Abstract
Free full text

Bacterial Cell Surface Display of Organophosphorus Hydrolase for Selective Screening of Improved Hydrolysis of Organophosphate Nerve Agents
Abstract
Organophosphorus hydrolase (OPH) is a bacterial enzyme that has been shown to degrade a wide range of neurotoxic organophosphate nerve agents. However, the effectiveness of degradation varies dramatically, ranging from highly efficient with paraoxon to relatively slow with methyl parathion. Sequential cycles of DNA shuffling and screening were used to fine-tune and enhance the activity of OPH towards poorly degraded substrates. Because of the inaccessibility of these pesticides across the cell membrane, OPH variants were displayed on the surface of Escherichia coli using the truncated ice nucleation protein in order to isolate novel enzymes with truly improved substrate specificities. A solid-phase top agar method based on the detection of the yellow product p-nitrophenol was developed for the rapid prescreening of potential variants with improved hydrolysis of methyl parathion. Two rounds of DNA shuffling and screening were carried out, and several improved variants were isolated. One variant in particular, 22A11, hydrolyzes methyl parathion 25-fold faster than does the wild type. Because of the success that we achieved with directed evolution of OPH for improved hydrolysis of methyl parathion, we believe that we can easily extend this method in creating other OPH variants with improved activity against poorly degraded pesticides such as diazinon and chlorpyrifos and nerve agents such as sarin and soman.
Organophosphates belong to a class of highly toxic neurotoxins that are commonly used as insecticides and chemical warfare agents. The continuous use of organophosphates in very large quantities throughout the world and their potential neurotoxicity to humans have led to the development of efficient and safe remediation strategies to deal with their wide dispersal in the ecosystem (3). Enzymatic degradation by organophosphorus hydrolase (OPH) has received considerable attention since it provides the possibility of both environmentally friendly and in situ detoxification (5, 6, 14). Identical opd genes coding for OPH were found in two soil microorganisms, Pseudomonas diminuta MG (18) and Flavobacterium sp. (13).
Although OPH hydrolyzes a wide range of organophosphates, the effectiveness of hydrolysis varies dramatically. For example, some widely used organophosphorus insecticides such as methyl parathion, chlorpyrifos, and diazinon are hydrolyzed 30 to 1,000 times slower than is the preferred substrate, paraoxon (5). This reduction in catalytic rate is due to the unfavorable interaction of these substrates with the active sites involved in catalysis and/or structural functions (10). Site-directed mutagenesis has been applied to the various residues involved in the active sites, resulting in OPH mutants with improved catalytic characteristics against paraoxon, sarin, and soman (4, 10, 11, 22). Since the kinetic characteristics of OPH can be altered with relatively few amino acid substitutions, it may be possible to create variants with improved activity against other poorly degraded insecticides or chemical warfare agents. Although the three-dimensional structure of OPH has been elucidated previously (1), identifying all the amino acids responsible for substrate specificity and those that might give rise to extended specificity remains an overwhelming challenge. To this end, in vitro directed evolution is perhaps the most useful way to sample this sequence flexibility in a simple and rapid fashion (16, 20).
Although improved variants can be evolved by screening a library of cells with OPH expressed intracellularly, this method is inadequate for organophospates, which are not readily taken up by cells (17). In order to provide free access to substrates and to screen for variants with truly improved kinetic properties, we have developed a generalized selection scheme using a surface-display OPH library for the isolation of improved variants. Enzymes with up to 25-fold-higher activity were generated after only two rounds of screening. With this approach, it is possible that novel OPH variants with improved activity against other organophosphorus pesticides such as malathion, chlorpyrifos, and diazinon and chemical warfare agents such as sarin and soman can be similarly created.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strain XL1-Blue (recA1 endA1 gyrA96 thi-1 hsdR17 [rK− mK+] supE44 relA1 lac [F′ proAB lacIq ZΔM15 Tn10 (Tetr)]) was used in all experiments. Plasmid pOPK132 harboring the lpp-ompA-opd fusion was used as the source of the opd gene (17). A low-copy-number plasmid, pK184 (9), was used for intermediate cloning. Plasmid pINCOP containing the truncated ice nucleation protein (INPNC) anchor was used to display OPH on the cell surface of E. coli (19).
DNA shuffling.
