Abstract
Free full text

CSN5/Jab1 Is Involved in Ligand-Dependent Degradation of Estrogen Receptor α by the Proteasome
Abstract
Here, we show that estrogen receptor α (ERα) coimmunoprecipitates with CSN5/Jab1, a subunit of the COP9 signalosome (CSN), and that overexpression of CSN5/Jab1 causes an increase in ligand-induced ERα degradation. Inhibition of either the kinase activity associated with the CSN complex by curcumin or of nuclear export by leptomycin B (LMB) impaired estradiol-induced ERα degradation by the proteasome. Degradation of ERα induced by the pure antagonist ICI 182,780 (ICI) was blocked by curcumin but not by LMB, indicating that in the presence of ICI, ERα is degraded by a nuclear fraction of the proteasome. In addition, we observed that curcumin inhibited estradiol-induced phosphorylation of ERα. The use of three inhibitors of ERα degradation that target different steps of the estrogen response pathway (inhibition of the CSN-associated kinase, nuclear export, and proteasome) suggests that a phosphorylation event inhibited by curcumin is necessary for ERα binding to its cognate DNA target. Our results demonstrate that transcription per se is not required for ERα degradation and that assembly of the transcription-initiation complex is sufficient to target ERα for degradation by the proteasome.
Estradiol (E2) regulates target cell proliferation and gene transcription through series of molecular events initiated by the hormone-dependent binding of the estrogen receptor α (ERα), a member of the nuclear receptor superfamily, to its cognate DNA target. In mammary cells, the effects of estradiol can be antagonized by compounds such as 4-hydroxy-tamoxifen (OH-Tam), a tamoxifen metabolite that is a selective estrogen receptor modulator, and ICI 182,780 (ICI), a pure antiestrogen. Both OH-Tam and ICI antagonize estrogen action primarily by competing with estradiol for receptor binding. However, OH-Tam has a partial agonist activity, depending on the tissue and response examined (14), and ERα-OH-Tam complexes accumulate in nuclei (12, 37). In contrast, ICI compounds are totally devoid of agonist activity in the models studied to date (32), and ICI treatment provokes a rapid relocalization of ERα-ICI complexes to a salt-resistant nuclear fraction, followed by rapid degradation by the proteasome (12, 36). A complex pathway leading to transcriptional activation of target genes involves ERα structural changes upon hormone binding, allowing dynamic interactions of the hormone-receptor complex with DNA and coactivators. This will allow chromatin remodeling and the subsequent recruitment of coactivators and the formation of preinitiation complexes (for a review, see reference 24). ERα is downregulated in the presence of E2, its cognate ligand, through the ubiquitin/proteasome (Ub/26S) pathway (1). It has been proposed that ERα-mediated transcription and proteasome-mediated degradation are linked (17) and act to continuously turn over ERα on responsive promoters (20). Recently, it was proposed that the p160 coactivator steroid receptor coactivator 3 (SRC3)/AIB1 mediates agonist-induced ERα degradation (29).
The 26S proteasome is a multiprotein complex composed of a 20S core complex and a 19S particle (base plus lid). It is involved in the degradation of short-lived proteins. The lid is thought to recognize ubiquitinated substrates that are then unfolded by the base and degraded in the proteolytic core complex. Protein ubiquitination is catalyzed by three critical factors: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a factor E3 or ubiquitin ligase (E3).
Ubiquitin ligases termed SKP1-Cullin/CDC53-F-box protein (SCF) complexes catalyze the polyubiquitination of a number of proteins to target them for degradation. The neddylation (addition of the Ub-like protein NEDD8) of the cul1 (cullin1) subunit of the SCF stimulates protein polyubiquitination (23). The COP9 signalosome (CSN), a complex of eight proteins (CSN1 to CSN8), removes NEDD8 from cul1 (18, 27, 40). This deneddylase activity has recently been attributed to the MPM plus motif of the CSN5/Jab1 subunit (8) of CSN. It stops the polyubiquitination process and allows the subsequent degradation of polyubiquitinated substrates by the proteasome. It has been proposed that CSN serves as an assembly and maintenance platform for the cullin-based ubiquitin ligases (38). In addition, this complex exhibits significant sequence homologies to the eight subunits of the 26S proteasome lid complex. It is involved in regulating the stability of proteins such as p27Kip1 (34), c-jun (22), and p53 (4), which are substrates of the Ub/26S proteasome pathway. A kinase activity, inhibited by curcumin, is associated to the CSN complex. Recent data demonstrated an association of inositol 1,3,4-trisphosphate 5/6-kinase (31), casein kinase II (CKII), and protein kinase D (35) with the CSN complex.
In the cell, two types of complexes containing CSN5/Jab1 have been described. One is the conventional 450-kDa CSN located in the nucleus and the other is a mini-CSN (approximately 100 kDa) containing only a subset of CSN components that is mainly located in the cytoplasm. The mini-CSN originates from the nuclear export of subunits CSN4 to -8 (33). As part of this mini-CSN, CSN5/Jab1 has been implicated in the nuclear export of the cyclin-dependent kinase inhibitor p27Kip1, playing the role of an adaptor between p27Kip1 and the nuclear transport protein CRM-1. The use of leptomycin B (LMB), a chemical inhibitor of CRM-1-dependent nuclear export, prevented p27Kip1 nuclear export and its CSN5/Jab1-mediated degradation (33). In addition, overexpression of CSN5/Jab1 caused the translocation of Smad7 from the nucleus to the cytoplasm, promoting its degradation (15).
Several results suggest that CSN could be involved in the mechanism of action of estrogens: CSN5/Jab1 has been described as a coactivator of nuclear receptors (6), and the neddylation pathway is involved in ERα degradation (9, 10).
The aim of our work was to investigate the contribution of CSN5/Jab1 to ERα ligand-dependent degradation and to the mechanism of transcription activation in response to estradiol. Here, we show that ERα and CSN5/Jab1 coimmunoprecipitate and that an increase in CSN5/Jab1 levels increases ligand-induced ERα degradation by the proteasome. Curcumin, which inhibits the kinase activity associated with CSN, prevents hormone-dependent ERα degradation, as well as estradiol-dependent phosphorylation of ERα. CSN5/Jab1 is also involved in the nuclear export of proteins. We show that LMB, an inhibitor of nuclear export, prevents estradiol-induced degradation of ERα, demonstrating that this degradation takes place in the cytoplasm. In contrast, for ERα complexed to ICI, a pure hormone antagonist, LMB had no effect, indicating that this degradation takes place in the nucleus. Finally, we investigated the effects of curcumin and LMB on estradiol-induced transcription. Curcumin, but not LMB, abolished estradiol-induced reporter gene activation. Curcumin prevented ERα binding to its cognate DNA target, indicating a probable involvement of CSN in a phosphorylation event, necessary for ERα binding. These results also demonstrate that ERα degradation is not required to sustain estradiol-induced transcription.
