be other yet unknown functions affected by this mutation. Taken together, our findings indicate that the functional NMTS motif is required to support intranuclear localization of the AML regulatory machinery in hematopoietic cells.
Requirement of AML1 Intranuclear Targeting for Early Events in Myeloid Differentiation. Because myeloid lineage is affected by the t(8;21) in AML patients, we determined the contributions of AML1 subnuclear targeting to differentiation of myeloid progenitors into the granulocytic lineage. All three 32D stable cell lines (EV, WT, and MT) were induced to differentiate into granulocytes by exposure to 25 ng/ml G-CSF for 12 days in the absence of IL-3. The morphological indicators of differentiation (nuclear/cytoplasmic ratio, nuclear shape, and degree of nuclear segmentation) were assessed. The EV and WT cells provided morphologic evidence of maturation to granulocytes. In contrast, MT cells were blocked at the promyelocytic stage of differentiation (Fig. 2 Left). These results were quantitated by scoring for blast cells (immature myeloid progenitors), metamyelocyte/band cells (early stages of myeloid differentiation), and segmented cells (mature granulocytes). As shown in Fig. 2 Right, a significantly higher percentage of MT cells was detected at the blast stage (60%) compared with 32-EV cells (8%) or 32-wt.A1 cells (23%). We also noted a slight maturational arrest in WT cells; ≈10% cells are morphologically delayed at the metamyelocytic stage. This maturational arrest may result from increased expression of wild-type AML1, which is normally downregulated in early differentiation (25). These results suggest that mutation of the AML1 NMTS, which abrogates subnuclear targeting, blocks granulocytic maturation at an early stage.
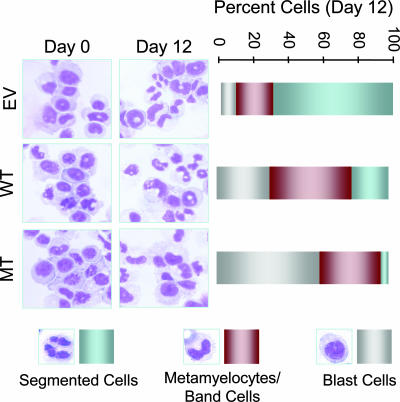
Myeloid progenitors expressing AML1 with compromised subnuclear targeting fail to differentiate into granulocytes. All three stable cell lines were differentiated to granulocytes with G-CSF. Photomicrographs of the morphology of Hema-3-stained 32D cell lines were taken at day 0 or day 12 of differentiation (Left). Right represents the percentage of cells for each cell line in three different stages of myeloid differentiation (i.e., blast or immature cells, metamyelocytes or band cells, and segmented or mature cells).
By using flow cytometry and cytochemistry, we determined the precise stage of granulocytic differentiation at which the MT cells are blocked. Markers for late stages of myeloid differentiation, Gr-1 and integrin M (CD11b) surface antigens (26, 27), were used to assess maturational status of the EV, WT, and MT cells. Consistent with morphological differences (Fig. 2), the CD11b up-regulation is significantly inhibited in MT cells; only 35% of MT cells express CD11b surface antigen as compared with 76% and 78% in EV and WT cells, respectively (Fig. 3 Upper). The Gr-1 expression is also compromised in MT cells but to a lesser extent (Fig. 3 Lower). These findings indicate that the MT cell line exhibits a blast-like phenotype.
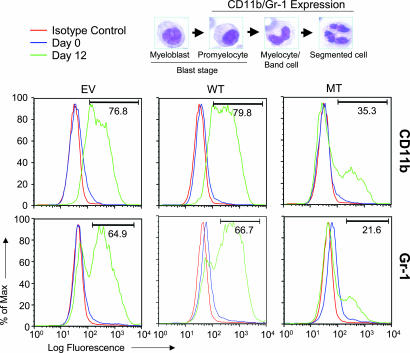
Expression patterns of markers for late myeloid differentiation reveal maturational arrest of myeloid progenitors with compromised AML1 subnuclear targeting. All three stable cell lines were differentiated to granulocytes, and samples were collected at days 0 and 12 for flow cytometric analysis of Gr-1 and CD11b, markers of late myeloid differentiation. The expression profiles of CD11b (Upper) and Gr-1 (Lower) are shown. The numbers in the graphs indicate the percentage of cells expressing CD11b and Gr-1. The micrographs at the top show the progression of myeloid progenitors and granulocytes. The expression of CD11b and Gr-1 is indicated.