The 1.1-kb opd gene from pOPK132 was amplified using primers 5′-AATTTCGGATCCCGGGATGC-3′ and 5′-GGGGAATTCAAGCTTCCAAAAAAAAGCCCGCTCATTAGGCGGGCTGCGTCATACGCCCAAGGTCGGTGACAG-3′. The amplified fragments were digested with restriction enzymes BamHI and HindIII and inserted into the vector pK184 to generate pKOP, which was used as a template for DNA shuffling. The procedures for DNA shuffling were performed as described previously (20). The opd fragments for the shuffling reactions were obtained by PCR using two primers, DB1 (5′-TGCGGGCCTCTTCGCTATTA-3′) and UH1 (5′-CCCCAGGCTTTACACTTTAT-3′), which flank the opd gene by approximately 100 bp. Following purification with the Wizard PCR purification kit (Promega, Madison, Wis.), the 1.3-kb fragments were digested with 0.01 U of DNase I (Boehringer Mannheim) at 15°C for 8 min. The reaction was stopped by heating the reaction mixture at 90°C for 10 min. DNA fragments of less than 50 bp were isolated from a 2% agarose gel with the DEAE cellulose membrane and subsequently purified by extraction with phenol and chloroform. Approximately 2 μg of DNA was mixed and reassembled in 100 μl of a primerless PCR mixture with the EasyStart (Molecular Bio-Products) mix and Taq DNA polymerase (Promega). Conditions for PCR were as follows: 5 min at 94°C and 50 cycles of 1 min at 94°C, 1 min at 45°C, and 1.5 min at 72°C, followed by 10 min at 72°C (12). After a 1:40 dilution of the primerless PCR products, DNA amplification was carried out in the presence of the M13/pUC sequencing primer and the M13/pUC reverse sequencing primer. A PCR program of 5 min at 95°C and 35 cycles of 1 min at 95°C, 1 min at 53°C, and 1.5 min at 72°C, followed by 10 min at 72°C, was used. The 1.1-kb amplification product was recovered using the Gene Clean II kit (QBIOGENE), digested with BamHI and HindIII, and subcloned into similarly digested pINCOP to generate a library of OPH variants fused to the INPNC anchor. The resulting plasmids were used to transform E. coli XL1-Blue by the CaCl2 method.
Screening of OPH variants.
A solid-phase top agar plate assay based on the formation of a yellow product (p-nitrophenol) from methyl parathion was developed for the selection of OPH with an improved hydrolysis rate. Single colonies of transformed E. coli were streaked onto M9 plates supplemented with 0.1% tryptone, 0.05% yeast extract, 0.1% Casamino Acids, 0.2% glucose, 100 μM ampicillin, and 10 μM CoCl2 to promote growth and OPH activity without increasing the background color. After a 48-h incubation at 30°C, a thin layer of 0.7% agarose containing 50 mM phosphate-citrate buffer (pH 8.0) and 0.5 mM methyl parathion was laid over the mixture. After 1 h of incubation at 37°C, colonies were selected based on the intensity of the yellow color from the hydrolyzed product p-nitrophenol. Any clones that appeared to have a larger yellow halo than that of the wild-type OPH were selected for rescreening. For each potential clone, cells were removed directly from the agar plate, resuspended in 200 μl of 50 mM phosphate-citrate buffer (pH 8.0) containing 0.5 mM methyl parathion and 10% methanol, and rescreened using a 96-well microplate reader (Bio-Rad 3550-UV) at 37°C. Hydrolysis of methyl parathion was measured at 405 nm for 30 min. The initial rate was divided by the turbidity measured at 595 nm of each well to provide a measure of specific activity. Clones with activity at least two times higher than that of the wild type were selected. The selected clones were cultured in Luria-Bertani medium for 45 h at 30°C, and the activity was measured using a Beckman model DU-640 spectrophotometer at 37°C. Cultured variants were resuspended in 50 mM phosphate-citrate buffer (pH 8.0) containing 0.5 mM methyl parathion and 10% methanol to an optical density at 600 nm (OD600) of 0.2. Whole-cell activity was expressed as the initial change in absorbance at 410 nm due to p-nitrophenol formation.
DNA sequencing.
Variants were sequenced in both forward and reverse directions with the automated dye terminator kit and an Applied Biosystems 282A sequencer. Small-scale isolation of plasmid DNA for sequencing was carried out with the Wizard Plus Minipreps DNA purification system (Promega).
Immunoblotting.
