Abstract
Free full text

RUNX3 Suppresses Gastric Epithelial Cell Growth by Inducing p21WAF1/Cip1 Expression in Cooperation with Transforming Growth Factor β-Activated SMAD
Abstract
RUNX3 has been suggested to be a tumor suppressor of gastric cancer. The gastric mucosa of the Runx3-null mouse develops hyperplasia due to enhanced proliferation and suppressed apoptosis accompanied by a decreased sensitivity to transforming growth factor β1 (TGF-β1). It is known that TGF-β1 induces cell growth arrest by activating CDKN1A (p21WAF1/Cip1), which encodes a cyclin-dependent kinase inhibitor, and this signaling cascade is considered to be a tumor suppressor pathway. However, the lineage-specific transcription factor that cooperates with SMADs to induce p21 expression is not known. Here we show that RUNX3 is required for the TGF-β-dependent induction of p21 expression in stomach epithelial cells. Overexpression of RUNX3 potentiates TGF-β-dependent endogenous p21 induction. In cooperation with SMADs, RUNX3 synergistically activates the p21 promoter. In contrast, RUNX3-R122C, a mutation identified in a gastric cancer patient, abolished the ability to activate the p21 promoter or cooperate with SMADs. Furthermore, areas in mouse and human gastric epithelium where RUNX3 is expressed coincided with those where p21 is expressed. Our results suggest that at least part of the tumor suppressor activity of RUNX3 is associated with its ability to induce p21 expression.
The RUNX family of transcription factors plays pivotal roles in normal development and neoplasias. In mammals, the RUNX family genes consist of RUNX1/AML1, RUNX2, and RUNX3 (see reference 37 for the nomenclature). All RUNX family members share the central Runt domain, which is well conserved and recognizes a specific DNA sequence, but each has relatively divergent N- and C-terminal regions. RUNX1 and RUNX2 are required for hematopoiesis and osteogenesis, respectively, and are genetically altered in leukemia and bone disease (15, 20, 26, 27, 29). In contrast, RUNX3 is involved in neurogenesis (10, 17) and thymopoiesis (36, 41) and functions as a tumor suppressor of gastric cancer (7, 18). About 60% of primary gastric cancer specimens do not express RUNX3 due to a combination of hemizygous deletion and DNA hypermethylation of the RUNX3 promoter region (7, 13, 18, 28). RUNX3-R122C, a mutation found in a gastric cancer case located in the conserved Runt domain, abolishes the tumor-suppressive activity of RUNX3 (18).
It is noteworthy that the RUNX3 gene is located on human chromosome 1p36, a region that has long been suggested to be a tumor suppressor locus of various cancers. Subsequent studies revealed that RUNX3 inactivation is not limited to gastric cancer. Frequent inactivation of RUNX3 due to DNA hypermethylation has been reported in various other cancers, including lung cancer (19, 45), hepatocellular carcinoma (42), breast cancer (13), colon cancer (6, 14), pancreatic cancer (38), bile duct cancer (38), prostate cancer (12), and larynx cancer (13). Unlike many tumor suppressors, such as p53, which are inactivated mainly by deletions and mutations, RUNX3 is unique in that it is inactivated primarily by epigenetic silencing, rather than mutations or deletions. Furthermore, RUNX3 can be reactivated and therefore considered to be a good drug target because mutations in its gene are rare (1).
RUNX3 activity is closely associated with transforming growth factor β (TGF-β) signaling since the gastric mucosa of the Runx3 knockout mouse is less sensitive to TGF-β, which induces both cell cycle arrest and apoptosis. To inhibit the growth of a given cell type, TGF-β can employ diverse mechanisms, such as down-regulating c-myc and CDK-2/CDK-4 activity by modulating the functions of p15INK4B, p21Waf1/Cip1, and p27Kip1 (9, 22, 32, 43). On the flip side, any genetic or epigenetic alteration of the TGF-β pathway can make normal cells vulnerable to tumorigenesis (4, 23). Indeed, TGF-β receptors and SMADs are the main targets that are altered in numerous types of cancers, including pancreatic and colon cancers (23). One of the downstream targets of TGF-β is p21, which has been particularly thoroughly studied due to the central role it plays in CDK inhibition and cell cycle control. The ability to modulate p21 expression and thus regulation of the cell cycle is often considered to be an intrinsic characteristic of many tumor suppressor proteins, including TP53 (p53), BRAC1, and SMADs (5, 30, 33-35).
Here we further investigated the role RUNX3 plays in the TGF-β-dependent cell growth inhibition of gastric cancer cell lines and observed that RUNX3 is the upstream regulator of p21 expression, through which it inhibits cell growth in response to TGF-β.
MATERIALS AND METHODS
Cell culture.
293 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin G, and 100 μg/ml streptomycin, while MKN28 and SNU16 cells were maintained in RPMI 1640 medium with the same supplements. The cells were grown at 37°C in a humidified atmosphere of 5% CO2.
Plasmids.
The pGL3-p21 luciferase reporter plasmid was constructed by transferring 2.3 kb of the p21 promoter from −2300p21-CAT (obtained from A. Moustakas, University of Crete, Heraklion, Greece) to plasmid pGL3Basic (Promega). The human RUNX3 cDNAs (nucleotides 1 to 1784; GenBank accession no. Z35278) was subcloned into the pEF-BOS expression vector (24) at the XbaI site to yield pEF-RX3. The reverse-oriented clone, which expresses antisense RUNX3, was named pEF-RX3-AS. To generate pOPI3-RX3 and pOIP3-RX3-R122C, RUNX3 and RUNX3-R122C cDNA fragments were, respectively, subcloned into the Lac-Switch-inducible mammalian expression vector pOPI3 (Stratagene). To generate the transient expression plasmids pCS4-myc-RX3 and pCS4-myc-RX3-R122C, the wild-type RUNX3 and RUNX3-R122C cDNAs were subcloned into pCS4-myc. Similarly, the SMAD3 and SMAD4 cDNAs were subcloned into pCS4-HA to generate the transient expression plasmids pCS4-HA-SMAD3 and pCS4-HA-SMAD4, respectively. pBabe-myc-RX3-R122C was constructed by subcloning the myc-RX3-R122C-encoding region of pCS4-myc-RX3-R122C into the BamHI/SalI sites of pBabe.