MATERIALS AND METHODS
Reagents and antibodies. (i) Chemicals.
MG132 was purchased from Calbiochem. Estradiol, N-acetyl-leucyl-leucyl-norleucinal (ALLN), lactacystin β-lactone, α-amanitin, 5,6-dichlorobenzimidazole riboside (DRB), H-7 dihydrochloride, curcumin, and E64 were purchased from Sigma. ICI 182,780 and 4-hydroxy-tamoxifen were purchased from Zeneca Pharmaceuticals.
(ii) Antibodies.
Anti-human ERα (HC20) was purchased from Santa Cruz. Anti-Jab1 (clone 2A10) was purchased from Abcam. Anti-c-myc (clone 9E10) and anti-green fluorescent protein (anti-GFP) (clones 7.1 and 13.1) were purchased from Roche. Anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) was purchased from Chemicon International. Anti-RNA polymerase II (anti-Pol II) (clone CTD4H8) was purchased from Upstate.
Constructs.
We used the following vectors: estrogen response element (ERE)-βglob-Luc, expressing luciferase under the control of a synthetic estrogen-regulated promoter; pHook-Jab1, expressing CSN5/Jab1 (5); SV4-luc and pCH110 expressing, respectively, luciferase or β-galactosidase under the control of a simian virus 40 promoter; GFP-C2, expressing GFP, purchased from Clontech; PSG5-HEG0, expressing wild-type ERα (21).
Cell lines and cell culture.
MCF7 and MELN cells were grown in Dulbecco's modified Eagle's medium F-12 containing 50 μg/ml gentamicin and 10% heat-inactivated fetal calf serum (Invitrogen) at 37°C in a humidified atmosphere containing 10% CO2. MELN cells were generated from MCF7 cells and express luciferase under the control of an estrogen-regulated promoter (3).
NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium containing 4.5 g/liter glucose, 50 μg/ml gentamicin, 1 mM sodium pyruvate, and 10% heat-inactivated fetal calf serum at 37°C in a humidified atmosphere containing 10% CO2.
To study the effects of estrogens, cells were grown for 3 days in medium without phenol red, without gentamicin, and containing 5% serum, depleted of endogenous steroids by three successive incubations with dextran-coated charcoal. Cells were treated or not with 10 nM estradiol (E2) or 100 nM ICI 182,780 for the indicated times.
Coimmunoprecipitation.
MCF7 cells from one subconfluent dish (10-cm diameter), grown for 3 days in steroid-free medium, were washed twice in phosphate-buffered saline (PBS), resuspended in 500 μl lysis buffer (50 mM Tris [pH 8], 0.4% NP-40, 300 mM NaCl, 10 mM MgCl2, antiprotease from Roche [1 mini-tablet/10 ml]) and incubated for 15 min at 4°C. After centrifugation for 20 min at 16,000 × g, the lysate was adjusted to 1 ml with 500 μl of dilution buffer (50 mM Tris [pH 8], 0.4% NP-40, 2.5 μM CaCl2, 1 μl DNase I). A total of 40 μl (vol/vol) protein A/protein G beads was added, and the lysate was incubated for 45 min at 4°C on a rotating wheel. The supernatant was collected by centrifugation, 30 μl was withdrawn for input assays, and the remaining material was incubated with 1 μg anti-CSN5/Jab1 or 1 μg anti-c-myc for 1 h at 4°C on a rotating wheel. A total of 20 μl (vol/vol) protein A/protein G beads was added again, and the incubation continued for 1 more hour. Beads were washed three times with wash buffer (50 mM Tris [pH 8], 0.4% NP-40, 150 mM NaCl, 5 mM MgCl2), and proteins were eluted by the addition of 20 μl H2O and 10 μl of 4× electrophoresis loading buffer (200 mM Tris [pH 6.8], 8% sodium dodecyl sulfate [SDS], 20% glycerol, 8 mM EDTA, 6% β-mercaptoethanol, 0.016% bromophenol blue). The totality of the immunoprecipitated material was electrophoresed on SDS gels and analyzed by Western blotting.
Western blots.
Western blots were performed as previously described (12). The quantity of extract loaded on the gels was normalized for total proteins content, assayed by the Amido Schwarz technique (26). In transient transfection experiments, cells were cotransfected with a vector expressing luciferase, and normalization of loaded samples was performed by measuring the luciferase activity. Antibodies were used at the following dilutions: anti-human ERα (HC20), 1/200; anti-GAPDH, 1/500; anti-CSN5/Jab1, 1/1,000; anti-GFP, 1/1,000.
Cell transfections.
NIH 3T3 cells (5 × 105 cells) were plated in 10-cm diameter dishes and grown for 3 days in steroid-free medium. Cells were transfected with the JetPEi kit from Qbiogen. At 48 h after transfection, the cells were treated or not with hormone or antagonist for the times indicated (see Results). After being washed twice with PBS, cells were lysed in 200 μl lysis buffer (Promega).
Luciferase assay.
Cells from a subconfluent 3.5-cm-diameter dish were lysed in 200 μl Promega lysis buffer. Luciferase activities were determined using luciferase assay reagent (Promega), and β-galactosidase activities were determined using the kit from Tropix. All measurements were performed using a Berthold Lumat LB 9501 luminometer.
Metabolic labeling.
