Abstract
Free full text

Characterization of the Filamentous Hemagglutinin-Like Protein FhaS in Bordetella bronchiseptica
Abstract
Filamentous hemagglutinin (FHA) is a large (>200 kDa), rod-shaped protein expressed by bordetellae that is both surface-associated and secreted. FHA mediates bacterial adherence to epithelial cells and macrophages in vitro and is absolutely required for tracheal colonization in vivo. The recently sequenced Bordetella bronchiseptica genome revealed the presence of a gene, fhaS, that is nearly identical to fhaB, the FHA structural gene. We show that although fhaS expression requires the BvgAS virulence control system, it is maximal only under a subset of conditions in which BvgAS is active, suggesting an additional level of regulation. We also show that, like FHA, FhaS undergoes a C-terminal proteolytic processing event and is both surface-associated and secreted and that export across the outer membrane requires the channel-forming protein FhaC. Unlike FHA, however, FhaS was unable to mediate adherence of B. bronchiseptica to epithelial cell lines in vitro and was not required for respiratory tract colonization in vivo. In a coinfection experiment, a ΔfhaS strain was out-competed by wild-type B. bronchiseptica, indicating that fhaS is expressed in vivo and that FhaS contributes to bacterial fitness in a manner revealed when the mutant must compete with wild-type bacteria. These data suggest that FHA and FhaS perform distinct functions during the Bordetella infectious cycle. A survey of various Bordetella strains revealed two distinct fhaS alleles that segregate according to pathogen host range and that B. parapertussishu most likely acquired its fhaS allele from B. pertussis horizontally, suggesting fhaS may contribute to host-species specificity.
Members of the gram-negative genus Bordetella cause respiratory tract infections. Bordetella pertussis and Bordetella parapertussishu strains infect humans, causing the disease known as whooping cough or pertussis, while Bordetella bronchiseptica infects a broad range of mammals, causing either asymptomatic infections or acute diseases such as kennel cough in dogs and atrophic rhinitis in pigs (9). Several Bordetella virulence factors, including those required for adherence to host tissues, induction and/or modulation of the immune response, and cytotoxicity, have been identified (9, 40). With the exception of tracheal cytotoxin, all are positively regulated by the two-component sensory transduction system BvgAS (39, 57, 62, 69). Under the appropriate growth conditions (37°C and relative lack of MgSO4 or nicotinic acid), BvgAS activates transcription of virulence genes (collectively called vag loci) and represses transcription of genes that are not required for or are detrimental to virulence (vrg loci) (2, 8, 10, 39). Growth at low temperature or in the presence of high concentrations of MgSO4 or nicotinic acid inactivates BvgAS. Under these conditions, virulence genes are not expressed and Bvg-repressed phenotypes are expressed (9). When bordetellae are grown in medium containing relatively low concentrations of MgSO4 or nicotinic acid, BvgAS controls the expression of a phase (the Bvgi phase), characterized by expression of some virulence factors (e.g., adhesins) but not others (e.g., toxins) and lack of expression of vrg loci (11, 39). Studies with both B. bronchiseptica and B. pertussis have shown that Bvg-mediated control of gene expression is critical for the development of respiratory infection (2, 8, 9, 39).
One of the best-characterized Bordetella virulence factors is filamentous hemagglutinin (FHA), a long, rod-shaped, highly immunogenic surface-associated and secreted protein that is a primary component of all acellular pertussis vaccines ( 53). FHA is prototypical of a class of proteins exported by the two-partner secretion (TPS) pathway (27, 36). It is synthesized as a >350-kDa precursor, FhaB. Following transport across the cytoplasmic membrane and loss of its unusually long signal sequence, FhaB is translocated across the outer membrane by the channel-forming protein FhaC (22, 31, 33, 35, 51). At some point during this process, FhaB undergoes a C-terminal proteolytic cleavage event, resulting in the formation of the mature ~250-kDa FHA protein. A serine protease of the autotransporter family, SphB1, has been implicated in the maturation process; however, its precise role has not been established (13, 14). The mechanism by which some FHA molecules remain surface localized while others are released is unknown.
FHA function has been investigated extensively in vitro. It mediates adherence to epithelial cells (5, 12) and has been shown to bind directly to ciliary membrane glycosphingolipids (59). FHA may modulate host immunity by influencing the expression of pro-inflammatory cytokines in epithelial cells and macrophages (1, 29). An Arg-Gly-Asp (RGD) triplet, located in the middle of the mature FHA protein, mediates interactions with leukocyte response integrin/integrin-associated protein (LRI/IAP) on monocytes/macrophages, resulting in upregulation of CR3 binding activity (28, 68). The RGD motif appears to also interact with very late antigen 5 (VLA-5) on epithelial cells, stimulating the upregulation of intercellular adhesion molecule 1 (ICAM-1) via an NF-κB signaling pathway (30). In vivo experiments with B. bronchiseptica and natural-host animal models have shown that FHA is absolutely required for tracheal colonization (12).
Determination of the complete nucleotide sequences of the genomes of B. pertussis Tohama 1, B. parapertussishu 12822, and B. bronchiseptica RB50 revealed two additional loci, fhaL and fhaS, with the potential to encode FHA-like proteins (48). The fhaL genes from all three subspecies are nearly identical to each other and potentially encode proteins that are approximately 19% similar to their cognate FhaB proteins. The fhaS genes of B. pertussis and B. parapertussishu are also nearly identical to each other and potentially encode proteins with 38% similarity to their cognate FhaB proteins. The fhaS gene of B. bronchiseptica, by contrast, is similar to the fhaS genes of B. pertussis and B. parapertussishu only at its 5′ and 3′ ends. Its central region, spanning 8 kb and constituting a majority of the open reading frame, is nearly identical (91% identity at the nucleotide level) to B. bronchiseptica fhaB. In this study, we characterized the B. bronchiseptica fhaS gene and its product. Our results show that while the FhaS protein appears to be processed and localized similarly to FhaB, its expression pattern and function(s) differ substantially.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
All strains and plasmids used in this study are listed in Table Table1.1. Bordetella strains were grown in Stainer-Scholte (SS) or Luria-Bertani (LB) broth as indicated or on Bordet-Gengou agar (Becton Dickinson Microbiology Systems) supplemented with 7.5% defribrinated sheep blood (Mission Laboratories). Escherichia coli strains were grown in LB broth or on LB agar. Relevant antibiotics were used at the following concentrations: streptomycin, 20 μg/ml; ampicillin, 100 μg/ml; gentamicin, 20 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype/description | Source or reference |