The expression of MPO is an early event in the granulocyte/monocyte differentiation pathway (28). We confirmed the blast-like phenotype of cells expressing the NMTS mutant AML1 protein by determining MPO activity. All three 32D stable cell lines exhibit blast characteristics in the proliferative stage, as evidenced by the lack of MPO enzymatic activity (Fig. 4, day 0). However, at day 12 of differentiation, MPO staining was observed only in the MT cell line, which expresses the mutant AML1 protein. These results confirm a block of differentiation at the promyelocytic stage for the mutated AML1 cell line (Fig. 4, day 12). Interestingly, M2 type (French-American-British nomenclature) leukemic cells expressing the AML1-ETO fusion protein, which lack the AML1 NMTS, also show strong MPO activity. Taken together, our data demonstrate that the targeting of AML1 to intranuclear gene regulatory sites is linked to early events in myeloid differentiation.
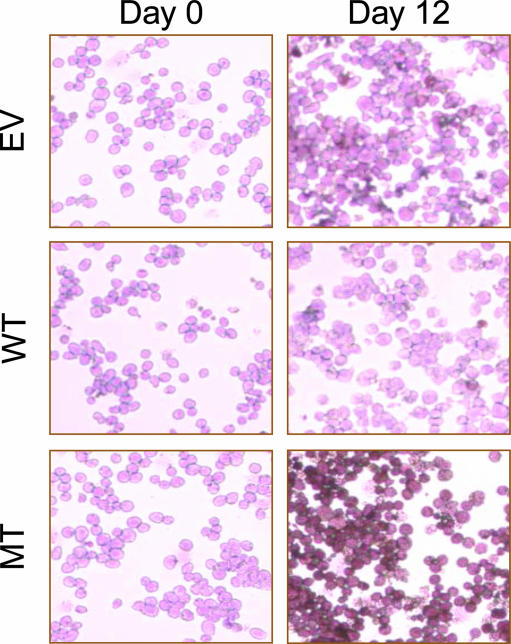
Abrogated AML1 subnuclear trafficking arrests myeloid progenitors at a very early stage of differentiation. Cells were stained for MPO activity at day 0 and day 12 of differentiation according to the supplier's instructions and counterstained with acid hematoxylin solution. The MPO activity was determined microscopically by the presence of brown-black granules.
Comparison of gene expression profiles for WT and MT cell lines after G-CSF treatment (day 0 and day 12) confirms differentiation block in MT cells. We note down-regulation of several genes, e.g., all three colony-stimulating factors (G-CSF, M-CSF, and GM-CSF), several interleukins (IL-2, IL-4, IL-10, and IL-12B), and leukocyte inhibitory factor (LIF), in cells expressing the NMTS mutant AML1. Additionally, AML1-responsive genes (e.g., IFN-γ, IL-13) that have been identified in other hematopoietic lineages show altered expression profiles (Fig. 8, which is published as supporting information on the PNAS web site). Promoters of these genes contain AML1 consensus elements, implicating AML1 in regulation of these genes during granulopoiesis.