MCF7 cells (2 × 106 cells) were plated in 10-cm-diameter dishes and grown for 3 days in steroid-free medium. After being washed three times, cells were incubated for 2 h in medium without hormone and without phosphate, and then 50 μM of curcumin was added or not. After 30 min, cells were treated or not with 10 nM E2 and 200 μCi/ml of [32P]phosphorus (Amersham) added to the medium. After 2 h, cells were washed two times with medium without phosphate and two times with PBS, and the pellet was resuspended in 150 μl lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.1], antiproteases, and antiphosphatases). After centrifugation for 10 min at 4°C, the cell lysate was diluted 10 fold in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris [pH 8.1]). The lysate was incubated for 2 h at 4°C on a rotating wheel with 40 μl (vol/vol) protein A beads and protein G beads. The supernatant was collected by centrifugation and incubated with 1 μg HC20 antibody overnight at 4°C on a rotating wheel. On the other hand, 20 μl (vol/vol) protein A beads and protein G bead suspension were saturated with 2 μg proteins of an SDS-MCF7 lysate for 1 h at 4°C on a rotating wheel. These saturated beads were collected by centrifugation and added to the lysate previously incubated with the antibody for 1 h at 4°C on a rotating wheel. Beads were washed in 1 ml buffer TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris [pH 8.1]) on a rotating wheel for 10 min, in 1 ml buffer TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris [pH 8.1]) for 10 min, and three times in 500 μl Tris-EDTA (pH 8.1) for 10 min. Proteins were eluted by addition of 20 μl H2O and 10 μl loading buffer 4X. A total of 85% of the sample was analyzed by gel electrophoresis, and the gel was dried and exposed on a film for 6 days at −80°C to determine ERα phosphorylation status. The remaining 15% of the sample served to determine ERα content by Western blot analysis.
Chromatin immunoprecipitation.
MCF7 cells were grown in a 14-cm-diameter dish for 3 days in steroid-free medium, followed by 24 h in medium containing 2.5% steroid-free fetal calf serum. Chromatin immunoprecipitations (ChIPs) were performed as described in reference 28, with the following modifications: immunoprecipitations were performed with 3 μg anti-ERα (HC20) or with 1 μg anti-Pol II antibodies. After cross-link reversion, 1/3 of DNA was purified on GFX columns (Amersham Biosciences) and 1/50 of this purified DNA was quantified by quantitative PCR. Quantitative PCR was performed with an iCycler (Bio-Rad) using the following oligonucleotides: PS2 CH1 (5′-GGCCATCTCTCACTATGAATCACTTCTGC-3′) and PS2 CH2 (5′-GGCAGGCTCTGTTTGCTTAAAGAGCG-3′) to amplify the TFF1/pS2 promoter fragment.
Amplification conditions were 3 min at 95°C, followed by 50 cycles, each cycle consisting of 20 s at 95°C, 30 s at 62.5°C, 20 s at 72°C, and 15 s at 82°C.
RESULTS
CSN5/Jab1 coimmunoprecipitates with ERα and is a coactivator of estradiol-induced transcription.
CSN5/Jab1 is a coactivator of ERα-dependent transcription (6), but no interaction of ERα with CSN5/Jab1 has been demonstrated. Figure Figure1A1A presents the result of a CSN5/Jab1-ERα coimmunoprecipitation experiment in MCF7 cells. Cells were pretreated with ALLN (a proteasome inhibitor) to block ERα degradation by the proteasome and treated or not with estradiol (E2). The extracts were immunoprecipitated with anti-CSN5/Jab1 antibodies, and ERα was analyzed by Western blotting. ERα coimmunoprecipitated with CSN5/Jab1 in cells treated with estradiol and/or ALLN. This result suggests an interaction (direct or indirect) between ERα and CSN5/Jab1. However, the immunoprecipitated band was always very faint compared to input material, suggesting either the involvement of a very small fraction of ERα in such an interaction or a transient interaction between ERα and CSN5/Jab1.
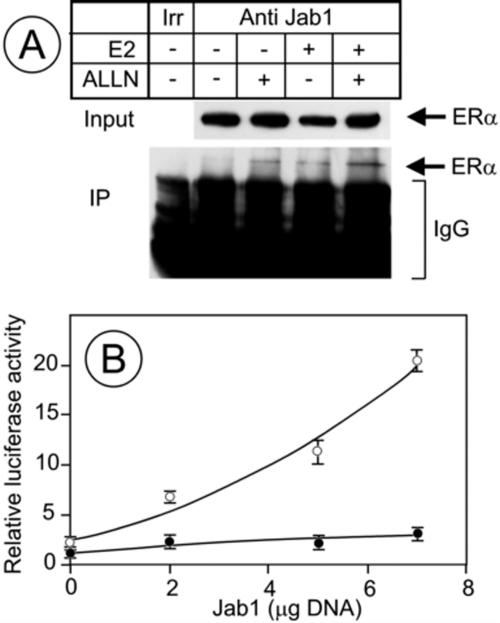
CSN5/Jab1 coimmunoprecititates with ERα and coactivates it. (A) MCF7 cells were pretreated for 30 min with a proteasome inhibitor (ALLN; 100 μM) and treated for 1 h with vehicle or E2. Extracts were immunoprecipitated with an anti-c-myc antibody (lane irr, irrelevant) or an anti-CSN5/Jab1 antibody, and ERα was analyzed by Western blotting in the immunoprecipitated fraction (IP) and in the inputs. The amount of material loaded for the immunoprecipitated fractions was 66-fold larger than the amount loaded for the input fractions. (B) NIH 3T3 cells were transfected with 1 μg of pSG5-HEG0, 1 μg of ERE-βglob-luc, 1 μg of pCH110, and the indicated amounts of pHOOK-CSN5/Jab1 (see Materials and Methods). The total amount of plasmid DNA was adjusted with empty vector to keep it identical in all samples. At 24 h after transfection, cells were treated with E2 (open circles) or vehicle (closed circles) for 24 h. Luciferase and β-galactosidase activities were measured, and the results were normalized for transfection efficiency using β-galactosidase activity.
To investigate the contribution of CSN5/Jab1 to estrogen-activated transcription, we transfected NIH 3T3 cells, which do not express ERα, with a constant amount of a vector expressing ERα and an ERE-luciferase reporter gene together with increasing amounts of a vector expressing CSN5/Jab1. We chose a cell line that does not express ERα to follow transcription in response to the increase of CSN5/Jab1 in transfected cells without masking this effect by nontransfected cells. Luciferase activity was measured after estradiol treatment or not. Figure Figure1B1B shows that an increase in luciferase activity followed the increase in the amount of the vector expressing CSN5/Jab1. The increase was modest in untreated cells and pronounced in cells treated with estradiol. Luciferase induction by estradiol was approximately twofold in the absence of the cotransfected CSN5/Jab1 expression vector. Estradiol-induced luciferase increased significantly with the increase in vector expressing CSN5/Jab1, up to 10 fold for the maximal amount of CSN5/Jab1 expression vector used in the experiment. In contrast, basal luciferase expression was only slightly changed. This resulted in an increase of up to two- to threefold in the hormone-induced level of luciferase, demonstrating that although an ERα-CSN5/Jab1 interaction was difficult to visualize, CSN5/Jab1 acts as a coactivator of ERα in NIH 3T3 cells.