|---|---|---|
| Strains | ||
    B. bronchiseptica B. bronchiseptica | ||
        RB50 RB50 | Wild type | 10 |
        RB50G RB50G | Gentamycin-resistant derivative of RB50 | 6 |
        RBX9 RBX9 | ΔfhaB | 12 |
        RBX11 RBX11 | ΔfhaS | This study |
        RBX20 RBX20 | ΔfhaB ΔfhaS | This study |
        RB50::pEG111(fhaB) RB50::pEG111(fhaB) | RB50 containing a chromosomal fhaB::lacZ fusion | This study |
        RB50::pEG111(fhaS) RB50::pEG111(fhaS) | RB50 containing a chromosomal fhaS::lacZ fusion | This study |
        RB50::pEG110 RB50::pEG110 | RB50 containing a chromosomal cyaA::lacZ fusion | This study |
        RB50::pTEN34 RB50::pTEN34 | RB50 containing a chromosomal bipA::lacZ fusion | 20 |
        RB53::pEG111 (fhaB) RB53::pEG111 (fhaB) | Bvg-constitutive containing a chromosomal fhaB::lacZ fusion | This study |
        RB53::pEG111(fhaS) RB53::pEG111(fhaS) | Bvg-constitutive containing a chromosomal fhaS::lacZ fusion | This study |
        RB53::pEG110 RB53::pEG110 | Bvg-constitutive containing a chromosomal cyaA::lacZ fusion | This study |
        RB53::pTEN34 RB53::pTEN34 | Bvg-constitutive containing a chromosomal bipA::lacZ fusion | 20 |
        RB54::pEG111(fhaB) RB54::pEG111(fhaB) | Bvg− locked containing a chromosomal fhaB::lacZ fusion | This study |
        RB54::pEG111(fhaS) RB54::pEG111(fhaS) | Bvg− locked containing a chromosomal fhaS::lacZ fusion | This study |
        RB54::pEG110 RB54::pEG110 | Bvg− locked containing a chromosomal cyaA::lacZ fusion | This study |
        RB54::pTEN34 RB54::pTEN34 | Bvg− locked containing a chromosomal bipA::lacZ fusion | 20 |
        RB53i::pEG111(fhaS) RB53i::pEG111(fhaS) | Bvgi locked containing a chromosomal fhaS::lacZ fusion | This study |
        RB53i::pTEN34 RB53i::pTEN34 | Bvgi locked containing a chromosomal bipA::lacZ fusion | 20 |
        RB50::pSJ22 RB50::pSJ22 | RB50 containing plasmid insertion (null mutation) in fhaC | This study |
        RBX9::pSJ22 RBX9::pSJ22 | ΔfhaB containing plasmid insertion (null mutation) in fhaC | This study |
        RBX11::pSJ22 RBX11::pSJ22 | ΔfhaS containing plasmid insertion (null mutation) in fhaC | This study |
        548 548 | Pig isolate | 64 |
        590 590 | Dog isolate | 64 |
        595 595 | Dog isolate | 64 |
        625 625 | Rat isolate | 64 |
        675 675 | Human isolate | 64 |
        676 676 | Pig isolate | 64 |
        704 704 | Rabbit isolate | 65 |
        Cb2 Cb2 | Dog isolate | 20 |
        JC100 JC100 | Human isolate | 20 |
    B. pertussis B. pertussis | ||
        Tohama l Tohama l | Wild type | 37 |
    B. parapertussishu B. parapertussishu | ||
        12822 12822 | Wild type | 26 |
    E. coli E. coli | ||
        DH5α DH5α | Molecular cloning strain | BRL; Gaithersburg, MD |
        SM10λpir SM10λpir | Conjugation strain | 47 |
| Plasmids | ||
    pEG110 pEG110 | pBR322-based Bordetella suicide vector containing 1.8 kb cyaA gene fragment 5′ to promoterless lacZ gene; Ampr, Gmr | This study |
    pEG111 pEG111 | pBR322-based Bordetella suicide vector containing 1.7 kb fhaB gene fragment 5′ to promoterless lacZ gene; Ampr, Gmr | This study |
    pSJ22 pSJ22 | pBR322-based Bordetella suicide vector containing 0.3 kb kb fhaC gene fragment 5′ to promoterless lacZ gene; Ampr, Gmr | This study |
Plasmid pEG110 was constructed by ligating a 1.9-kb EcoRI-XmaI fragment corresponding to bp 2585 to 4465 of the B. bronchiseptica cyaA gene into EcoRI-XmaI-digested pEGZ, a pBR322-based suicide vector containing a promoterless lacZ gene (39). In a similar manner, a 1.6-kb XhoI-BamHI fragment corresponding to bp 1316 to 2927 of the B. bronchiseptica fhaB gene was ligated into pEGZ to create plasmid pEG111. Plasmid pSJ22 was constructed by amplifying a 323-bp product corresponding to bp 7 to 330 from the B. bronchiseptica fhaC gene by use of primers fhaC.fwd (5′-GATTGAATTCGACGCAACGAACCGTTTCCGG-3′) and fhaC.rev (5′-GATTGGATCCAACGGCGCGGGGTCGAAC-3′) (Integrated DNA Technologies, Coralville, IA). The primers were designed such that EcoRI and BamHI sites would be incorporated at the ends of the PCR product. The PCR product was digested with EcoRI and BamHI, purified, and ligated to EcoRI-BamHI-digested pEGZ.
DNA methods.
Isolation of plasmid DNA and genomic DNA, restriction endonuclease digestions, agarose gel electrophoresis, and DNA ligations were performed using standard methods (52). Restriction enzymes, T4 DNA ligase, Taq polymerase, and high-fidelity Phusion polymerase were purchased from Promega Corp. (Madison, WI), New England Biolabs (Beverly, MA), or MJ Research (Waltham, MA) and were used according to the manufacturers' instructions.
Construction of bacterial strains.
To create an in-frame (nonpolar) deletion in the B. bronchiseptica fhaS gene, an allelic exchange system employing the sacB gene was used as previously described (2, 39). Initially, an allelic exchange plasmid, pEG7SΔfhaS, was constructed as follows. Primers EBPSFHA3x (5′-AATCTAGAACCCGCAACCAGTCCGAGAAGTACG-3′) and EBPSFHA4x (5′-AAGGTACCCGCGTTCATGTTGTCTCCTGATGTCG-3′) were used to amplify a 672-bp fragment corresponding to the 5′ region and containing the first three codons of the fhaS coding sequence. Primers were designed such that XbaI and KpnI sites are introduced at the 5′ and 3′ ends of the PCR product, respectively. Primers EBPSFHA1x (5′-AAGGTACCCTGCATCGCTAGCGCGAGG-3′) and EBPSFHA2 (5′-AAGCATGCCGCAGGACATCGACATAGGG-3′) were used to amplify a 679-bp fragment including the last three codons of the fhaS coding sequence and downstream sequences. Primers were designed such that KpnI and SphI sites were introduced at the 5′ and 3′ ends of the PCR product, respectively. These fragments were purified, ligated into TA cloning vectors (Invitrogen), digested with the appropriate restriction enzymes, and cloned into BamHI-XbaI-digested pEG7S, a derivative of pEG7 (2), resulting in an in-frame deletion of all but the first three and last three codons of the fhaS gene. Sequence analysis of the cloned DNA (Laragen, Los Angeles, CA) was performed to confirm the fidelity of B. bronchiseptica sequences. The resulting plasmid (pEG7SΔfhaS) was transformed into E. coli SM10 λpir and then mobilized into the RB50 chromosome by conjugation. Cointegrants were selected on medium containing streptomycin (B. bronchiseptica strains are naturally streptomycin resistant) and gentamicin. Recombinants that had excised the plasmid from the chromosome were selected on media containing 10% sucrose (the sacB gene encodes for levansucrase, which is toxic to cells grown in the presence of high concentrations of sucrose) (19), and PCR was used to screen for mutants that had undergone a recombination event resulting in a chromosomal deletion of the fhaS gene. A ΔfhaB ΔfhaS double mutant was constructed by the same method by mobilizing pEG7SΔfhaS into RBX9.