Our findings support the necessity of AML1 intranuclear targeting for granulocyte differentiation as demonstrated by in vivo genetic models. For example, the AML1 knock-in mouse model, in which the entire AML1 C terminus containing the NMTS is replaced by the galactosidase reporter (29), exhibits a phenotype analogous to that of AML1-deficient mice. Thus, the AML1 C terminus and associated regulatory functions are required for in vivo biological activity of the protein. Furthermore, loss of the AML1 C terminus in the predominant (8;21) translocation in AML patients results in the arrest of myeloid progenitor cells at the blast stage, a diagnostic morphological feature of this leukemia (30). Taken together, these results demonstrate the requirement of AML1 intranuclear targeting for myeloid differentiation and suggest that its disruption is involved in the onset of leukemogenesis.
Impaired Subnuclear Targeting of AML1 Alters Myeloid Progenitor Proliferation and Promotes a Transformation-Like Phenotype. Our findings demonstrate that the mutation that impairs intranuclear targeting of AML1 also arrests myeloid differentiation at an immature blast stage (Figs. (Figs.2,2, ,3,3, ,4).4). A block in differentiation is often linked to deregulation of cell proliferation as evident by the altered cell cycle in cancer cells (31). We therefore measured the growth rates of 32D stable cell lines expressing the wild-type and mutant proteins. These cells, as well as the control EV cell line, were cultured at an equal density in either the absence or the presence of IL-3, which is required for 32D cell growth (32), or G-CSF, which induces granulocytic differentiation (33).
As expected, all three cell lines continue to grow in the presence of IL-3 and cease to proliferate when both IL-3 and G-CSF are removed (Fig. 9, which is published as supporting information on the PNAS web site). When grown in G-CSF alone, the MT cells continue to proliferate (Fig. 5A), consistent with the defective granulocyte maturation of this cell line (Figs. (Figs.2,2, ,3,3, ,4).4). These findings are supported by the observation that IL-3, which contains five AML1 consensus binding sites in its promoter, is increased in MT cells relative to 32-wt.A1 cells (Fig. 8). In addition, gene profiling also reveals an up-regulation of the S-phase accelerator Cdc21 and the mitotic exit protein Cdc16 that parallels the increased proliferation of the MT cells (Fig. 8). The TGFβ/BMP signaling cascade is another important pathway that regulates proliferation of 32D cells (34). We find that several components of the TGFβ/BMP signaling pathway (ligands such as TGFβ, BMP5, GDF6; receptors such as BMP receptor type IB and IIB; and downstream effectors such as Smad 2, 5, and 9) are highly induced in the mutant cell line (Fig. 8). However, this pathway may not be functional in MT cells because the AML1 intranuclear targeting is required for completion of this signaling cascade (18).
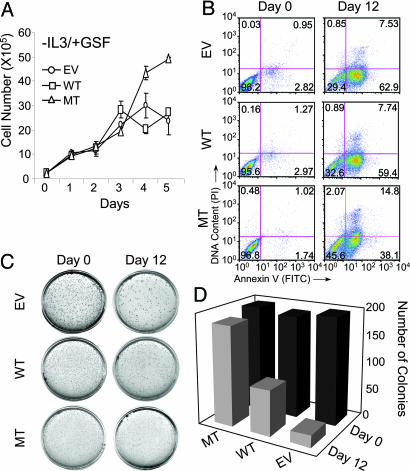
The mutant AML1 protein results in altered growth properties and a transformation-like phenotype. (A) Increased proliferative potential of cells expressing the NMTS mutant of AML1. Cells were counted at indicated days of differentiation. The MT cells expressing the mutant AML1 protein continue to proliferate when placed in the differentiating agent G-CSF, whereas the EV and WT cell lines stopped dividing 3 days after GCSF treatment. Error bars represent standard deviation (n = 3). (B) Compromised AML1 subnuclear targeting leads to decreased apoptosis. Equal numbers of cells from either actively proliferating (day 0) or differentiated (day 12) cell lines were stained for annexin V (a marker of apoptotic cells) and DNA content (propidium iodide, PI). Cells were then analyzed by flow cytometry to determine the presence of apoptotic cells. The numbers in each quadrant represent the percentage of cells with different survival status. (C) Transformation-like phenotype of cells with impaired AML1 subnuclear targeting. (Left) The growth of 32D stable cell lines in methyl cellulose is shown. Cells at day 0 or day 12 of differentiation were plated in methyl cellulose medium containing IL-3. The plates were then incubated at 37°C for 14 days. (D) Quantitation of the colonies for each experimental condition.