Tumor samples.
Sixteen surgically resected primary gastric cancer specimens (16 primary gastric cancers, including 2 that had metastasized to the liver) were examined. Their distribution according to their clinical stages is as follows: stage I, two cases; stage II, four cases; stage III, six cases; stage IV, four cases. The clinicopathological factor of each patient was classified according to the International Union against Cancer classification system.
cDNA microarray.
cDNA microarray and data analyses were performed as previously described (16). The Twi-Chip Human 10K Digital Genomics cDNA microarray was used for hybridization.
Reporter assays.
Cells were transfected using Lipofectamine (Gibco/BRL) according to the manufacturer's instructions. Each transfection assay was performed with 0.1 μg of the pGL3-p21 reporter plasmid and the β-galactosidase-expressing plasmid pCMV-β-Gal as an internal control together with the combination of effector plasmids indicated in Fig. Fig.4C4C and and5.5. The amount of exogenous DNA that was transfected in each transfection was made equal by adding the appropriate amount of empty vector DNA as required. Forty-eight hours after transfection, the cells were harvested and subjected to the luciferase assay using the Luciferase Reporter assay kit according to the manufacturer's instructions (Promega). The luciferase activity that was measured was normalized according to the β-galactosidase activity.

p21 is a direct target of RUNX3. (A) Schematic depiction of the RUNX-binding sites in the p21 promoter (2.3-kb region). (B) SNU16 cells were subjected to ChIP analysis. Their chromatin was immunoprecipitated with the polyclonal anti-RUNX3 antibody, and the p21 promoter region was amplified by PCR. GAPDH was amplified as a negative control. (C) 293 cells were cotransfected with the wild-type or mutant pGL3-p21 promoter construct (50 ng/well in a 24-well plate) and various amounts (0, 0.25, and 0.5 μg) of pCS4-3myc-RX3. Reporter activities were measured by a luminometer.
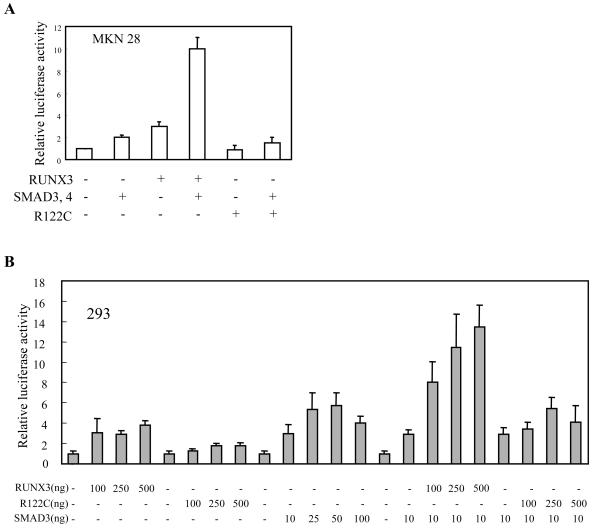
RUNX3 induces p21 expression in cooperation with SMADs. (A and B) MKN28 (A) and 293 (B) cells were cotransfected with pGL3-p21 reporter plasmids and the indicated combination of plasmids. For each transfection, 100 ng/well (24 wells) of each plasmid was used for panel A and 100 ng/well of reporter plasmids was used in all transfections.
Stable cell lines.
SNU16-RX3 cell lines were obtained by transfecting SNU16 cells with pEF-RX3, and G418-resistant clones were selected with G418 (1 mg/ml). Exogenous RUNX3 expression was confirmed by Northern blotting (see Fig. Fig.2B).2B). Similarly, SNU16-AS cell lines were obtained by transfecting SNU16 cells with pEF-RX3-AS. SNU16-R122C cell lines were obtained by transfecting SNU16 cells with pBabe-myc-RX3-R122C, and puromycin-resistant clones were selected with puromycin (0.4 μg/ml). Exogenous RUNX3 expression was confirmed by Western blotting (data not shown). SNU16 cells that were stably transfected with empty vector were used as a control. The MKN28-Lac cell line was obtained by stably transfecting MKN28 cells with the p3SS repressor plasmid (Stratagene). MKN28-Lac-RX3 and MKN28-Lac-R122C cell lines were then obtained by stably transfecting MKN28-Lac cells with pOPI3-RX3 and pOPI3-RX3-R122C, respectively. Induction of RUNX3 and RUNX3-R122C in response to isopropyl-β-d-thiogalactopyranoside (IPTG) was confirmed by Northern blotting (see Fig. 3A and B). Transfections were performed with Lipofectamine according to the manufacturer's instructions (Gibco/BRL).

Involvement of RUNX3 in TGF-β-dependent p21 induction and cell growth inhibition. (A) SNU16 and SNU16-RX3 cells were treated with the indicated concentration of TGF-β1, and the RUNX3 and p21 expression levels were measured by Northern blotting. The GAPDH probe was used as a loading control. (B) Similarly, SNU16, SNU16-RX3, SNU16-R122C, and SNU16-AS cells were treated with TGF-β1 and the p21 expression levels were measured by Northern blotting. (C) Densitometric analysis of the p21 mRNA band intensities in panel B. (D) SNU16, SNU16-RX3, SNU16-R122C, and SNU16-AS cells were cultured in the presence or absence of 0.03 ng/ml TGF-β1 for 24 h, and the cells were counted. The percentage of growth inhibition of the cells in the presence of TGF-β1 is shown.