To construct strains that harbor chromosomal lacZ reporter plasmids or fhaC null mutations, pEG110, pEG111, or pSJ22 was transformed into SM10 λpir and then conjugated into the appropriate B. bronchiseptica strain. Cointegrants were selected on media containing streptomycin and gentamicin. PCR was used to verify integration of the plasmid at the proper chromosomal locus. For strains containing pEG111, PCR was used to distinguish between integration events at either the fhaB or fhaS locus, since the 1.7-kb fhaB fragment in pEG111 is nearly identical to sequences present in the fhaS gene.
β-Galactosidase assays.
Stationary phase cultures from strains harboring lacZ transcriptional fusions were used to measure β-galactosidase activity. Bacteria were grown overnight in either LB broth or SS media and permeabilized by the addition of sodium dodecyl sulfate (SDS) and CHCl3, and β-galactosidase activity was determined essentially as described previously (39), except that measurements were taken using a Victor3 1420 microplate reader (PerkinElmer Life Sciences, Boston, MA).
Bacterial cell fractionation and immunoblot analysis.
Stationary-phase cultures were used for all bacterial cell preparations. Whole-cell lysates were prepared as described previously (39). Outer membrane preparations were isolated from bacterial cell sonicates as the Triton X-100 insoluble fraction. Briefly, clarified cell sonicates were spun at 16,000 × g, the pellet was resuspended in 2% Triton X-100 and spun again, and the pellet was resuspended in an appropriate volume of 10 mM HEPES (pH 7.5). Secreted proteins were concentrated by addition of 10% TCA to the culture supernatant and then recovered by centrifugation. Proteins were resuspended in an equal mixture of 1× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (52) and saturated Tris solution (to neutralize excess acid).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (38) using denaturing 3 to 8% linear gradient gels (acrylamide-bisacrylamide, 29:1). Proteins from whole cells (derived from approximately 8.0 × 108 CFU), outer membranes (derived from 5.0 × 109 CFU), or supernatant fractions (derived from 1.5 × 1010 CFU) were run on SDS-PAGE, transferred to nitrocellulose membranes (Schleicher and Schuell BioScience, Dassel, Germany), and incubated with a 1:10,000 dilution of chicken polyclonal FHA antibody (raised against amino acids [aa] 1048 to 1385 of B. bronchiseptica FHA, which are identical [except for one amino acid difference] with the corresponding sequence in FhaS). Antigen-antibody complexes were detected by incubating with a 1:20,000 dilution of goat anti-chicken IRdye 800-conjugated secondary antibody (Rockland, Gilbertsville, PA) and visualized using the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE).
In vitro adherence assay.
Adherence to rat lung epithelial (L2) cells was performed as described previously (12), with the exception that LB broth was used as the bacterial growth medium as well as the diluent for addition of bacteria to cell monolayers.
In vitro competition experiment.
Wild-type (RB50G) and ΔfhaS strains were grown overnight in either SS or LB and then diluted by coinoculating equivalent numbers of each (based on measurements of optical density at 600 nm) into fresh media to an optical density at 600 nm of approximately 0.1. Cultures were then incubated on a roller at 37°C for 8 h (approximately five cell divisions). The relative proportion of wild-type to ΔfhaS bacteria was determined by plating aliquots onto media with and without gentamicin.
In vivo competition experiment.
Three- to 4-week-old female Wistar rats (Charles River Laboratories, Wilmington, MA) were coinoculated intranasally with approximately 250 CFU of the wild-type and 250 CFU of the ΔfhaS strain (from cultures grown overnight in SS media) essentially as previously described (41). Animals were sacrificed 7, 14, and 28 days postinoculation, and colonization levels in the nasal cavity and trachea were determined as described previously (41).
Phylogenetic analysis of fhaS alleles.
Total genomic DNA was prepared from various B. bronchiseptica strains, B. pertussis strain Tohama I, and B. parapertussishu strain 12822 and used as templates for PCR. Primers PfhaSOUT1 (5′-TGGCGGCCCGCTATATCGA-3′) and CD64 (5′-GGCTGAGCGGGCCGCGCTGG-3′) were used to amplify B. bronchiseptica RB50-specific fhaS sequences, and primers PfhaSOUT1 and fhaSBpBpp1086.rev (5′-GCCGGCATGCAGCGACAAGGCGTC-3′) were used to amplify fhaS sequences specific to B. pertussis and B. parapertussishu. Cycling conditions were as follows: 94°C for 2 min followed by 30 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 2 min, with a final extension step of 72°C for 10 min.
Statistical analyses.
For animal experiments, significant differences between the ratio of mutant to wild-type CFU present in the inocula and the ratio of mutant to wild-type CFU recovered from host tissues were examined using a Student's t test. A P value of <0.05 was considered statistically significant.
RESULTS
The fhaS gene of B. bronchiseptica, but not those of B. pertussis or B. parapertussishu, codes for a protein that is nearly identical to FHA.
The annotated genome sequences of B. pertussis, B. parapertussishu and B. bronchiseptica indicate the presence of a locus, named fhaS, in all three subspecies that is predicted to encode a protein with similarity to the well-characterized Bordetella virulence factor FHA (48). Comparison of the predicted FhaS proteins from the three subspecies, however, revealed that the amino acid sequence predicted for FhaS from B. bronchiseptica (FhaSBb) differs substantially from those predicted for FhaS from B. pertussis and B. parapertussishu (FhaSBp and FhaSBpp, respectively). The predicted amino acid sequences of FhaSBp and FhaSBpp are ~90% identical over their entire length (Fig. (Fig.1A).1A). By contrast, FhaSBb is predicted to be significantly larger (3206 aa compared to 2601 and 2554 for FhaSBp and FhaSBpp, respectively) and its similarity with FhaSBp and FhaSBpp is confined primarily to the N and C termini: the sequences of the predicted N-terminal 148 aa and C-terminal 933 aa of FhaSBb and FhaSBpp are 97% and 98% identical, respectively. The central region of FhaSBb, however, is only 36% identical to that of FhaSBpp. Comparison of FhaSBb with FhaBBb, however, revealed striking similarity (Fig. (Fig.1B).1B). From aa 149 to aa 2658, which constitutes nearly 80% of the predicted protein, FhaSBb is nearly identical to FhaBBb: exclusive of a gap of eight 19-aa repeat segments in FhaSBb, these sequences are 95% identical. This region of near identity includes several functional domains previously identified in FHA, including the heparin binding domain (HBD), carbohydrate recognition domain (CRD), RGD, and C-terminal maturation site (14, 44, 49, 50, 54). Conversely, the extreme N and C termini of FhaSBb and FhaBBb exhibit little similarity (Fig. (Fig.1B).1B). Overall, therefore, the predicted amino acid sequence of FhaSBb is much more closely related to that of FhaBBb than it is to those of FhaSBp and FhaSBpp.