CSN5/Jab1 overexpression decreases ERα content in NIH 3T3 cells.
Turnover of estrogen receptor and coactivator has been proposed to contribute to ERα transcriptional activity (17). On another hand, CSN5/Jab1, which is an ERα coactivator, regulates the proteasome-dependent degradation of cellular proteins, mainly those involved in cell cycle regulation, such as p27Kip and p53 (4, 34). Accordingly, the effect of CSN5/Jab1 on estrogen-induced transcription may reflect a change in ERα content as a result of CSN5/Jab1 overexpression. We investigated the influence of CSN5/Jab1 overexpression on ERα stability in the presence of hormone or antihormone in NIH 3T3 cells cotransfected with vectors expressing ERα and CSN5/Jab1 (Fig. (Fig.2).2). The half-life of ERα is 2 to 3 h when it is complexed with estradiol and much shorter when it binds the pure antagonist ICI. Thus, we treated NIH 3T3 cells with E2 for 1 h 30 and with ICI for 1 h to be able to clearly show differences in receptor stability. In the presence of E2, ERα levels decreased when CSN5/Jab1 expression increased (Fig. 2A and B), whereas the increase in CSN5/Jab1 did not significantly affect GFP stability (Fig. (Fig.2B).2B). In cells treated with ICI, CSN5/Jab1 overexpression also caused a decrease in ERα cellular content (Fig. (Fig.2C).2C). We note that CSN5/Jab1 did not accumulate when the amount of CSN5/Jab1 expression vector transfected was increased (Fig. 2A and C). This is due to the fact that only a small number of cells (<10%) were transfected so that the newly expressed CSN5/Jab1 was diluted in bulk CSN5/Jab1 from nontransfected cells. The situation is different for ERα and GFP, which are not expressed in NIH 3T3 cells. Any change in ERα or GFP content is a result of events occurring only in the transfected cell population. Thus, in this cell population, overexpression of CSN5/Jab1 decreases ERα stability.

CSN5/Jab1 overexpression increases ERα degradation. NIH 3T3 cells were transfected as described in the legend to Fig. Fig.11 with 1 μg of pSG5-HEG0, 0.2 μg of GFP-C2, 0.8 μg of SV4-luc, and the indicated amounts of pHOOK-CSN5/Jab1. At 48 h after transfection, cells were treated for 1 h 30 with E2 (A and B) or treated for 1 h with ICI (C). ERα, CSN5/Jab1, and GFP were analyzed in the extracts by Western blotting. The amounts of proteins loaded on the gels were normalized for transfection efficiency with luciferase activities. The quantification of the number of independent experiments (n) is shown (B). Solid bars, ERα; open bars, GFP. The values are expressed as percentages of the value of the control sample corresponding to the cells that were not transfected with the CSN5/Jab1 vector. The bars indicate standard deviations for ERα and mean errors for GFP.
Curcumin, an inhibitor of the CSN-associated kinase, blocks ERα degradation and E2-dependent ERα phosphorylation.
Our experiments point towards a role for CSN5/Jab1 in ERα degradation. CSN5/Jab1 is part of the CSN complex, suggesting an involvement of this complex in the process. All the models proposed for the mechanism of action of CSN in protein degradation involve curcumin-sensitive kinase activity associated with the CSN complex. We compared the effect of curcumin to that of either protease inhibitors or other kinase inhibitors on ligand-dependent ERα degradation. In these experiments, we choose to treat cells with hormone or antihormones for 2 h, because under such conditions estradiol causes a 50% decrease in ERα levels. As previously described by others (36), the pure antagonist ICI induces a rapid degradation of ERα. In contrast, the partial antagonist OH-Tam had no effect on ERα degradation. Three different inhibitors of the proteasome (ALLN, MG132, and lactacystin β-lactone) blocked estradiol- and ICI-induced degradation of ERα. In contrast, the cysteine protease inhibitor E64 did not block the process (unpublished data). Figure Figure3A3A shows that curcumin blocked E2- or ICI-induced ERα degradation as efficiently as ALLN, suggesting that a kinase activity possibly associated with the CSN complex could be involved in the ligand-dependent degradation of ERα. To control the specificity of this process, we tested the effect of two other kinase inhibitors on ERα degradation. Neither H7, which inhibits protein kinase A (PKA), protein kinase G (PKG), and protein kinase C (PKC), nor DRB, which inhibits the phosphorylation of RNA Pol II by CDK7 and CDK9, blocked ERα degradation (Fig. (Fig.3A3A and unpublished data). We then investigated the effect of curcumin on hormone-induced ERα phosphorylation (Fig. (Fig.3B).3B). MCF7 cells pretreated or not with curcumin were treated or not with E2 in the presence of 32P. Figure Figure3B3B shows that curcumin inhibits estradiol-dependent phosphorylation of ERα. This supports the hypothesis that the effect of curcumin on ERα degradation is mediated by ERα phosphorylation.
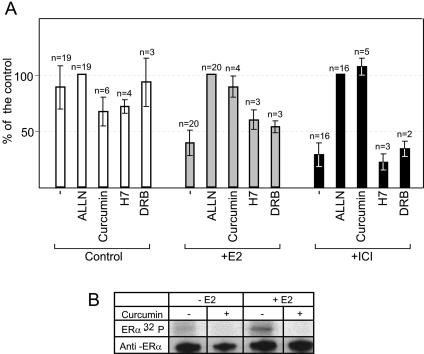
Curcumin, an inhibitor of the CSN-associated kinase, blocks ERα degradation and ERα phosphorylation. (A) MCF7 cells were pretreated for 30 min with either 100 μM ALLN (proteasome inhibitor), 20 μM H7 (inhibitor of PKA, PKC, and PKG kinases), 50 μM DRB (inhibitor of CDK7 and CDK9 kinases), or 50 μM curcumin (inhibitor of the CSN-associated kinase) and treated for 2 h with either vehicle (Control), E2, or ICI. ERα was analyzed in cellular extracts by Western blotting with anti-ERα antibodies. One microgram of total protein was loaded per lane. The figure presents the quantification of the number of independent experiments (n). The values are expressed as percentages of the value of the sample treated with ALLN (100%). (B) MCF7 cells were treated for 30 min with 50 μM curcumin (inhibitor of the CSN-associated kinase) and grown for 2 h in the presence or not of 10 nM E2 and 200 μCi/ml of [32P]phosphorus. Extracts were immunoprecipited with anti-ERα antibodies. Proteins were separated by electrophoresis on an 8% SDS-polyacrylamide gel. First line, autoradiogram of the gel; second line, control of immunoprecipitation by Western blotting with the anti-ERα antibody.