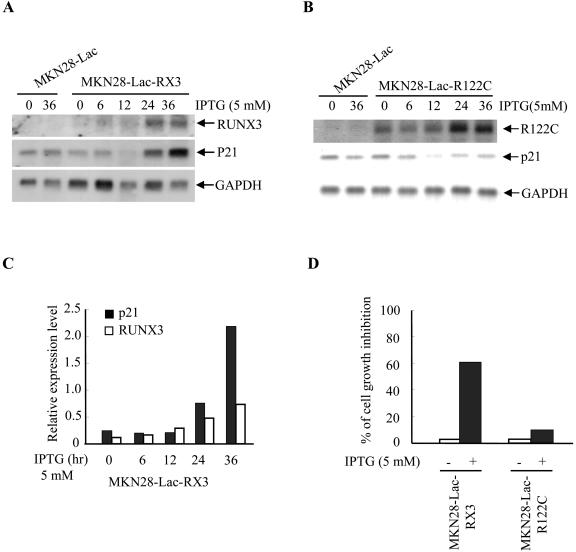
Effect of IPTG-inducible RUNX3 expression on p21 transcription. (A and B) MKN28-Lac, MKN28-Lac-RX3 (A), and MKN28-Lac-R122C (B) cells were treated with IPTG (5 mM) for the indicated periods, and exogenous RUNX3 and endogenous p21 expression levels were examined by Northern blotting. (C) Densitometric analysis of the p21 mRNA band intensities in panel A. (D) MKN28-Lac-RX3 and MKN28-Lac-R122C cells were cultured in the presence or absence of IPTG (5 mM) for 48 h and counted. The percentage of growth inhibition of the cells is shown.
Electrophoretic mobility shift assay (EMSA).
Nuclear protein extracts were prepared from 293 cells transfected with pCS4-myc, pCS4-myc-RX3, or pCS4-myc-RX3-R122C. Protein concentrations of the extracts were determined with the Bradford assay (Bio-Rad). A double-stranded DNA probe, RXE, was prepared and used for EMSA. m1 was used as a specific competitor, while m2 served as a nonspecific competitor. The oligonucleotide sequences of the sense strands of the double-stranded oligonucleotides used in the EMSAs are as follows (the underlined nucleotides indicate mutations; the perfect match of the RUNX-binding site in the oligonucleotide is indicated by capital letters): RXE, 5′-gatccaccacagccaGACCACAggcagacatgagga-3′; m1, 5′-gatccaggacagccaGACCACAggcagacatgagga-3′; m2, 5′-gatccaccacagccaGAggACAggcagacatgagga-3′.
All binding assays were performed at 30°C for 30 min in 20 μl binding buffer [20 mM HEPES (pH 7.6), 4% (wt/vol) Ficoll type 400, 50 mM KCl, 2 mM EDTA, 2 μg of poly(dI-dC)] containing 1 nM 32P-end-labeled probe (20,000 to 30,000 cpm) and nuclear protein extract. Samples were electrophoresed on a 4% polyacrylamide gel in 0.25× Tris-borate-EDTA buffer at 250 V for 1 h and autoradiographed for 16 h at −70°C using two sheets of intensifying screens.
Chromatin immunoprecipitation (ChIP) assays.
ChIP assays were performed by following the Upstate Biotechnology ChIP assay kit protocol (catalog no. 17-295). About 106 SNU16 cells were used per ChIP reaction. The cells were treated with 1% formaldehyde at 37°C for 10 min and washed twice with ice-cold phosphate-buffered saline containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml pepstatin A). Cells were harvested and resuspended in 200 μl of sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 10 mM EDTA, 15 mM Tris, pH 8.1). DNA was fragmented to an average length of about 1 kb by sonication and centrifuged. The supernatant was diluted 10-fold with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl), and chromatin was immunoprecipitated by using an anti-RUNX3 polyclonal antibody. The genomic DNA pellets were resuspended in 50 μl water, and the p21 promoter region was amplified by PCR with p21-5′ (5′-CACCAGACTTCTCTGAGCCCCAG-3′) and p21-3′ (5′-GCACTGTTAGAATGAGCCCCCTTTC-3′).
Immunohistochemical analysis.
Stomach samples from newborn mice were fixed with 4% paraformaldehyde in phosphate-buffered saline overnight, embedded in paraffin, and sectioned at a 6-μm thickness. The sections were used for hematoxylin-and-eosin (HE) staining and immunohistochemical detection of RUNX3 and p21. Anti-bacterial β-galactosidase (NE056/Bio; Nordic Immunological Laboratories) and anti-p21 (sc-397; Santa Cruz) antibodies were used as primary antibodies. These antibodies were visualized by employing a biotin-conjugated anti-rabbit antibody, followed by incubation with avidin-fluorescein isothiocyanate.
In situ hybridization.
To detect RUNX3 and p21 expression in human gastric cancer specimens, in situ hybridization was performed with RNA probes as described previously (44) using sense and antisense digoxigenin-labeled probes consisting of RUNX3 nucleotides 550 to 848 and p21 nucleotides 42 to 584. The hybridized, digoxigenin-labeled probes were visualized with a nucleic acid detection kit (Boehringer Mannheim).
Western blotting and immunoprecipitation analysis.
293 cells were transfected using the calcium phosphate method and, 15 μg of protein extract from each transfection was subjected to SDS-polyacrylamide gel electrophoresis, followed by Western blot analysis using anti-myc (9E10; Santa Cruz) and anti-hemagglutinin (HA) antibodies (12CA5; Roche). For immunoprecipitation experiments, 500 μg of each protein extract was incubated with an anti-myc antibody and precipitated with protein G beads. The immunoprecipitates were then analyzed by Western blotting using appropriate antibodies.
RESULTS AND DISCUSSION
Involvement of RUNX3 in TGF-β-dependent induction of endogenous p21 expression.