Similarity of the predicted B. bronchiseptica FhaS protein (FhaSBb) to related Bordetella proteins. (A) Comparison of FhaSBb to the predicted FhaS protein of B. parapertussis (FhaSBpp) and comparison of FhaSBpp with the predicted FhaS protein of B. pertussis (FhaSBp). (B) Comparison of FhaSBb to full-length B. bronchiseptica FHA (FhaBBb). The entire FhaSBb protein and regions in other proteins similar to FhaSBb are depicted in gray. White and stippled segments denote regions that are not significantly similar to FhaSBb. Thin dashed lines between two proteins denote the boundaries of regions of similarity, and the percentage amino acid identity within that region is shown. The amino acid number at the junctions of the various regions and at the end of each protein is indicated. Thick vertical dashed lines represent the predicted N and C termini of mature proteins. A horizontal dashed line in FhaSBb depicts the absence of eight 19-amino acid repeat segments that are present in FhaBBb. HBD, heparin binding domain; RGD, Arg-Gly-Asp integrin binding domain; CRD, carbohydrate recognition domain.
Expression of fhaS.
Expression of all Bordetella virulence genes characterized so far is positively regulated by the BvgAS phosphorelay and is maximal in cells grown at 37°C in SS broth (Bvg+ phase conditions) and minimal in cells grown at room temperature in SS broth or at 37°C in SS broth supplemented with ≥20 mM MgSO4 or nicotinic acid (Bvg− phase or “modulating” conditions). Expression of the Bvgi phase gene bipA is both positively and negatively regulated by BvgAS; it is maximal in cells grown at intermediate temperatures in SS broth or at 37°C in SS broth containing “semimodulating” concentrations of MgSO4 or nicotinic acid (Bvgi phase conditions) (71). To determine the expression pattern of fhaS in B. bronchiseptica, we constructed a strain containing a chromosomal fhaS-lacZ fusion and measured β-galactosidase activity in cultures grown at 37°C ± 50 mM MgSO4. fhaS-lacZ expression was approximately 2.5-fold greater in cells grown in SS broth than in cells grown in SS broth + MgSO4. This expression pattern was similar in profile, but not magnitude, to Bvg-activated genes such as fhaB and cyaA (Fig. (Fig.2).2). To determine whether fhaS expression is controlled by BvgAS, we introduced the fhaS-lacZ fusion into Bvg+ and Bvg- phase-locked derivatives of RB50. Expression of fhaS-lacZ in the Bvg+ phase-locked strain grown in SS broth with or without added MgSO4 was similar to fhaS-lacZ expression in the wild-type strain grown in SS broth. fhaS-lacZ expression was the same in the Bvg- phase-locked strain grown in SS ± MgSO4 as in the wild-type strain grown in SS + MgSO4 (Fig. (Fig.2).2). These results demonstrate that fhaS is positively regulated by BvgAS.

Comparison of fhaS expression patterns with those of known Bvg-regulated genes. An fhaS::lacZ, fhaB::lacZ, bipA::lacZ, or cyaA::lacZ transcriptional fusion was introduced into the chromosome of wild type (WT), Bvg+ phase locked (Bvgc), Bvgi phase locked (Bvgi) or Bvg− phase locked (Bvg−) B. bronchiseptica strains as indicated. Bacteria were grown overnight in Stainer-Scholte (SS) or L broth (LB) in the presence (+) or absence (-) of 50 mM MgSO4. β-galactosidase activity is reported in Miller units, and data represent the average of three independent experiments done in duplicate. Error bars indicate one standard deviation from the mean.
We discovered serendipitously that fhaS expression is approximately fivefold greater in cells grown in LB broth than in cells grown in SS broth (Fig. (Fig.2).2). Since we had previously observed that wild-type B. bronchiseptica showed differences in colony morphology and motility when grown on LB agar, consistent with modulation to the Bvgi or Bvg- phase (not shown), we hypothesized that LB might be semimodulating. Consistent with this hypothesis, expression of the gene encoding adenylate cyclase, cyaA, (which is minimal under Bvgi phase conditions) (11), was decreased in LB-grown cultures compared with SS-grown cultures, and expression of bipA (which is maximal under Bvgi phase conditions) (18) was slightly greater in LB-grown cultures than in SS-grown cultures (Fig. (Fig.2).2). These results suggested that fhaS, like bipA, might be expressed maximally under Bvgi phase conditions. However, fhaS-lacZ expression was greater in the Bvg+ phase-locked strain than in the Bvgi phase-locked strain, and expression in both of these strains was greater when cells were grown in LB than in SS (Fig. (Fig.2).2). fhaS therefore does not appear to be a Bvgi phase gene like bipA (compare fhaS-lacZ expression with bipA-lacZ expression in the various strain backgrounds and media in Fig. Fig.2).2). Instead, fhaS appears to be expressed differently than all previously characterized Bvg-regulated genes; it is expressed maximally only under a subset of Bvg-activating conditions. Inspection of the 246-nucleotide sequence between the end of the gene upstream of fhaS (purL) and the fhaS initiation codon revealed no discernible BvgA binding sites, suggesting Bvg-mediated control of fhaS expression is indirect.
Cellular localization of FhaS.
In Bordetella strains grown in vitro, mature FHA can be detected in concentrated culture supernatants, outer membrane fractions, and whole-cell lysates, while preprocessed FhaB can be detected only in whole-cell lysates. Because of the extensive similarity between FhaS and FhaB, we hypothesized that FhaS is processed and localized in a manner similar to FhaB. To test this hypothesis, we performed Western blot analysis on proteins derived from whole-cell, outer membrane, and supernatant fractions from wild-type, ΔfhaB, ΔfhaS, and ΔfhaB ΔfhaS B. bronchiseptica strains grown overnight in either LB or SS. Membranes were probed with an anti-FHA antibody generated against amino acids 1048 to 1385 of B. bronchiseptica FHA, a region surrounding the RGD motif in which FhaS and FHA are predicted to differ by only one amino acid. In whole-cell lysates, this antibody detected many polypeptides, with one of ~350 kDa and one of ~260 kDa being most prominent in the wild-type and ΔfhaS strains grown in either LB or SS (Fig. (Fig.3).3). Because only the highest-molecular-weight polypeptides were detected when an antibody against the C terminus of FhaB was used (data not shown), we can conclude that the ~350-kDa and ~250-kDa polypeptides correspond to unprocessed FhaB and mature FHA, respectively. In whole-cell lysates of the wild-type and ΔfhaB strains, prominent polypeptides of approximately 230 to 240 kDa were detected when the cells were grown in LB. Similarly sized polypeptides were only minimally detectable when these strains were grown in SS (Fig. (Fig.3).3). The absence of these polypeptides in lysates of the ΔfhaB ΔfhaS double mutant strongly suggests that they are FhaS. Cleavage within the amino acids that are identical to those in FhaB that are predicted to represent the C-terminal maturation site would result in maturation of the 328-kDa full-length FhaS protein into a mature protein of ~233 kDa. It therefore appears that the 230-to-240-kDa polypeptides present in lysates of the wild-type and ΔfhaB strains, but absent in the ΔfhaS and ΔfhaS ΔfhaB strains, represent a processed, mature form of FhaS.