Increased cell number in the MT cell line may be due to enhanced proliferation or decreased apoptosis. Our data suggest that both possibilities are in part responsible for the increased MT cell number. For example, the MT cell line expressing the mutated AML1 also exhibits enhanced survival in the absence of IL-3 (Fig. 9). Consistent with these results, we observe an increase in the number of MT cells that have not entered an apoptotic pathway (Fig. 5B). This decreased rate of apoptosis is reflected by down-regulation of proapoptotic genes that include caspase 7 and TNF receptor 2 (Fig. 8). In addition, we find increased number of MT cells in the S phase, whereas fewer cells exhibit apoptosis (data not shown). Thus, the increased growth of MT cells correlates with enhanced cell survival. However, the mutation that impaired AML1 subnuclear targeting is not sufficient to result in cytokine independence, a characteristic of leukemic cells (35). The leukemia-related AML1-ETO fusion protein shows a similar effect; AML1-ETO, which exhibits a misdirected intranuclear trafficking, is also insufficient to render myeloid cells cytokine-independent (36). Thus, interference with AML1 intranuclear targeting, either by introducing a point mutation or because of chromosomal translocation, alters the growth and survival characteristics of myeloid progenitors.
Next, we determined whether the mutation that abrogated targeting of AML1 to subnuclear regulatory domains contributes to acquisition of transformed phenotypic properties. We determined the colony-forming ability of the 32D cell lines in methyl cellulose. As shown in Fig. 5C, all three cell lines exhibit similar colony-forming ability when isolated at day 0 (proliferating cells). However, after treatment with G-CSF (day 12), the MT cell line forms significantly more colonies than either EV or WT. However, as reported in ref. 37, there is a slight increase in the number of WT cell colonies relative to the control cell line. The enhanced colony formation in the mutant cell line is consistent with an increased number of blast cells (Fig. 2 Right) and sustained MPO activity of this cell line (Fig. 4 Bottom). Our findings are reinforced by the induction of FGF4 and EVI1, key regulators of cell transformation, in the MT cell line (Fig. 9 and refs. 38 and 39). Taken together, our findings indicate that the mutation compromised AML1 with altered subnuclear targeting results in a transformation-like phenotype of myeloid progenitors that is coupled with related changes in gene expression.
The increased colony-forming ability, sustained MPO activity, and decreased apoptosis associated with the AML1 NMTS mutant protein are indicative of transformation and tumorigenesis. Our results are compatible with the recently described effects of the AML1-ETO translocation-fusion protein on human myeloid progenitors. AML1-ETO inhibits maturation of these CD34+ cells into granulocytes, with a concomitant increase in blast-like cells and enhanced colony-forming potential; however, these cells remain cytokine-dependent for their growth (36). We conclude that the functions altered by the AML point mutation, including subnuclear targeting of the AML1 protein to gene-regulatory sites, are required for normal myeloid differentiation. The perturbed functions, including altered intranuclear trafficking, increase proliferative potential and result in acquisition of a transformation-like phenotype, although competency to cause leukemia in vivo remains to be established.
Conclusions
During hematopoiesis and leukemogenesis, cells exhibit striking modifications in nuclear morphology concomitant with changes in the subnuclear organization of nucleic acids and regulatory proteins that constitute the machinery for gene expression (1-9). For example, the transition of a myeloid progenitor cell to a granulocyte is characterized by progressive changes from a large ovoid nucleus to a lobulated structure (40). Similarly, leukemia is typically diagnosed by a predominance of primitive hematoprogenitor-like cells that retain the characteristic blast-like nuclear morphology and additionally acquire functional changes (41). These modifications are characteristics of transformation and include genomic instability, perturbed gene expression, and impaired signaling pathways, as well as altered nucleolar organization, composition, and function (42). It is therefore apparent that nuclear structure-function interrelationships are compromised during the progressive acquisition of the transformed phenotype.