To determine whether RUNX3 is a critical downstream effector in TGF-β-mediated growth inhibition, we initially employed DNA microarray technology to screen for genes whose transcription is critically regulated by TGF-β1. The cell line used for this analysis was gastric cancer-derived SNU-16, whose growth is partially inhibited by TGF-β1, unlike most other gastric tumor lines (31). To understand the mechanism by which TGF-β-inhibited cell growth, SNU-16 mRNAs were harvested at five different time points following TGF-β1 stimulation and screened with the Human 10K Digital Genomics cDNA array as previously described (16). The result revealed that various genes encoding cyclins and cyclin-dependent kinase (CDK) inhibitors were induced by TGF-β1 treatment. Among the CDK inhibitors, p21 was the most dramatically up-regulated gene in the array, and its mRNA levels peaked 2 h after stimulation (Fig. (Fig.1A).1A). In contrast, relatively little change was observed in the expression of other CDK inhibitors such as p27kip1, p57kip2, p15, p18, and p19 (Fig. (Fig.1A).1A). TGF-β1 stimulation of p21 protein levels was confirmed by Western blotting (Fig. (Fig.1B1B).
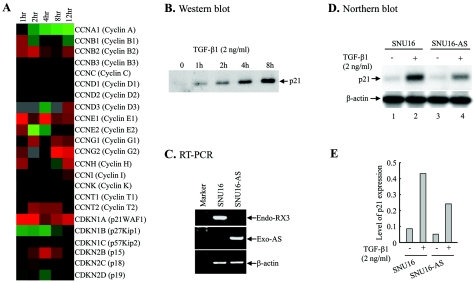
TGF-β1-dependent induction of p21 expression in SNU16 cells and its requirement for RUNX3. (A) SNU16 cells were stimulated with TGF-β1 (2 ng/ml) for various periods, and their gene expression profile was examined by microarray analysis. Changes in relative expression at the indicated times after TGF-β1 stimulation are presented in graduated color patterns. Red, expression induced by TGF-β1 stimulation; green, expression reduced by TGF-β1 stimulation; black, unchanged expression. (B) SNU16 cells were treated with TGF-β1 (2 ng/ml) for various periods, and the p21 protein levels were analyzed by Western blotting. (C) Suppression of endogenous RUNX3 expression in the SNU16-AS cell line. After reverse transcription (RT), the endogenous RUNX3 mRNA in the SNU16-AS cell line (Endo-RX3) was amplified with RX3-p (5′-CTACGGGACATCCTCTGGCTCC-3′) and RX3-cn (5′-CATCTCTGCCAGCAGCGTGCTG-3′), resulting in an 840-bp fragment. Exogenous antisense RUNX3 mRNA (Exo-AS) was amplified with EF-p (5′-CTTCGCCTCGTGCTTGAGTTGAG-3′) and RX3-p, resulting in a 1,570-bp fragment. The EF-p primer was based on the vector sequence (24), while the RX3-p and RX3-cn primer was based on the RUNX3 cDNA sequence. (D) SNU16 and SNU16-AS cells were cultured for 2 h in the presence or absence of TGF-β1 (2 ng/ml), and p21 expression was analyzed by Northern blotting. The β-actin probe was used as a loading control. (E) The band intensities in panel D were analyzed by densitometry, and the p21 mRNA levels are shown as a graph.
To examine whether RUNX3 is involved in the TGF-β-dependent induction of p21 expression, SNU16 cells were transfected with an antisense RUNX3 construct and the stably transfected cell line SNU16-AS was isolated. Analysis by reverse transcription-PCR using two RUNX3-specific primers revealed greatly reduced endogenous RUNX3 transcript levels in SNU16-AS cells (Fig. (Fig.1C).1C). The transfected antisense RUNX3 gene was confirmed to be transcribed by reverse transcription-PCR using primers specific for the plasmid vector and antisense RUNX3 (Fig. (Fig.1C).1C). Northern blot analysis of SNU16 with or without TGF-β1 treatment showed the TGF-β treatment elevated endogenous p21 mRNA levels (Fig. (Fig.1D,1D, compare lanes 1 and 2, and E), thus confirming the microarray data (Fig. (Fig.1A).1A). However, TGF-β1 induced a lower level of p21 mRNA when RUNX3 expression was inhibited by antisense RUNX3 (Fig. (Fig.1D,1D, compare lanes 2 and 4, and E). This result suggests that RUNX3 participates in TGF-β-dependent p21 induction.
The induction of p21 expression was examined further by using stable cell lines expressing wild-type RUNX3, RUNX3-R122C, and antisense RUNX3. Interestingly, SNU16 cell lines that express RUNX3 at high levels could not be obtained, whereas many RUNX3-R122C-overexpressing clones were isolated without much difficulty. We assumed that high levels of RUNX3 expression alone would be sufficient to induce p21 gene expression and inhibit growth. Therefore, stably transfected clones expressing RUNX3 and RUNX3-R122C at low levels were chosen for the present study; these were designated SNU16-RX3 and SNU16-R122C, respectively. First, we examined the effect of exogenous expression of RUNX3 on TGF-β-dependent p21 induction. The SNU16 and SNU16-RX3 cell lines were treated with increasing concentrations of TGF-β, and the mRNA level of p21 was measured by Northern blotting. As shown in Fig. Fig.2A,2A, mRNA levels of p21 were increased by TGF-β treatment, while those of endogenous and exogenous RUNX3 were unchanged. Notably, SNU16-RX3 cells showed a higher level of p21 induction than SNU16 cells at low doses of TGF-β1 (Fig. (Fig.2A,2A, compare lanes 3 and 4). Next, we compared SNU16, SNU16-RX3, SNU16-R122C, and SNU16-AS cells. In the absence of TGF-β1, all these cell lines expressed p21 at similarly low levels (Fig. (Fig.2B,2B, lanes 1, 4, 7, and 10). However, when the cells were treated with low doses of TGF-β1, the induction profiles of the three cell lines differed significantly. SNU16-RX3 cells required only 0.03 ng/ml TGF-β1 to attain high p21 expression levels, while the control SNU16 cells needed 10 times more TGF-β1 (i.e., 0.3 ng/ml) to generate the same p21 mRNA levels (Fig. (Fig.2B,2B, compare lanes 3 and 5). Thus, the SNU16-RX3 cell line is markedly more sensitive to TGF-β1 than its parent line and SNU16-AS cells inhibited the induction of p21 expression at the same concentration of TGF-β1 (Fig. (Fig.2B,2B, lanes 10 to 12). Densitometric analysis of the band intensities in this Northern analysis is shown in Fig. Fig.2C.2C. Since the vectors used for the SNU16-RX3 and SNU16-R122C cell lines are different (see Materials and Methods), the sizes of the RUNX3 and RUNX3-R122C transcripts could be different due to the different 3′ noncoding regions provided by the vectors (Fig. (Fig.2B,2B, lanes 4 to 6 and 7 to 9). It is unclear why several species of transcript are seen in the SNU16-AS line (Fig. (Fig.2B,2B, lanes 10 to 12). The transcripts are not due to multiple integrations of the transgene, because two other independent clones showed the same species of transcripts (data not shown). We think that these transcripts may be caused by inappropriate transcriptional termination.