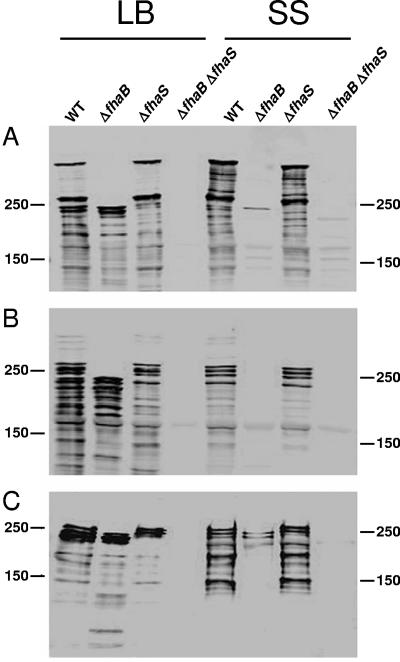
FhaS localizes to the outer membrane and is released into the culture supernatant. Proteins from whole cell (A), outer membrane (B), and supernatant (C) fractions were prepared from the wild-type (WT) and the indicated mutant strains grown in either LB or Stainer-Scholte (SS) broth and subjected to immunoblot analysis using a polyclonal antibody to B. bronchiseptica FHA, which also recognizes FhaS. The positions of molecular weight markers (in kDa) are shown.
In outer membrane preparations, the anti-FHA antibody detected distinct polypeptides from 230 to 260 kDa in the wild-type and ΔfhaS strains grown in SS, corresponding to FHA. Polypeptides with the same mobility were present in these strains grown in LB; however, several additional prominent polypeptides ranging in size from ~180 to 240 kDa were detected in the wild-type and ΔfhaB strains grown in this medium (but not in SS). Because these polypeptides were absent in the ΔfhaB ΔfhaS double mutant, they most likely correspond to FhaS. Moreover, these data suggest that, like FHA, several distinct forms of FhaS are localized to the outer membrane, or alternatively, there is significant proteolysis of FHA and FhaS in outer membrane preparations.
The anti-FHA antibody also detected several polypeptides in concentrated culture supernatants. At least five polypeptides ranging from ~150 to 260 kDa were detected in supernatants prepared from the wild-type and ΔfhaS strains when the cells were grown in SS (Fig. (Fig.3).3). The top three bands were also present in these strains when grown in LB, indicating that FHA is efficiently secreted from the cell under both growth conditions. In addition, polypeptides of ~230 to 240 kDa were detected in the wild-type and ΔfhaB strains when the cells were grown in LB (and were only minimally detected from the ΔfhaB strain when the cells grown in SS), demonstrating that FhaS is also secreted from the cell, and in amounts similar to FHA when the cells are grown in LB.
Export of FhaS is dependent on FhaC.
FhaC is an outer membrane channel-forming protein required for the export of FhaB to the cell surface (22, 31, 33, 51). Sequences within FhaB required for FhaC-dependent export are located within the N-terminal 300 aa (the so-called “secretion domain”) (7, 36). Although the first ~150 aa of FhaS and FhaB are not identical, they are moderately similar and two amino acid motifs (NPGL and NPNG) that have been shown to be critical for export of FhaB (32) are present in the same relative location in FhaS. Moreover, a search of the B. bronchiseptica genome revealed no other open reading frames with similarity to FhaC. We therefore hypothesized that FhaS is exported via FhaC. Previous experiments have shown that FhaB/FHA is undetectable in strains containing loss-of-function mutations in fhaC, presumably because it is degraded in the periplasm (32, 70). Using Western blot analysis, we found that polypeptides corresponding to FHA and/or FhaS were detectable in whole-cell lysates of the wild-type, ΔfhaB, and ΔfhaS strains grown in LB but were undetectable in lysates of strains containing disruption mutations in fhaC (Fig. (Fig.4).4). These results demonstrate that like FHA, FhaS is dependent on FhaC, and strongly suggest that FhaS is exported across the outer membrane by FhaC. B. bronchiseptica, therefore, appears to use the same protein for the export of both FHA and FhaS.
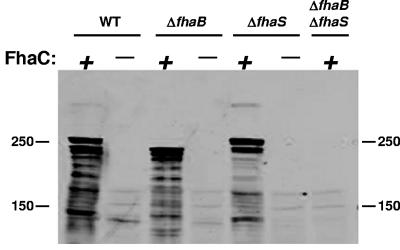
Export of FhaS is dependent on FhaC. Whole cell lysates were prepared from LB-grown wild-type (WT) and ΔfhaS, ΔfhaB, and ΔfhaB ΔfhaS mutant strains in isogenic fhaC+ (+) and fhaC− (−) backgrounds. Proteins were subjected to immunoblot analysis and probed with anti-FHA antibody, which also recognizes FhaS. The positions of molecular mass markers (in kDa) are shown.
Evaluation of FhaS-mediated adherence of B. bronchiseptica to epithelial cells.
The role of B. pertussis FHA as an adhesin is well documented (4, 25, 28, 43, 44, 49, 50, 54, 59-61). Additionally, it has been demonstrated that FHA is both necessary and sufficient to mediate B. bronchiseptica adherence to a rat lung epithelial (L2) and other cell lines in vitro (12, 41). FhaS shares a high degree of similarity with FHA in regions that have been implicated in mediating adherence to eukaryotic cells or cell surface components, including the CRD (>99% identical), which directs binding to glycolipids and ciliated cells (49), and the HBD (99% identical), which promotes binding to highly sulfated polysaccharides, structures commonly found on eukaryotic cell surfaces (23). The integrin-binding RGD motif is also conserved between FhaS and FHA. To test the hypothesis that FhaS can mediate binding of B. bronchiseptica to eukaryotic cells, we utilized L2 cells in a standard in vitro adherence assay with a ΔfhaB strain grown overnight in LB in order to induce maximal expression of FhaS in the absence of FHA. Under these growth conditions, FhaS is produced and localized to the cell surface in amounts similar to FHA (Fig. (Fig.3).3). Although wild-type RB50 grown in LB adhered efficiently to the L2 cells (Fig. (Fig.5A),5A), the ΔfhaB strain was severely defective in adherence (Fig. (Fig.5B),5B), indicating that, at least under these experimental conditions, FhaS was not able to mediate binding of B. bronchiseptica to L2 cells. To determine whether FhaS contributes to adherence in the presence of FHA (i.e., that both FHA and FhaS are required for adherence of LB-grown bacteria to epithelial cells), we compared the binding ability of the ΔfhaS strain with that of wild-type B. bronchiseptica. The ΔfhaS strain adhered to L2 cells as efficiently as RB50, suggesting that binding of the wild-type strain to L2 cells, even when grown in LB, is mediated primarily by FHA (Fig. (Fig.5C).5C). We obtained similar results using a human bronchial epithelial (BEAS-2B) cell line (data not shown). FhaS, therefore, does not appear to be important for mediating adherence to epithelial cells—at least not in a manner that we can detect with these cultured cell assays.