Leptomycin B, an inhibitor of CRM1-dependent nuclear export, blocks the degradation of the ERα-estradiol complex but not of the ERα-ICI complex.
CSN5/Jab1 is part of two different complexes, the eight-subunit CSN complex and a mini-CSN involved in CRM-1-dependent nuclear export (33). The effect of CSN5/Jab1 on ligand-dependent ERα degradation could be due to increased nuclear export of ERα. To investigate this possibility, MCF7 cells were treated with LMB, an inhibitor of CRM-1-dependent nuclear export, or with ALLN. Figure Figure44 shows that both LMB and ALLN blocked estradiol-induced ERα degradation. In contrast, LMB treatment had no effect on ICI-induced ERα degradation while ALLN prevented it. This suggests that ERα is degraded by the proteasome in the cytoplasm in the presence of estradiol and in the nucleus by a nuclear proteasome in the presence of ICI.
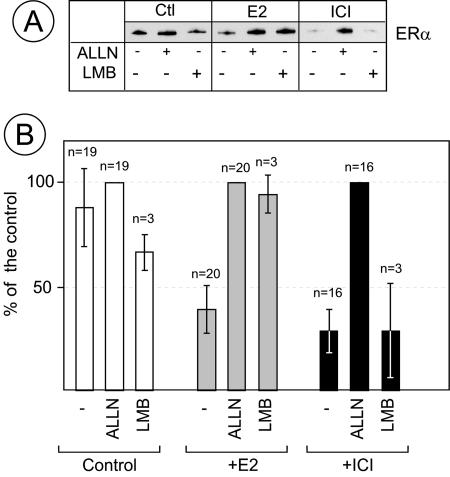
Leptomycin B, an inhibitor of CRM1-dependent nuclear export, blocks the degradation of the ERα-estradiol complex but not of the ERα-ICI complex. MCF7 cells were pretreated with 100 μM ALLN or 10 nM LMB (an inhibitor of CRM1-dependent nuclear export) for 30 min and treated with vehicle (Ctl), E2, or ICI for 2 h. (A) ERα was analyzed in cellular extracts by Western blotting with anti-ERα antibodies. One microgram of total protein was loaded per lane. (B) Quantification of three independent experiments. The values are expressed as percentages of the value of the sample treated with ALLN (100%). The bars represent standard deviations.
Curcumin stabilizes the CSN5/Jab1-ERα interaction.
The faint band of ERα coimmunoprecipitated with CSN5/Jab1 (Fig. (Fig.1A)1A) suggested that their interaction is either weak or transient. We reasoned that since a CSN-associated kinase and nuclear export are involved in ERα degradation, inhibition of either the kinase activity or of nuclear export may influence the CSN5/Jab1-ERα interaction. Figure Figure5A5A shows the results of a coimmunoprecipitation experiment in which the effects of ALLN, curcumin, and LMB on CSN5/Jab1-ERα interaction were compared in the presence of estradiol. We found that curcumin dramatically increased the amount of ERα that coimmunoprecipitated with CSN5/Jab1. That the amount of material loaded on this lane was lower than for the other samples (as revealed by the lower amount of ERα in the input fraction) increased this effect. Figure Figure5B5B shows the quantification of three independent experiments in which the amount of immunoprecipitated material was expressed as the percentage of the amount of material precipitated in the presence of curcumin. It shows, as in Fig. 1A, a slight increase in the amount of coimmunoprecipitated ERα in samples treated with E2. ALLN had no significant effect, while LMB slightly increased the amount of immunoprecipitated ERα. However, this increase was modest compared to the dramatic increase in the presence of curcumin and E2. This suggests that the inhibition by curcumin of a kinase may stabilize an interaction, direct or indirect, between CSN5/Jab1 and ERα.
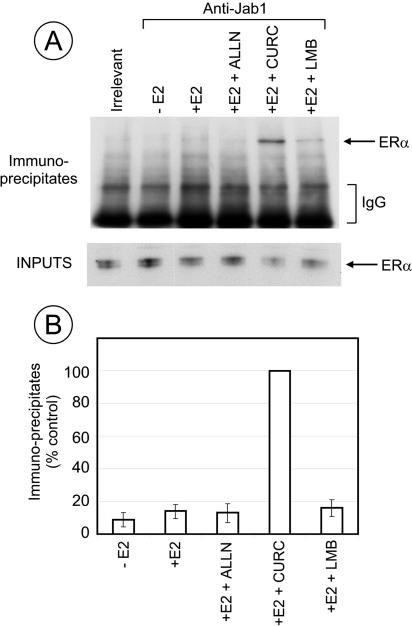
Curcumin stabilizes the interaction between CSN5/Jab1 and ERα. MCF7 cells were pretreated for 30 min with 100 μM ALLN, 50 μM curcumin, or 10 nM LMB and treated with E2 for 1 h. Extracts were immunoprecipitated with anti-c-myc (Irrelevant) or anti-CSN5/Jab1 antibodies. (A) ERα was analyzed by Western blotting in the immunoprecipitated fraction and in the inputs. The amount of material loaded for the immunoprecipitated fractions was 66-fold larger than the amount loaded for the input fractions. (B) Quantification of three independent experiments. The values are expressed as percentages of the value of the sample treated with E2 and curcumin (100%). The bars represent standard deviations.
Curcumin inhibits estrogen-induced transcription in a dose-dependent manner that parallels its effect on ERα degradation.