Thus, our results demonstrate that TGF-β-induced p21 expression is mediated by RUNX3. Interestingly, in the absence of TGF-β1, low exogenous RUNX3 expression did not affect p21 expression, and the effect of RUNX3 expression on p21 expression was only observed in the presence of TGF-β1. It is known that TGF-β-activated SMADs activate the p21 promoter (25) and that RUNX3 physically interacts and functionally cooperates with TGF-β-activated SMADs (8). These observations suggest that p21 promoter activation by TGF-β1 requires cooperation between TGF-β1-activated SMADs and RUNX3. This possibility was examined as described below.
It is important to note that, unlike wild-type RUNX3 expression, RUNX3-R122C, which lacks the tumor suppressor activity, failed to increase TGF-β-dependent p21 induction. This result suggests that the role RUNX3 plays in TGF-β-dependent p21 induction is related to the tumor suppressor activity of RUNX3.
Involvement of RUNX3 in TGF-β-dependent cell growth inhibition.
We examined whether the increased sensitivity of p21 to TGF-β1 due to exogenous RUNX3 is also associated with enhanced cell growth inhibition. When SNU16-RX3, SNU16-R122C, and SNU16-AS cells were treated with very low concentrations of TGF-β1 (0.03 ng/ml) and counted, only SNU16-RX3 cells showed dramatically reduced growth in the presence of 0.03 ng/ml TGF-β1 (Fig. (Fig.2D).2D). In contrast, the growth of SNU16, SNU16-R122C, and SNU16-AS cells was only marginally affected by TGF-β1 at the same concentration (Fig. (Fig.2D2D).
To examine whether high levels of RUNX3 expression alone may be sufficient to induce p21 gene expression and inhibit growth, we established RUNX3-inducible cell lines using the MKN28 human gastric cancer cell line, which lacks RUNX3 expression and sensitivity to TGF-β1 (18). We generated the MKN28-Lac-RX3 and MKN28-Lac-R122C lines that, respectively, express RUNX3 and RUNX3-R122C upon IPTG treatment. IPTG-induced RUNX3 expression coincided with elevated endogenous p21 expression (Fig. (Fig.3A),3A), but RUNX3-R122C expression failed to induce p21 (Fig. (Fig.3B).3B). Densitometric analysis of the band intensities in Fig. Fig.3A3A is shown in Fig. Fig.3C.3C. In agreement with these observations, IPTG treatment inhibited the growth of MKN28-Lac-RX3 but not MKN28-Lac-R122C (Fig. (Fig.3D).3D). Taking these findings together, we conclude that RUNX3, but not RUNX3-R122C, is capable of inducing p21 expression and inhibiting cell growth.
The data presented in Fig. Fig.22 and and33 suggest that RUNX3 participates in TGF-β1-dependent cell growth suppression by inducing p21 expression. These observations also explain why we could not obtain cells expressing RUNX3 at high levels.
RUNX3 binds to and activates the p21 promoter.
The p21 promoter contains five putative RUNX-binding sites, namely, three perfect matches to the consensus binding site called sites a to c and two imperfect matches called sites d and e (Fig. (Fig.4A)4A) (21). When we performed a ChIP assay, we found that the DNA fragment of the p21 promoter was immunoprecipitated by an anti-RUNX3 antibody (Fig. (Fig.4B).4B). Thus, endogenous RUNX3 binds to the p21 promoter in vivo. We then examined whether RUNX3 must bind to the p21 promoter to induce p21 expression by subcloning the 2.3-kb-long DNA fragment of the p21 promoter upstream of the luciferase gene in the pGL3Basic vector (Promega) and performing a transient luciferase expression assay in the 293 cell line. Cotransfection with increasing amounts of the RUNX3 expression plasmid resulted in a dose-dependent increase in reporter activity (Fig. (Fig.4C).4C). Mutation of each RUNX-binding site in the p21 promoter significantly decreased its responsiveness to RUNX3, except for site e. Mutation of all three perfectly matching RUNX-binding sites did not further reduce the responsiveness to RUNX3. This result indicates that sites a to d in the p21 promoter are mainly responsible for RUNX3-mediated p21 expression and that all four sites are required for full responsiveness. Site e also appears to be functional since weak responsiveness to RUNX3 remained even after sites a to d were all mutated. Mutations in both sites d and e abolished responsiveness to RUNX3; however, the basal activity of the reporter gene was also decreased. The mutation of all five RUNX-binding sites completely abrogated responsiveness to RUNX3 and even suppressed the basal activity of the reporter gene. One possible explanation is that the mutation of sites d and e may inactivate other essential transcription factor-binding sites. Further analysis is required to understand the role sites d and e play. Taken together, our results demonstrate that RUNX3 must bind the p21 promoter in order to stimulate its expression.
RUNX3 induces p21 expression in cooperation with SMADs.