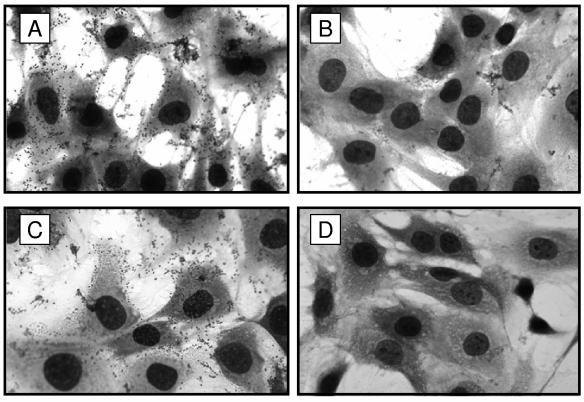
FhaS does not mediate adherence of B. bronchiseptica to rat lung epithelial (L2) cells. Bacteria were grown overnight in LB and added to a monolayer of L2 cells in a standard adherence assay. L2 cells were incubated with wild-type RB50 (A); ΔfhaB (B); ΔfhaS (C); or L broth alone (D). Data are representative of similar results obtained from three independent experiments. Magnification is ×1,000.
Role of FhaS in respiratory tract infection.
To evaluate the contribution of fhaS to respiratory tract infection, we performed both single-infection and coinfection experiments. When inoculated into separate animals, the ΔfhaS strain did not differ from RB50 in its ability to colonize the nasal cavities and tracheas of Wistar rats at days 14 and 28 post infection (data not shown). However, when coinoculated into the same animal with RB50G, a derivative of our wild-type strain expressing a gentamicin resistance gene (6), a defect for the ΔfhaS strain was revealed. In these experiments, groups of rats were inoculated intranasally with 5 μl of phosphate-buffered saline containing approximately 250 CFU each of wild-type and mutant bacteria, and the number of wild-type and mutant bacteria recovered from the nasal cavities and tracheas of infected animals was determined 7, 14, and 28 days postinoculation. The competitive index (CI) was calculated as the ratio of mutant to wild-type bacteria recovered divided by the ratio of mutant to wild-type bacteria administered; thus, a CI of less than 1 indicates a virulence defect for the mutant strain. On days 7 and 14 postinoculation, although the total CFU recovered (shown at the top of each panel in Fig. Fig.6)6) was comparable to that from animals infected with wild-type bacteria alone, the ΔfhaS strain was significantly outcompeted by the wild-type strain in both the nasal cavity and trachea (Fig. (Fig.6).6). By day 28, however, although in the majority of animals the number of CFU of the ΔfhaS mutant recovered was far lower than that seen with the wild-type strain, in a few cases the number of ΔfhaS CFU isolated was close to or higher than the number of wild-type CFU isolated. The ΔfhaS strain was not outcompeted by RB50G when coinoculated into SS or LB broth and grown for several generations in vitro (see Materials and Methods). Together, these data indicate that fhaS contributes to bacterial fitness in vivo in a manner revealed when the ΔfhaS strain must compete with wild-type bacteria.
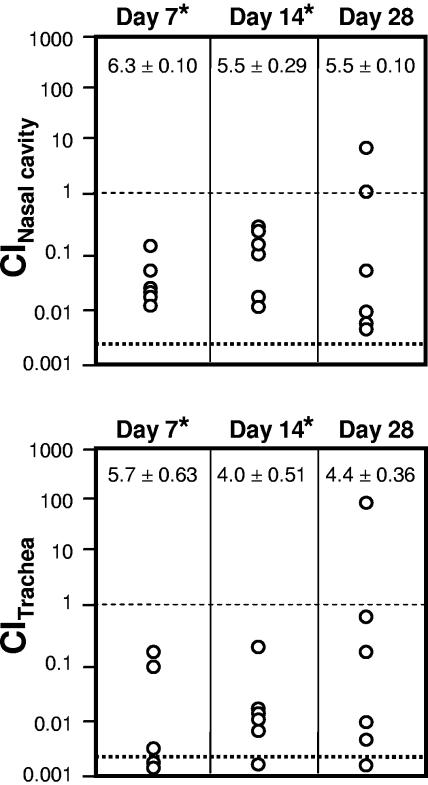
ΔfhaS strains are out-competed by wild-type B. bronchiseptcia in vivo. Wistar rats were inoculated intranasally with approximately 250 CFU of both the wild-type (RB50G) and ΔfhaS strains. Animals were sacrificed on the indicated day postinoculation, and the number of wild-type and ΔfhaS bacteria present in the nasal cavity and trachea was determined. Each symbol represents the competitive index (CI) from the indicated tissue of a single animal, which was calculated as the ratio of mutant to wild-type bacteria recovered divided by the ratio of mutant to wild-type bacteria inoculated. A CI value of 1 (thin dashed line) indicates equal numbers of wild-type and ΔfhaS bacteria were recovered. Symbols below the thick dashed line represent cases where no ΔfhaS bacteria were present in at least 500 total colonies recovered. The average number of CFU recovered from each tissue ± 1 standard error of the mean (log10) is shown at the top of each panel. *, P < 0.05.
Comparison of fhaS alleles among B. bronchiseptica strains.
Consistent with the predicted amino acid sequence comparison described above, nucleotide sequences at the 5′ and 3′ ends of the fhaS loci of B. bronchiseptica RB50, B. pertussis Tohama 1, and B. parapertussishu 12822 are nearly identical, while those corresponding to a majority of the central portion of the open reading frame in RB50 differ substantially from those of Tohama 1 and 12822 (Fig. (Fig.7,7, top panel). Sequences similar to the central region of fhaS from Tohama 1 and 12822 are not present anywhere within the RB50 genome (48). Because B. pertussis and B. parapertussishu strains appear to have diverged independently from B. bronchiseptica or a B. bronchiseptica-like ancestor (21, 64), these observations suggest that either B. pertussis and B. parapertussishu strains acquired their fhaS alleles independently after diverging from B. bronchiseptica or that both subspecies evolved from a B. bronchiseptica(-like) strain that possessed an fhaS allele similar to those present in Tohama 1 and 12822. To investigate these possibilities, we used PCR to identify the fhaS alleles present in B. bronchiseptica strains representing various clades within the B. bronchiseptica cluster (20, 64). Interestingly, although nearly identical purL homologs are present 5′ to fhaS in RB50, Tohama I, and 12822, the intergenic sequences differ substantially in Tohama I compared to RB50 and 12833 (Fig. (Fig.7,7, top panel). Therefore, PCR was performed using a primer designed to hybridize to sequences at the 3′ end of purL. Genomic DNA was prepared from the various B. bronchiseptica strains, B. pertussis Tohama I, and B. parapertussishu 12822 and used as template DNA for PCR. Using primers 1 and 2 (Fig. (Fig.7,7, top panel), a 1.4-kb DNA fragment was amplified from RB50 and all other B. bronchiseptica strains tested (except strain 675, in which only a slightly larger faint band could be detected that was determined by DNA sequence analyses to not correspond to fhaSBb), but no PCR product was amplified from Tohama 1 or 12822 (Fig. (Fig.7,7, bottom panel). Conversely, PCR using primers 1 and 3 (Fig. (Fig.7,7, top panel) resulted in the amplification of a 1.6-kb DNA fragment only from Tohama 1 and 12822 (Fig. (Fig.7,7, bottom panel). These results suggest that all B. bronchiseptica strains contain a common fhaS allele (or no fhaS allele at all) and support the hypothesis that the fhaS allele present in B. pertussis and B. parapertussishu was acquired by these strains independently after they diverged from a B. bronchiseptica(-like) ancestor.