A link between ERα degradation by the proteasome and transcription has been proposed (17). This led us to investigate the effects of ALLN, curcumin, and LMB, which all prevent estradiol-induced ERα degradation, on estrogen-induced transcription. For these experiments, we used the MELN cell line (3) derived from MCF7 cells in which a luciferase reporter gene is under the control of an estrogen-regulated promoter. Cells were treated with either curcumin, ALLN, or LMB, and luciferase activity was measured in cells treated or untreated with estradiol for 3 h (Fig. (Fig.6).6). We selected this duration for hormone treatment because it was the shortest one with sufficient luciferase accumulation to perform accurate enzyme activity measurements. Figure Figure6A6A shows luciferase activity at different concentrations of curcumin. At a concentration of 50 μM, curcumin had a dramatic effect on hormone-induced luciferase activity. The effect on basal luciferase activity was much less pronounced. To compare the effect of curcumin on hormone-induced transcriptional activity and hormone-induced degradation, we investigated the effect of increasing concentrations of curcumin on ERα stability in the presence of hormone. Figure Figure6B6B shows the analysis of ERα contents by Western blotting, and Fig. Fig.6C6C presents a quantification of the results shown in panel B after normalization for unequal loads with an internal GAPDH control. ERα degradation was blocked for curcumin doses in the same range as those that inhibited luciferase expression, supporting the hypothesis that ERα degradation is required for transcriptional activation. Figure Figure6D6D shows the results of an experiment in which MELN cells were treated with LMB or ALLN and luciferase activity was measured before or after estradiol treatment. In contrast to the results obtained in the presence of curcumin, LMB treatment only increased luciferase expression slightly and ALLN had a minor effect on luciferase expression. Figure Figure6E6E shows that, in the same experiment, LMB and ALLN completely blocked estrogen-induced ERα degradation. These two results indicate that the inhibitory effect of curcumin on E2-induced luciferase activity is not a direct result of the absence of degradation of ERα.

Curcumin inhibits estrogen-induced transcription in a dose-dependent manner. MELN (A, D, and E) or MCF7 (B and C) cells were pretreated or not with different amounts of curcumin for 30 min and treated for 3 h with vehicle (open circles) or E2 (closed circles). Luciferase activity measurement (A); ERα and GAPDH (internal control) detection by Western blotting (B); quantification of the data presented in panel B, after normalization to GAPDH levels (C). The value of the intensity of the ERα band from the sample treated with E2 in the absence of curcumin was subtracted from all values. Results are expressed as percentages of the untreated control. (D and E) MELN cells were pretreated or not with 100 μM ALLN or 10 nM LMB for 30 min and treated for 3 h with vehicle or E2. Cell extracts were analyzed for luciferase activity (D) or for ERα and GAPDH content by Western blotting (E). Panel D represents the quantification of two independent experiments each performed in triplicate. The values are expressed as percentages of the value of the sample treated with E2 (100%). The bars represent standard deviations.
Curcumin changes the kinetics of ERα binding to the TFF1/pS2 promoter.
The inhibitory effect of curcumin on luciferase expression can result from either a specific effect on ERα or coactivators associated with ERα or a general effect on the transcription machinery. To obtain new insights into the effect of curcumin on estrogen-induced transcriptional activation, we investigated ERα and RNA Pol II binding to the estrogen-regulated TFF1/pS2 promoter by the ChIP technique. Kinetic analyzes have shown periodic binding of both ERα and Pol II on this promoter (25, 28). Figure 7A and B show the binding kinetics of ERα and Pol II on the TFF1/pS2 promoter in MCF7 cells. Promoters were synchronized by a treatment with α-amanitin as described in reference 25. After being released from the α-amanitin block, cells pretreated with curcumin were treated with E2 for different times. ERα and Pol II binding was then analyzed by ChIP. Figure 7A and B show that, as expected upon hormone treatment, both ERα and Pol II displayed periodic binding to the TFFI/pS2 promoter. In contrast, curcumin dramatically decreased the binding of ERα and abolished Pol II recruitment to the promoter. We verified that in curcumin-treated cells, immunoprecipitation of ERα and Pol II was complete (Fig. (Fig.7C).7C). Curcumin did not affect the immunoprecipitation. We conclude that inhibition of the CSN-associated kinase activity prevents the binding of ERα on the TFFI/pS2 promoter, consistent with the inhibition of transcription observed in the presence of curcumin.

Curcumin prevents hormone-induced ERα binding to the TFF1/pS2 promoter. MCF7 cells were pretreated for 1 h with 2.5 μM α-amanitin to clear ERα and the transcription machinery from the promoter. The cells were then washed and treated or not with 50 μM of curcumin for 30 min and then with 10 nM E2 for the indicated times; t = 0 corresponds to the addition of E2. The amount of TFF1/pS2 promoter DNA coimmunoprecipitated with anti-ERα (A) or with anti-Pol II (B) antibodies was quantified by real-time PCR. Results are expressed as percentages of the maximum value obtained in each experiment in the absence of curcumin. Solid lines, no curcumin treatment; dotted lines, treatment with curcumin. The results are the means of two independent experiments in which DNA quantification was performed in duplicate. The bars correspond to the mean error. (C) Immunoprecipitation of ERα and Pol II in the presence of curcumin was controlled by Western blotting with anti-ERα and anti-Pol II antibodies. A total of 1.8-fold more material was loaded in input lanes than in immunoprecipitate lanes.
DISCUSSION
CSN5/Jab1, one of the subunits of the CSN complex, has been described as a nuclear receptor coactivator (6). One essential function of CSN could be in the cellular response to the environment (8) as a mediator between kinase signaling and protein degradation (13). This complex plays a key role in sustaining the activity of SCF and other cullin-based ubiquitin ligases. These ligases are activated by neddylation of the cullin subunit. CSN could be involved in its deneddylation, a role attributed at least in part to CSN5/Jab1, which contains an NEDD8 isopeptidase active site.