Although TGF-β1 induces endogenous p21 expression and exogenous expression of RUNX3 activates the p21 promoter, we did not detect an increase in RUNX3 mRNA levels after TGF-β1 stimulation (Fig. (Fig.2A).2A). Since p21 expression can be induced by TGF-β-activated SMADs (3) and the physical interaction of RUNX3 with SMADs has been reported (8), we speculated that the ability of RUNX3 to transactivate the p21 promoter may be turned on by TGF-β-activated SMADs. We tested this by cotransfecting MKN28 cells with the pGL3-p21 luciferase reporter plasmid and plasmids expressing SMAD3 or SMAD4 in the presence or absence of the plasmid expressing RUNX3. SMAD3 and SMAD4 could transactivate the p21 promoter by about twofold on their own (Fig. (Fig.5A),5A), which is similar to what has been reported previously (25). RUNX3 on its own also transactivated p21 promoter activity by about threefold (Fig. (Fig.5A).5A). However, when RUNX3 was cotransfected with SMAD3 and SMAD4, the transactivation activity was increased by up to 10-fold (Fig. (Fig.5A).5A). Similar observations were made when the luciferase assay was performed with 293 cells (Fig. (Fig.5B).5B). Thus, RUNX3 transactivates p21 expression in collaboration with SMADs. In contrast, RUNX3-R122C only weakly transactivated the p21 promoter and poorly cooperated with SMADs (Fig. 5A and B). These observations further substantiate the above findings, namely, that RUNX3, but not RUNX3-R122C, increases TGF-β1-dependent p21 expression (Fig. (Fig.2).2). Collectively, these results indicate that RUNX3 induces p21 expression in collaboration with TGF-β1-specific SMADs. This explains why the SNU16-RX3 cell line showed much higher TGF-β1-induced p21 expression levels compared to its parental cell line and why SNU16-R122C cells failed to show any TGF-β1-induced p21 expression (Fig. (Fig.22).
Recently, it was reported that RUNX1 and RUNX2 suppress p21 expression in cooperation with mSin3A (21) and histone deacetylase 6 (39), respectively. However, the activation of p21 expression by RUNX3 in cooperation with SMADs is not surprising because RUNX family members function as both transcriptional activators and repressors, depending on the partner proteins involved.
Recently, Seoane and others reported that Forkhead transcription factor FoxO, together with SMAD3 and SMAD4, binds to the promoter of the p21 gene and activates its transcription when HaCaT cells are treated with TGF-β (33). It is known that Akt in the phosphoinositol 3 (PI3) kinase pathway phosphorylates FoxO to exclude it from the nucleus. Therefore, these results suggest that FoxO is a node for integration of TGF-β and PI3/Akt pathways. In their study, they also showed that in human keratinocyte and glioblastoma cells, TGF-β did not alter the level of phospho-Akt or the subcellular localization of FoxO, suggesting that involvement of FoxO in activation of the p21 gene varies from cell type to cell type. Although we did not see a significant effect of FoxO on the overall p21 gene expression in human gastric cancer cell lines, it would be worth examining further whether any other signals would coregulate p21 gene expression together with SMADs in stomach epithelial cells.
Differential DNA-binding affinity of wild-type RUNX3 and the R122C mutant.
As shown above, the RUNX3-R122C mutant neither induced p21 expression nor cooperated with SMADs as efficiently as wild-type RUNX3. We reasoned that the mutant RUNX3 protein may be defective in binding to its cognate target DNA sequences. Therefore, we examined whether RUNX3-R122C bind to the consensus RUNX-binding DNA sequence. As shown in Fig. Fig.6A,6A, the RUNX3 protein exhibited higher affinity toward the 32P-labeled DNA probe bearing the RUNX-binding site than the RUNX3-R122C protein. A quantitative EMSA using serial amounts of nuclear protein extract (Fig. (Fig.6B)6B) and densitometric analysis (data not shown) revealed that the RUNX3-R122C mutant has only one-fourth of the DNA-binding affinity of the wild-type molecule. That RUNX3 and RUNX3-R122C were expressed at similar levels in the transfected cells was confirmed by Western blotting (Fig. (Fig.6C6C).
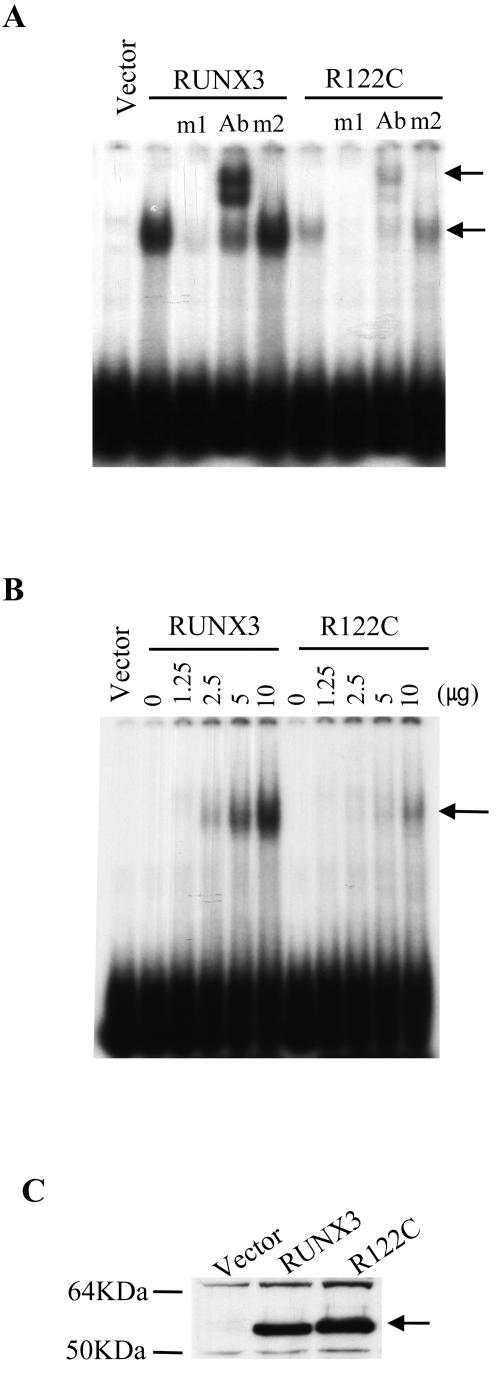
Comparison of the DNA-binding affinities of RUNX3 and RUNX3-R122C by EMSA. (A) Similar amounts of nuclear extracts obtained from 293 cells transfected with pCS4-myc, pCS4-myc-RUNX3, and pCS4-myc-RX3-R122C were subjected to EMSA using the 32P-labeled RUNX binding site-bearing double-stranded oligonucleotide RXE as the probe. The m1 and m2 oligonucleotides were used as specific and nonspecific competitors, respectively (see Materials and Methods). The arrows indicate the DNA-RUNX3 and DNA-RUNX3-antibody complexes. (B) EMSA was performed using a fixed amount of probe DNA and increasing amounts of the nuclear extract obtained from 293 cells transfected with pCS4-myc-RUNX3 or pCS4-myc-RX3-R122C. The arrow indicates the DNA-RUNX3 and DNA-RUNX3-R122C complexes. (C) RUNX3 and RUNX3-R122C protein levels in the transfected cells described in panels A and B were determined by using an anti-myc antibody. The arrow indicates RUNX3 and RUNX3-R122C.