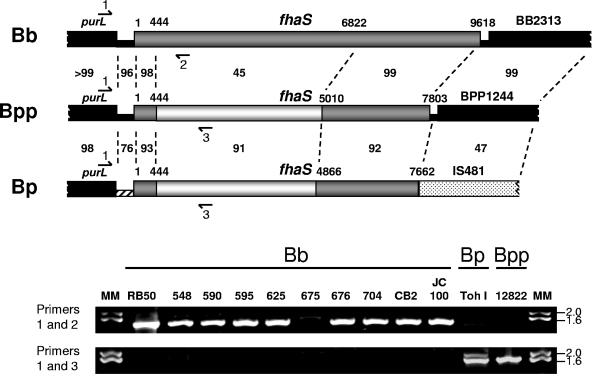
Distribution of fhaS alleles among B. bronchiseptica strains. (Upper panels) Genetic organization of the fhaS loci in B. bronchiseptica (Bb), B. parapertussis (Bpp), and B. pertussis (Bp). Open reading frames (thick lines) and intergenic DNA (thin lines) are shown with nucleotide numbering relative to the first codon of fhaS. The intergenic region upstream of fhaS in B. pertussis is hatched to indicate that its sequence is significantly different from those of B. bronchiseptica and B. parapertussishu. The percentage of identity of nucleotide sequences between various regions (thin dashed lines) is shown. The entire B. bronchiseptica fhaS gene and regions of nucleotide similarity of the B. parapertussis and B. pertussis fhaS genes to B. bronchiseptica fhaS are shown in dark gray. White segments denote nucleotide sequences in the B. parapertussis and B. pertussis fhaS genes that are similar to each other but dissimilar to B. bronchiseptica fhaS. The purL gene is located upstream of fhaS in all three subspecies. Highly similar open reading frames are located downstream of fhaS in B. bronchiseptica and B. parapertussis. By contrast, an open reading frame encoding an insertion sequence transposase (IS481) is located immediately downstream of fhaS in B. pertussis. Approximate binding sites of primers used to amplify fhaS sequences are shown. Primer PfhaSOUT1 (Primer 1) hybridizes to the 3′ end of purL in all three subspecies. Primer CD64 (Primer 2) hybridizes to RB50-specific fhaS sequences, and primer fhaSBpBpp1086rev (Primer 3) hybridizes to B. pertussis- and B. parapertussis-specific fhaS sequences. (Lower panels) 0.8% agarose gel electrophoresis of PCR products obtained from Bordetella genomic DNA templates. Bb, various B. bronchiseptica strains; Bp, B. pertussis strain Tohama I; Bpp, B. parapertussishu strain 12822. The upper gel shows PCR results using primers 1 and 2; the lower gel shows PCR results using primers 1 and 3. Molecular mass (MM) markers (in kilobases) are shown at the right.
DISCUSSION
FHA is prototypical of proteins secreted by the two-partner secretion (TPS) pathway (36). Many of these large exoproteins, which can be surface-associated, secreted into the extracellular milieu, or both, have postulated or proven roles in virulence (34, 36). In some cases, two exoproteins are encoded within the same genome, e.g., the HMW1 and HMW2 adhesins from Haemophilus influenzae and the LspA1 and LspA2 proteins from Haemophilus ducreyi (55, 67). Although in each pathogen these proteins share a high degree of similarity to one another, they are thought to play distinct roles in virulence, due to differences in expression profile, protein functionality, or both (16, 17, 55, 56, 63, 66, 67). In this study, we characterized the fhaS gene from B. bronchiseptica, which encodes a protein that is nearly identical to that encoded by the fhaB gene. We show that fhaS is expressed maximally under different growth conditions than have been described for all other characterized Bordetella virulence genes. We also show that FhaS is exported (via FhaC), processed, and localized in a manner similar to that of FhaB. However, despite the extremely high degree of amino acid similarity between mature FhaS and FHA, FhaS did not mediate bacterial adherence to cultured epithelial cells in vitro and was apparently dispensable for tracheal colonization in vivo. We hypothesize, therefore, that similarly to what has been proposed for the HMW and Lsp proteins, FHA and FhaS may play distinct roles in the Bordetella infectious cycle.
Although fhaS expression requires BvgAS, the fact that it is much greater in LB than SS suggests that fhaS belongs to a class of Bordetella genes different from those that have been characterized thus far. Since inspection of sequences upstream of the start of translation failed to reveal a discernible consensus or near-consensus BvgA binding site(s), we hypothesize that BvgAS control of fhaS expression is indirect. Although other possibilities exist, the simplest explanation for these data is that BvgAS controls a regulator(s) that controls fhaS expression and that this intermediate regulator(s) is differentially active in LB and SS. Indirect control of gene expression by BvgAS in Bordetella is not unprecedented. For example, BvgR controls the expression of vrg loci (45, 46), FrlAB is a master regulator required for the synthesis and assembly of flagella in the Bvg− phase of B. bronchiseptica (3), and the regulators encoded by the btr locus control expression of the type III secretion system (42). However, fhaS is to our knowledge the first example of a gene that is potentially subject to intermediate-regulatory control and is expressed in response to a different set of environmental conditions than all other known Bvg-regulated genes. An intriguing possibility is that fhaS belongs to a subset of virulence genes whose coordinate expression is activated in response to specific temporal or spatial environmental conditions in vivo that are different from those that induce the expression of known Bordetella virulence genes. We are currently attempting to identify the hypothesized fhaS intermediate regulator and other gene(s) under its control in order to determine whether such a regulatory pathway exists.
In addition to revealing differences in expression pattern, our in vitro analyses also suggest that FhaS functions differently than FHA. FhaS failed to mediate or contribute to binding of B. bronchiseptica to two different epithelial cell lines, under conditions in which FHA was proficient at promoting bacterial adherence. This result was surprising given the high degree of similarity between FhaS and FHA, especially within the domains that have been implicated in binding to cell surface components (i.e., the HBD, CRD, and RGD). These data suggest that the amino acids within FHA that are important for binding eukaryotic cells are those that are different between FhaS and FHA. Interestingly, most of the amino acids that are different between the two mature proteins are localized to the extreme C termini, a region of FHA that has not previously been implicated as a functional domain. Although it is possible that FhaS does not function as an adhesin, it is also possible that FhaS is an adhesin with a different cellular binding specificity than FHA. Such a scenario would suggest FhaS and FHA recognize different host cell receptors, as has been proposed for the HMW1 and HMW2 adhesins, which, despite their high degree of similarity, have been shown to mediate adherence to different cell types (55). We are currently performing targeted mutagenesis and constructing strains expressing chimeric FHA and FhaS proteins to investigate the role of amino acids near the C terminus of FHA and FhaS in host cell adherence.