Here we have used the AML1 transcription factor to study the mechanistic linkage of changes in nuclear organization with hematopoietic differentiation and/or leukemogenesis coupled transformation. AML1 is a scaffolding protein that assembles and organizes the machinery for hematopoietic gene expression at multiple sites in target genes as well as at subnuclear domains (5). AML1 protein integrates and executes signaling pathways, interacts with proteins involved in chromatin remodeling and histone modifications, and mediates transcriptional activation and repression through the multifunctional C terminus that contains the NMTS (14, 18-20). Furthermore, the AML1 gene is a frequent target of leukemia-associated chromosomal translocations (12, 13). The predominant (8;21) translocation results in loss of the AML1 C terminus containing the NMTS and thus alters the composition, content, and intranuclear localization of regulatory machinery for AML1-responsive transcription (5, 17).
Our study shows that AML1 intranuclear targeting is involved in granulopoiesis. By combining biochemical, immunocytochemical, and functional approaches, we establish that myeloid progenitors expressing AML1 with a mutated subnuclear targeting signal display a blast-like phenotype and arrest at the promyelocytic stage. These cells show an immature phenotype and enhanced competency to form colonies in methyl cellulose but do not exhibit cytokine-independent growth. Whereas an NMTS point mutation that blocks subnuclear targeting of AML1 is sufficient to inhibit myeloid maturation, we cannot exclude the loss or gain of additional functions/activities. However, expression of either AML1 with impaired subnuclear targeting or AML1-ETO fusion protein with intranuclear misrouting results in strikingly similar effects on growth and phenotypic properties of myeloid progenitors (36), suggesting that impaired AML1 intranuclear trafficking is functionally linked with transformation-coupled arrest of granulocytic maturation. Our findings link the architectural organization of gene-regulatory machinery in the nucleus with the regulation of normal hematopoiesis as well as the onset and progression of leukemia (Fig. 6).
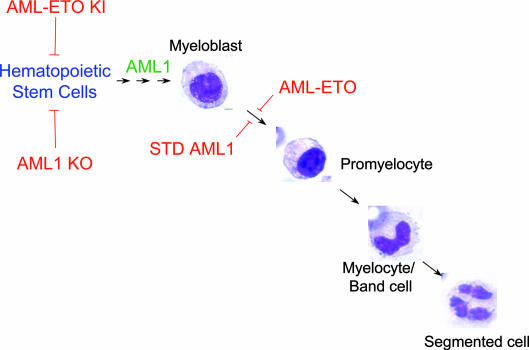
Compromised intranuclear targeting of the AML1 hematopoietic transcription factor inhibits myeloid differentiation and promotes a transformation-like phenotype. Myeloid precursor cells originate from the hematopoietic stem cell (HSC). The commitment of HSCs to any hematopoietic lineage requires AML1 in vivo. Homozygous deletion of AML1 results in the failure of HSCs to differentiate and loss of definitive hematopoiesis. Myeloblasts, characterized by an oval nucleus, differentiate into granulocytes in the presence of G-CSF. The granulocytic differentiation of myeloblasts is characterized by alterations in nuclear shape from oval (promyelocyte) to horseshoe shape (myelocyte/band cell) to multilobulate (segmented cell). AML1-ETO, the product of the (8;21) chromosomal translocation, results in maturational arrest of the myeloblast to granulocyte lineage (red block arrow). Our findings demonstrate that compromised AML1 intranuclear targeting is sufficient to result in transformation-coupled maturational arrest of myeloid progenitors into granulocytes. Thus, the subnuclear targeting of AML1 to regulatory domains within the nucleus is required for myeloid differentiation, and abrogation of AML1 intranuclear targeting can result in transformation blast-like phenotype, a hallmark of leukemogenesis. KO, knockout; KI, knock-in.