Differential SMAD-binding affinity of wild-type RUNX3 and the R122C mutant.
Since RUNX3-R122C binds to the cognate target DNA sequence, albeit more weakly than wild-type RUNX3, we asked whether there are other defects in RUNX3-R122C that would help explain its profound inability to mediate TGF-β1-induced p21 expression. We thus compared the binding affinities of wild-type RUNX3 and its R122C mutant for SMADs by cotransfecting 293 cells with myc-tagged RUNX3 or RUNX3-R122C and HA-tagged SMAD3 and/or SMAD4. Coimmunoprecipitation analysis revealed that RUNX3-R122C binds more weakly to SMAD3 and SMAD4 than wild-type RUNX3 (Fig. (Fig.7).7). The expression levels of the transfected SMAD, RUNX3, and RUNX3-R122C genes were confirmed to be comparable by Western blotting.
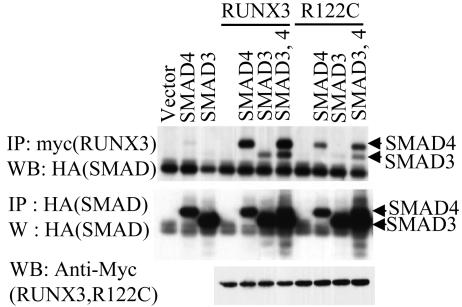
Interaction between RUNX3 and TGF-β-specific SMADs. (A) 293 cells were transfected with a fixed amount of pCS4-myc-RUNX3, pCS4-myc-RX3-R122C, pCS4-HA-SMAD3, and/or pCS4-HA-SMAD4 as indicated. (Upper panel) The physical interactions between RUNX3 or RX3-R122C and SMADs were examined by immunoprecipitation (IP) analyses using an anti-myc antibody, followed by Western blotting (WB) using the anti-HA antibody. (Middle panel) Expression levels of SMAD3 and SMAD4 as measured by immunoprecipitation with an anti-HA antibody, followed by Western blotting using the same antibody. (Lower panel) Expression levels of RUNX3 and RUNX3-R122C as measured by Western blotting using an anti-myc antibody.
These observations demonstrate that the ability of RUNX3 to induce p21 expression is controlled by SMADs which are activated by TGF-β1 and that RUNX3-R122C lacks the ability to activate p21 expression due to the combination of reduced DNA- and SMAD-binding affinities.
Requirement of RUNX3 for p21 expression in vivo.
Our finding that RUNX3 is required for p21 expression in gastric epithelial cell lines prompted us to examine whether RUNX3 expression is also required for p21 expression in mouse and human gastric epithelia. Immunohistochemical staining of the newborn Runx3+/− mutant stomach showed that Runx3 is expressed in epithelial cells of the glandular stomach (Fig. (Fig.8C)8C) as reported previously (18). Interestingly, staining of serial sections with an anti-p21 antibody revealed that the cells expressing Runx3 and those expressing p21 overlap (compare Fig. 8C and D). Furthermore, p21 expression in Runx3−/− stomach epithelial cells was significantly lower than that in normal stomach epithelial cells (compare Fig. 8E and F). The low level of p21 expression in the Runx3−/− mouse gastric epithelium is consistent with the hyperplasia observed in these mutant mice.

RUNX3 and p21 expression patterns in mouse and human gastric epithelium overlap. (A to F). Pyloric areas of stomach tissues were obtained from newborn normal, Runx3+/−, and Runx3−/− mutant mice, and the expression levels of Runx3 and p21 were detected by immunohistochemical staining. A to D are adjacent sections. A and B are control staining and HE staining, respectively. Runx3 was detected using the anti-bacterial β-galactosidase antibody since Runx3 was fused with the bacterial β-galactosidase (β-gal) gene and expressed as the Runx3-β-galactosidase fusion protein in Runx3 mutant mice (18). IgG, immunoglobulin G. (G to L). Expression pattern of RUNX3 and p21 in human gastric epithelium. HE staining (G, H), in situ hybridization with RUNX3 (I, J), and in situ hybridization with p21 (K, L) are shown. Arrows and arrowheads indicate normal and cancerous regions, respectively. Enlargement (×200 magnification) of the boxed regions in panels G, I, and K are shown in panels H, J, and L, respectively.
Since RUNX3 is frequently inactivated in primary human gastric cancer specimens, we examined whether p21 expression was also suppressed in RUNX3-inactivated gastric cancer tissues. Thus, 16 surgically resected primary gastric cancer specimens were subjected to in situ hybridization analysis. Typical results are shown in Fig. 8G to L. These data reveal that p21 expression is restricted to the gastric epithelial cells that express RUNX3 (compare Fig. 8J and L). In the cancerous regions that do not express RUNX3, p21 expression is also significantly decreased. These results together suggest that RUNX3 expression is required for the in vivo expression of p21.