Our rat experiments indicated that FhaS is also not functionally redundant with FHA in vivo. Unlike the case for fhaB, which is absolutely required for tracheal colonization (12), our fhaS deletion strain was indistinguishable from wild-type B. bronchiseptica in its ability to establish respiratory infection in rats. When compared in coinfection experiments, however, the ΔfhaS strain was outcompeted by the wild-type strain, suggesting that fhaS is expressed in vivo and that its role is revealed when the mutant must compete with wild-type bacteria. Thus, like cyaA (24), fhaS appears to play a role in the infectious cycle that is not revealed in single-infection rat colonization experiments. For example, fhaS may be involved in the initial establishment of infection, modulation of the host immune response, and/or transmission. We are in the process of developing sensitive in vivo assays using natural infection models to test each of these possibilities.
Whatever the function of FhaS in the B. bronchiseptica life cycle, it is apparently not essential, since our PCR-based assay indicated that at least one B. bronchiseptica strain does not appear to contain an fhaS gene. Moreover, the fhaS alleles present in B. pertussis Tohama I and B. parapertussis 12822, while very similar to each other, are very different from fhaS of B. bronchiseptica. This allele distribution suggests the possibility that fhaS may contribute to a phenotype that distinguishes B. bronchiseptica strains from B. pertussis and B. parapertussishu strains. One such phenotype is host specificity. One can envisage that the FhaS protein encoded by B. bronchiseptica promotes respiratory colonization of a broad range of mammals whereas those encoded by B. pertussis and B. parapertussishu allow, or facilitate, infection of humans specifically. Alternatively, it may be that expression of the B. bronchiseptica fhaS allele is somehow disadvantageous for human infection. If the B. pertussis and B. parapertussishu alleles play similar roles, it would seem likely that their expression would be regulated similarly. However, although we did not measure fhaS expression in these subspecies, comparison of the Tohama I and 12822 genomes revealed that the nucleotide sequence of the intergenic region upstream of fhaSBp differed significantly from that of fhaSBpp (which was nearly identical to that of fhaSBb), suggesting that fhaSBp may be regulated differently. We are currently conducting experiments to compare the expression profiles of fhaSBp, fhaSBpp and fhaSBb and are also constructing strains expressing heterologous fhaS genes to investigate these hypotheses.
The distribution of fhaS alleles among the bordetellae is also interesting from an evolutionary perspective. How did B. pertussis and B. parapertussishu, which diverged independently from a B. bronchiseptica-like ancestor (15, 21, 64), evolve to contain nearly identical fhaS alleles that are not present in the B. bronchiseptica genome? While it is of course possible that B. pertussis and B. parapertussishu strains diverged from an ancestral B. bronchiseptica(-like) strain that did contain an fhaS allele similar to fhaSBp and fhaSBpp, the fact that nucleotide sequences at the 5′ and 3′ ends of fhaSBpp are more similar to those of B. bronchiseptica than B. pertussis suggests that B. parapertussishu acquired the central region of fhaS (the region that is similar to fhaSBp and different from fhaSBb) by horizontal transfer from a B. pertussis strain. The original source of the fhaSBp allele is more mysterious, as a BLAST search of the sequences unique to fhaSBp revealed no significant database matches. We showed previously that a similar allele distribution and genomic organization exists for the bipA genes of B. pertussis, B. parapertussishu, and B. bronchiseptica (20). bipA encodes a large outer membrane protein, and its similarity to intimin of enteropathogenic and enterohemorrhagic Escherichia coli and invasin of Yersinia spp. suggests it may play a role in adherence to host tissues (58). Together, these data suggest a mechanism by which B. bronchiseptica, which can infect humans but which rarely (if ever) causes acute disease in this host, may have evolved rapidly into B. parapertussishu via horizontal acquisition of specific alleles from B. pertussis. Additional comparison of Bordetella genomes, together with direct investigation of the roles of genes with allele distribution patterns like those of fhaS and bipA, will allow us to test this hypothesis and may shed light on the molecular bases of host specificity and mechanisms underlying the establishment of acute versus chronic infections.
Acknowledgments
We thank Carol Inatsuka for helpful discussions and technical assistance, Robin Hulbert for critical review of the manuscript, and Joseph Mazar for technical assistance.
This research was supported by grants from the National Institutes of Health (AI43896), U.S. Department of Agriculture (National Research Initiative of the USDA Cooperative State Research, Education and Extension Service grant 2003-35204-13555), and the University of California Superfund Basic Research and Education Program to P.A.C.
REFERENCES
Articles from Infection and Immunity are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/iai.73.8.4960-4971.2005
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1201180?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/iai.73.8.4960-4971.2005
Article citations
Bordetella filamentous hemagglutinin and adenylate cyclase toxin interactions on the bacterial surface are consistent with FhaB-mediated delivery of ACT to phagocytic cells.
mBio, 15(5):e0063224, 27 Mar 2024
Cited by: 0 articles | PMID: 38534159 | PMCID: PMC11077949
Relaxed specificity of BcpB transporters mediates interactions between Burkholderia cepacia complex contact-dependent growth inhibition systems.
mSphere, 8(4):e0030323, 27 Jul 2023
Cited by: 0 articles | PMID: 37498085 | PMCID: PMC10449530
DegP Initiates Regulated Processing of Filamentous Hemagglutinin in Bordetella bronchiseptica.
mBio, 12(3):e0146521, 29 Jun 2021
Cited by: 6 articles | PMID: 34182780 | PMCID: PMC8263021
The Two TpsB-Like Proteins in Anabaena sp. Strain PCC 7120 Are Involved in Secretion of Selected Substrates.
J Bacteriol, 203(4):e00568-20, 25 Jan 2021
Cited by: 0 articles | PMID: 33257527 | PMCID: PMC7847546
BrlA and AbaA Govern Virulence-Required Dimorphic Switch, Conidiation, and Pathogenicity in a Fungal Insect Pathogen.
mSystems, 4(4):e00140-19, 09 Jul 2019
Cited by: 53 articles | PMID: 31289140 | PMCID: PMC6616149
Go to all (26) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Molecular characterization of Bordetella bronchiseptica filamentous haemagglutinin and its secretion machinery.
Microbiology (Reading), 146 ( Pt 5):1211-1221, 01 May 2000
Cited by: 31 articles | PMID: 10832649
New Insight into Filamentous Hemagglutinin Secretion Reveals a Role for Full-Length FhaB in Bordetella Virulence.
mBio, 6(4):e01189-15, 18 Aug 2015
Cited by: 20 articles | PMID: 26286694 | PMCID: PMC4542190
Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity.
Proc Natl Acad Sci U S A, 102(51):18578-18583, 09 Dec 2005
Cited by: 60 articles | PMID: 16339899 | PMCID: PMC1317942
The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp.
Mol Microbiol, 9(4):653-660, 01 Aug 1993
Cited by: 134 articles | PMID: 8231801
Review