Acknowledgments
We thank Drs. Ricardo Medina, Hayk Hovhannisyan, Jitesh Pratap, and Daniel Young for helpful suggestions and Dr. Gerard P. Zambetti for the gift of IL-3. This study was supported by Grant P01 CA82834 from the National Cancer Institute, National Institutes of Health and Grant DK 32520 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Notes
Author contributions: D.V., S.K.Z., J.B.L., A.J.v.W., J.L.S., and G.S.S. designed research; D.V. and S.K.Z. performed research; D.V. and S.K.Z. contributed new reagents/analytic tools; D.V., S.K.Z., J.B.L., A.J.v.W., J.L.S., and G.S.S. analyzed data; and D.V., S.K.Z., J.B.L., A.J.v.W., J.L.S., and G.S.S. wrote the paper.
Abbreviations: AML, acute myeloid leukemia; BMP, bone morphogenetic protein; AML1, acute myelogenous leukemia transcription factor 1; Runx1, Runt-related transcription factor 1; NMTS, nuclear matrix targeting signal; MPO, myeloperoxidase; G-CSF, granulocyte colony-stimulating factor; Gr-1, granzyme 1.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0502130102
Read article for free, from open access legal sources, via Unpaywall:
http://www.pnas.org/content/102/20/7174.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.0502130102
Article citations
Unrelated hematopoietic stem cell transplantation for familial platelet disorder/acute myeloid leukemia with germline RUNX1 mutations.
Int J Hematol, 118(3):400-405, 10 Mar 2023
Cited by: 1 article | PMID: 36897502
OGP46 Induces Differentiation of Acute Myeloid Leukemia Cells via Different Optimal Signaling Pathways.
Front Cell Dev Biol, 9:652972, 04 Mar 2021
Cited by: 4 articles | PMID: 33748146 | PMCID: PMC7969801
PML/RARA destabilization by hyperthermia: a new model for oncogenic fusion protein degradation?
Blood Cancer Discov, 2(4):300-301, 01 Jun 2021
Cited by: 5 articles | PMID: 34230915 | PMCID: PMC7611121
Inhibition of the RUNX1-CBFβ transcription factor complex compromises mammary epithelial cell identity: a phenotype potentially stabilized by mitotic gene bookmarking.
Oncotarget, 11(26):2512-2530, 30 Jun 2020
Cited by: 10 articles | PMID: 32655837 | PMCID: PMC7335667
Tracing Hematopoietic Progenitor Cell Neutrophilic Differentiation via Raman Spectroscopy.
Bioconjug Chem, 29(9):3121-3128, 06 Sep 2018
Cited by: 11 articles | PMID: 30148625 | PMCID: PMC6346746
Go to all (32) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The t(8;21) chromosomal translocation in acute myelogenous leukemia modifies intranuclear targeting of the AML1/CBFalpha2 transcription factor.
Proc Natl Acad Sci U S A, 96(26):14882-14887, 01 Dec 1999
Cited by: 61 articles | PMID: 10611307 | PMCID: PMC24742
Rearrangements of the AML1/CBFA2 gene in myeloid leukemia with the 3;21 translocation: in vitro and in vivo studies.
Leukemia, 11 Suppl 3:273-278, 01 Apr 1997
Cited by: 4 articles | PMID: 9209363
New mechanisms of AML1 gene alteration in hematological malignancies.
Leukemia, 17(1):9-16, 01 Jan 2003
Cited by: 81 articles | PMID: 12529654
Review
Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene.
Nat Genet, 15(3):303-306, 01 Mar 1997
Cited by: 221 articles | PMID: 9054947
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P01 CA082834
Grant ID: P01 CA82834
NIDDK NIH HHS (2)
Grant ID: P30 DK032520
Grant ID: DK 32520