Here, we show that CSN5/Jab1 coimmunoprecipitates with ERα, indicating that they interact directly or indirectly and that the increase in hormone-induced transcription resulting from increased amounts of CSN5/Jab1 is accompanied by an increase in hormone-induced ERα degradation. These results, taken together with the role of CSN on cullin-based ubiquitin ligases, support the recent observation that disruption of the NEDD8 pathway impairs ligand-ERα degradation (9). In addition, we show that the amount of coimmunoprecipitated ERα and CSN5/Jab1 is increased in the presence of curcumin, suggesting a stabilization of the interaction by this inhibitor of a kinase activity associated with CSN (4). Curcumin also blocks ligand-dependent ERα phosphorylation and ligand-dependent ERα degradation, suggesting that, as described for p53, CSN-dependent phosphorylation of ERα could target it for degradation. The effect of kinases involved in cell signaling was recently investigated to address the question of the link between ERα phosphorylation and degradation. An earlier study (19) described an enhanced degradation of ERα by PKC, regardless of its ligand binding status. In contrast, PKA, mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase impeded the degradation of ERα, whether ligand free or complexed to estradiol. No correlation between the capacity of kinase inhibitors to affect ERα stability and their effect on basal or estradiol-induced transcription was found (19). However, there is as yet no clear evidence that ERα phosphorylation is involved in regulating its ligand-induced degradation (16).
ERα is phosphorylated on multiple amino acids, mainly Ser residues located within its N-terminal activation domain. Ser-118, the site predominantly phosphorylated in vivo, is phosphorylated in response to the activation of the MAPK pathway and to estradiol. The identity of the kinases responsible for Ser-118 phosphorylation in response to estradiol binding is controversial. One possible candidate is the TFIIH cyclin-dependent kinase (7). Ser-167 is also phosphorylated in response to the activation of the MAPK pathway. In addition, Ser-167 is phosphorylated in vitro by CKII and AKT (2). To address a possible role of ERα phosphorylation in its degradation and/or cycling on estrogen-responsive promoters, we investigated the effect of kinase inhibitors on ligand-induced ERα degradation. H7, which inhibits a wide spectrum of kinases (PKA, PKG, and PKC), had no effect on ERα stability. We also investigated the effect of DRB, an inhibitor of CDK7, which blocks transcription by RNA Pol II, and again we found no effect on ERα stability. These results, obtained for short treatment times with DRB, agreed with data obtained with pituitary cells (1). However, for long treatment times with DRB in the presence of trichostatin A, ERα degradation was prevented (25; unpublished data). This suggests that the effect of DRB on ERα degradation is indirect and it demonstrates that transcription is not required for ligand-dependent ERα degradation.
Recently, it was demonstrated that to activate ERα-dependent transcription, SRC3/AIB1, a coactivator of ERα, must be phosphorylated (39). In addition, it was shown that in presence of estradiol, SRC3/AIB1 is necessary for ERα binding on the TFF1/pS2 promoter and for ERα degradation by the proteasome (29). SRC3/AIB1 itself is degraded in a estradiol-dependent fashion. The effect of curcumin on ERα degradation may result from phosphorylation inhibition of ERα and/or of a coactivator such as SRC3/AIB1. In the presence of ICI, SRC3/AIB1 does not interact with ERα whereas curcumin blocks the ICI-induced degradation of ERα. Three kinases, inositol 1,3,4-triphosphate 5/6 kinase (31), CKII, and protein kinase D (35), have been described as being associated with CSN. Of these, CKII is a good candidate for ERα phosphorylation, since as mentioned above CKII was found to be capable of phosphorylating ERα on Ser-167 (2). These three kinases have not been proposed as candidate kinases for SRC3/AIB1 phosphorylation. Taken together with the observation that curcumin inhibits E2-dependent phosphorylation, these data suggest that the target of the curcumin-sensitive kinase would be ERα itself rather than a coactivator.
Another role for CSN is to facilitate the nuclear export of proteins that do not have a nuclear export signal, allowing degradation by the proteasome. Inhibition of nuclear export by LMB on ERα degradation resulted in a block of the estrogen-dependent degradation of ERα, indicating that estradiol-induced ERα-degradation by the proteasome occurs in the cytoplasm. In contrast, ICI-induced ERα degradation was not affected by LMB, indicating that in this case degradation takes place in the nucleus. This conclusion is in agreement with our previous finding that ICI induces the sequestration of ERα in a salt-resistant nuclear compartment in which it is rapidly degraded (12). Our results are also compatible with data from experiments with fluorescence recovery after photobleaching, which show a decreased mobility of ERα in the nuclei in the presence of ICI (30). ERα complexed to ICI is degraded by a different pathway than ERα complexed to E2. It is possible that in the presence of ICI, ERα adopts a conformation not recognized by the cell. In this case, the antagonist activity of ICI should result from the rapid degradation of ERα.
An important question is whether ligand-induced ERα degradation and transcription activation are interlinked processes. Our results on the effect of DRB on ligand-induced ERα degradation demonstrate that transcription by itself is not required for ERα degradation. To obtain further insights in this process, we compared, for the same time of treatment, the effect of three types of compounds that inhibit ERα degradation on ERα stability and transcription activation. ALLN, curcumin, and LMB completely prevented ERα degradation, but they had very different effects on transcription. Curcumin totally blocked the expression of a target reporter gene at a dose similar to that inhibiting ERα degradation. ALLN had a minor effect on expression of a reporter gene while LMB increased it slightly. These results differed from previous reports describing a decrease in hormone-induced transcription by treatment of the cells with proteasome inhibitors (17, 25). The differences in the duration of treatment with the proteasome inhibitor cannot account for this discrepancy. We found that increasing the length of treatment with ALLN increased the expression of the reporter gene (unpublished data). Our results agree with a very recent report demonstrating that proteasome inhibition increases hormone-induced activation (11). The differences in proteasome inhibitors on hormone-induced transcription probably result from differences in the promoters, as has been suggested elsewhere (11, 17). All together, these results demonstrate that there is no direct relation between hormone-induced ERα degradation and transcription. Inhibition of ERα nuclear export that results in ERα accumulation in the nucleus is accompanied by increased transcription. Curcumin had a very different effect, blocking both ERα degradation and transcription. ChIP experiments clearly show that the lack of transcription activation is due to a dramatic decrease in ERα binding to its target on an estradiol-regulated promoter. A similar decrease in ERα binding to the TFF1/pS2 promoter was observed when SRC3/AIB1 expression was abolished with a small interfering RNA (29). One possible mechanism of coactivation by CSN5/Jab1 could be a stabilization of the interaction between ERα and SRC3/AIB1, as was shown for another nuclear receptor and coactivator, the progesterone receptor and SRC1 (6).