Targeted inactivation of Runx3 in mice resulted in gastric epithelial hyperplasia due to stimulated proliferation and suppressed apoptosis accompanied by a decreased sensitivity to TGF-β1. However, the role Runx3 plays in the TGF-β signaling pathway has not been established. In this investigation, we studied the molecular mechanism by which RUNX3 inhibits cell growth and found that RUNX3 inhibits gastric epithelial cell growth by inducing p21 gene expression in response to TGF-β1. The involvement of RUNX3 in TGF-β-dependent p21 induction is revealed by the fact that exogenous expression of RUNX3 increased endogenous p21 expression in response to TGF-β1. Moreover, suppression of RUNX3 expression decreased TGF-β1-induced endogenous p21 expression (Fig. (Fig.2).2). Furthermore, in mouse and human gastric epithelia, RUNX3 expression coincided with p21 expression (Fig. (Fig.8).8). That p21 is a direct target of RUNX3 is indicated by the observations that the p21 promoter is activated by RUNX3 and that mutation of RUNX-binding sites within the promoter significantly decreased this activity (Fig. (Fig.4).4). The p21 promoter was synergistically activated when RUNX3 and SMAD3 were coexpressed. This is the first report that shows RUNX3 is an essential component of the TGF-β-mediated p21 induction that is critical for TGF-β-mediated cell growth inhibition.
p21 plays a key role in cell cycle arrest at the G1 stage by inhibiting CDKs and was originally identified as an important transcriptional target of p53. One of the critical targets of G1 cyclin CDKs is the retinoblastoma protein (pRb), which, in its hypophosphorylated state, binds and sequesters transcription factors required for the entry of the cell into S phase (2, 11). Our results demonstrate that RUNX3 also directly influences the cell cycle machinery by inducing p21 expression in response to TGF-β. The ability to induce p21 expression and to inhibit cell growth is essential for the tumor suppressor activity of p53. The TGF-β signaling cascade also induces p21 gene expression. Therefore, our finding that RUNX3 directly regulates p21 expression in response to TGF-β stimulation suggests that the tumor suppressor activity of RUNX3 is at least partly associated with its ability to induce p21 expression. This claim is further supported by the observation that the RUNX3-R122C mutant, which lacks tumor suppressor activity, failed to induce p21 expression in response to TGF-β1. Reduced DNA- and SMAD-binding affinity appears to be responsible for this loss of activity. It is not known whether the loss of transactivation activity of RUNX3-R122C is specific to the p21 promoter. However, the R122C mutation may not abolish all target gene expression since the runt mutant in Drosophila that lacks all DNA-binding activity still retains significant in vivo activity (40).
It is worth mentioning that p53 controls the cell cycle by transcriptionally activating p21 expression, which then inhibits pRb phosphorylation. Interestingly, RUNX3, which is frequently inactivated in human cancers, controls the cell cycle by a similar mechanism, although the stimulating signals differ, namely, cellular stress for p53 and TGF-β stimulation for RUNX3. Well-known tumor suppressors such as pRb, p53, p16INK4a, and p14ARF directly influence the cell cycle machinery, and RUNX3 is a new addition to this list.
Acknowledgments
This work was supported by Creative Research grant R16-2003-002-01001-0 from the Korea Science and Engineering Foundation to S.-C. Bae. Y. Ito was supported by the Agency for Science Technology and Research (A*STAR), Singapore, to Y. Ito.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.25.18.8097-8107.2005
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1234316?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Receptor-activated transcription factors and beyond: multiple modes of Smad2/3-dependent transmission of TGF-β signaling.
J Biol Chem, 300(5):107256, 02 Apr 2024
Cited by: 2 articles | PMID: 38569937 | PMCID: PMC11063908
Review Free full text in Europe PMC
An Imbalance in Histone Modifiers Induces tRNA-Cys-GCA Overexpression and tRF-27 Accumulation by Attenuating Promoter H3K27me3 in Primary Trastuzumab-Resistant Breast Cancer.
Cancers (Basel), 16(6):1118, 11 Mar 2024
Cited by: 1 article | PMID: 38539453 | PMCID: PMC10968641
Differentially expressed microRNAs in aneuploid preimplantation blastocysts: a systematic review.
Front Reprod Health, 6:1370341, 14 Mar 2024
Cited by: 0 articles | PMID: 38550247 | PMCID: PMC10973143
Review Free full text in Europe PMC
RUNX3 inhibits KSHV lytic replication by binding to the viral genome and repressing transcription.
J Virol, 98(2):e0156723, 10 Jan 2024
Cited by: 0 articles | PMID: 38197631 | PMCID: PMC10878072
Aberrant Upregulation of RUNX3 Activates Developmental Genes to Drive Metastasis in Gastric Cancer.
Cancer Res Commun, 4(2):279-292, 01 Feb 2024
Cited by: 1 article | PMID: 38240752 | PMCID: PMC10836196
Go to all (134) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - Z35278
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Growth regulation of gastric epithelial cells by Runx3.
Oncogene, 23(24):4330-4335, 01 May 2004
Cited by: 38 articles | PMID: 15156189
Review
Tumor suppressor, AT motif binding factor 1 (ATBF1), translocates to the nucleus with runt domain transcription factor 3 (RUNX3) in response to TGF-beta signal transduction.
Biochem Biophys Res Commun, 398(2):321-325, 25 Jun 2010
Cited by: 27 articles | PMID: 20599712
Possible involvement of RUNX3 silencing in the peritoneal metastases of gastric cancers.
Clin Cancer Res, 11(18):6479-6488, 01 Sep 2005
Cited by: 39 articles | PMID: 16166423
Suppression of tumorigenesis and induction of p15(ink4b) by Smad4/DPC4 in human pancreatic cancer cells.
Clin Cancer Res, 8(11):3628-3638, 01 Nov 2002
Cited by: 43 articles | PMID: 12429655