We propose a model (Fig. (Fig.8)8) in which the first event after hormone addition would be ERα and/or coactivator phosphorylation by a curcumin-sensitive kinase associated with the CSN complex (Fig. (Fig.8A).8A). This would allow ERα to interact productively with the TFF1/pS2 promoter (Fig. (Fig.8B).8B). Once the transcription complex is assembled, ERα would be polyubiquitinated (Fig. (Fig.8C)8C) and released from the DNA (D). This complex containing polyubiquitinated ERα would then be exported to the cytoplasm (Fig. (Fig.8E)8E) to be degraded by the proteasome (F). CSN5/Jab1 could also be involved at that step, as has been described for p27Kip1, permitting the export of ERα, which does not have an export signal sequence. Alternatively, ERα could be deubiquitinated and recycled to activate transcription (Fig. (Fig.8A).8A). This hypothesis is supported by the results obtained with LMB, which prevents ERα degradation and increases transcription at the same time. In the presence of ICI, ERα would also be phosphorylated by a kinase (Fig. (Fig.8G),8G), but this does not result in DNA binding. It is targeted to a nuclear subcompartment and is rapidly degraded by a nuclear proteasome (Fig. (Fig.8H).8H). In our experiments, curcumin blocks ERα degradation in the presence of E2 and ICI, impairing its polyubiquitination. According to our model, ERα degradation should not be required to sustain transcription, which as already discussed is indeed slightly enhanced by increased amounts of nuclear ERα. Degradation would be the consequence of one of the dynamic events that lead to transcription initiation rather than of transcription per se.
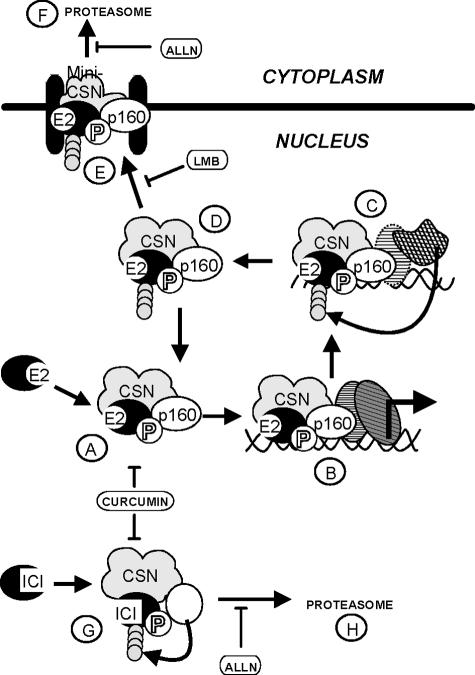
Model. (A) In the nucleus, a kinase associated with CSN would phosphorylate ERα complexed to estradiol and/or a coactivator such as SRC3/AIB1, a member of the p160 family. (B) This would result in the binding of the ERα-SRC3/AIB1 complex to its cognate DNA target, recruiting the preinitiation complex. Then, polyubiquitination of ERα could take place (C), inducing the release of the ERα-SRC3/AIB1 complex from DNA (D). The polyubiquitinated REα should then either be exported to the cytoplasm by the mini-CSN (E) and degraded by the proteasome (F) or recycled after deubiquitination (A). In the presence of ICI, a kinase associated with CSN could phosphorylate ERα or another cofactor (G). ERα would then be ubiquitinated and degraded by a proteasome in the nucleus (H).
Acknowledgments
This work was partially supported by Ligue contre le Cancer (Tarn et Garonne), Association pour la Recherche contre le Cancer, ARECA, and Région Midi-Pyrénées. M.C. was supported by Association pour la Recherche contre le Cancer.
We thank P. Balaguer, E. Bianchi, P. Chambon, H. Loosfelt, and H. Gronemeyer for the gift of constructs and expression vectors used in this study. We are grateful to E. Käs, K. Bystricky, and M. Grigoriev for their constructive suggestions and their critical reading of the manuscript. We thank D. Villa for electronic illustrations.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.25.11.4349-4358.2005
Read article for free, from open access legal sources, via Unpaywall:
https://mcb.asm.org/content/25/11/4349.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.25.11.4349-4358.2005
Article citations
Regulatory mechanisms and therapeutic potential of JAB1 in neurological development and disorders.
Mol Med, 29(1):80, 26 Jun 2023
Cited by: 1 article | PMID: 37365502 | PMCID: PMC10291812
Review Free full text in Europe PMC
The crosstalk between ubiquitination and endocrine therapy.
J Mol Med (Berl), 101(5):461-486, 24 Mar 2023
Cited by: 2 articles | PMID: 36961537
Review
Pro-Apoptotic and Anti-Invasive Properties Underscore the Tumor-Suppressing Impact of Myoglobin on a Subset of Human Breast Cancer Cells.
Int J Mol Sci, 23(19):11483, 29 Sep 2022
Cited by: 2 articles | PMID: 36232784 | PMCID: PMC9570501
Ligand dependent gene regulation by transient ERα clustered enhancers.
PLoS Genet, 16(1):e1008516, 06 Jan 2020
Cited by: 17 articles | PMID: 31905229 | PMCID: PMC6975561
Increased Jab1/COPS5 is associated with therapeutic response and adverse outcome in lung cancer and breast cancer patients.
Oncotarget, 8(57):97504-97515, 27 Oct 2017
Cited by: 6 articles | PMID: 29228627 | PMCID: PMC5722579
Go to all (55) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex.
J Biol Chem, 277(3):2302-2310, 09 Nov 2001
Cited by: 190 articles | PMID: 11704659
Various phosphorylation pathways, depending on agonist and antagonist binding to endogenous estrogen receptor alpha (ERalpha), differentially affect ERalpha extractability, proteasome-mediated stability, and transcriptional activity in human breast cancer cells.
Mol Endocrinol, 17(10):2013-2027, 10 Jul 2003
Cited by: 83 articles | PMID: 12855746
COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system.
EMBO J, 20(7):1630-1639, 01 Apr 2001
Cited by: 245 articles | PMID: 11285227 | PMCID: PMC145508
JAB1/CSN5 and the COP9 signalosome. A complex situation.
EMBO Rep, 2(2):96-101, 01 Feb 2001
Cited by: 113 articles | PMID: 11258719 | PMCID: PMC1083824
Review Free full text in Europe PMC