One milliliter of cells (OD600 = 1) was sonicated for 2 min. After the debris was removed by centrifugation, 100 μl of the supernatant was combined with 20 μl of disruption buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2% [wt/vol] sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 0.05% [wt/vol] bromophenol blue) and boiled for 10 min. Twenty microliters of samples was then electrophoresed through SDS-12.5% (wt/vol) polyacrylamide gels prior to immunoblotting analysis. Immunoblotting was performed using a Bio-Rad Immun-Blot GAR-AP kit (Bio-Rad, Hercules, Calif.). Antisera against INPNC were used as the first antibody (19). Prestained low-range protein markers (Bio-Rad) were utilized for estimation of protein molecular weights.
Purification of intracellularly expressed variants.
To determine the specific activities of the OPH variants, the opd genes were excised with BamHI and HindIII and ligated into pPROEXHTa (GIBCO BRL) to generate pHOP. This plasmid allowed the intracellular expression of the OPH variant as an N-terminal fusion to a hexahistidine tag (His6). For the purification of the variants, plasmid-bearing bacterial cultures were grown in 200 ml of Terrific broth supplemented with ampicillin to a final concentration of 100 μg/ml at 30°C and induced with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an OD600 of 1.0. Cell pellets were resuspended in 30 ml of binding buffer (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, and 1% Triton X-100) with 0.1 mM CoCl2 added. Benzonase (Novagen) and the protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride (Calbiochem) were added at 37°C for 20 min. Cells were cooled to 0°C and passed through a French pressure cell (15,000 to 20,000 lb/in2). The solution was centrifuged for 10 min at 20,000 × g. The resulting solution was added to a His·Bind Quick column (Novagen), which was prewashed with the binding buffer. The column was washed with 25 ml of binding buffer, followed by 25 ml of washing buffer (50 mM phosphate [pH 8.0], 500 mM NaCl, 10% glycerol, and 25 mM imidazole). Enzymes were eluted with 5 ml of elution buffer (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, 5% glycerol) containing 200 mM imidazole. All eluted fractions were tested for OPH activity and protein purity by SDS-polyacrylamide gel electrophoresis. Fractions with the highest OPH activity were pooled, dialyzed in 50 mM phosphate-citrate buffer (pH 8.0) supplemented with 0.1 mM CoCl2, and used for specific activity analyses.
OPH assay.
Purified variants were resuspended in 1 ml of 50 mM phosphate-citrate buffer (pH 8.0) containing 0.5 mM methyl parathion and 10% methanol in 1.5-ml disposable methacrylate cuvettes (Fisher). The change in absorbance (410 nm, e410 = 16,500 M−1 cm−1 for p-nitrophenol) was measured with a Beckman DU-60 spectrophotometer for 5 min at 37°C. All assays were preformed in triplicate. Enzyme concentrations were determined according to the Bradford method (Bio-Rad) using bovine serum albumin as the standard. Specific activities were expressed as units (micromoles of p-nitrophenol produced per minute) per milligram of protein.
RESULTS AND DISCUSSION
Experimental strategy for evolving OPH.
Methyl parathion, which is used extensively on cotton, corn, peaches, wheat, barley, soybeans, and rice throughout the world and is poorly degraded by OPH, was chosen as the target (15). The initial task was to generate a library of OPH variants with improved activity against methyl parathion. Plasmid pINCOP, a pUC18 derivative carrying the inpnc-opd fusion gene on an EcoRI-HindIII fragment, was used to target OPH to the surface of E. coli. For this study, only the opd gene coding for OPH was subjected to DNA shuffling.
Directed evolution requires the screening of a large number of variants, since most mutations or recombinations are either neutral or deleterious. In this regard, colorimetric (visual) assays are convenient for rapid and sensitive screening. A solid-phase top agar plate assay based on the formation of a yellow product (p-nitrophenol) from methyl parathion (Fig. (Fig.1A)1A) was developed for the selection of OPH with an improved hydrolysis rate. Transformants were grown on M9 plates, and a thin layer of agarose containing 0.5 mM methyl parathion was then laid on top of the plate. The intensity of the yellow color formed after 1 h was used as a preliminary screen for improved activity (Fig. (Fig.1B).1B). All potential clones identified were rescreened in a 96-well microplate assay using citrate-phosphate buffer (pH 8) containing 0.5 mM methyl parathion. Because the methyl parathion concentrations (0.5 mM) used in the screening assay are comparable to the reported Km value of 0.8 mM, the assay should favor the selection of variants with improved kcat rather than improved kcat/Km.
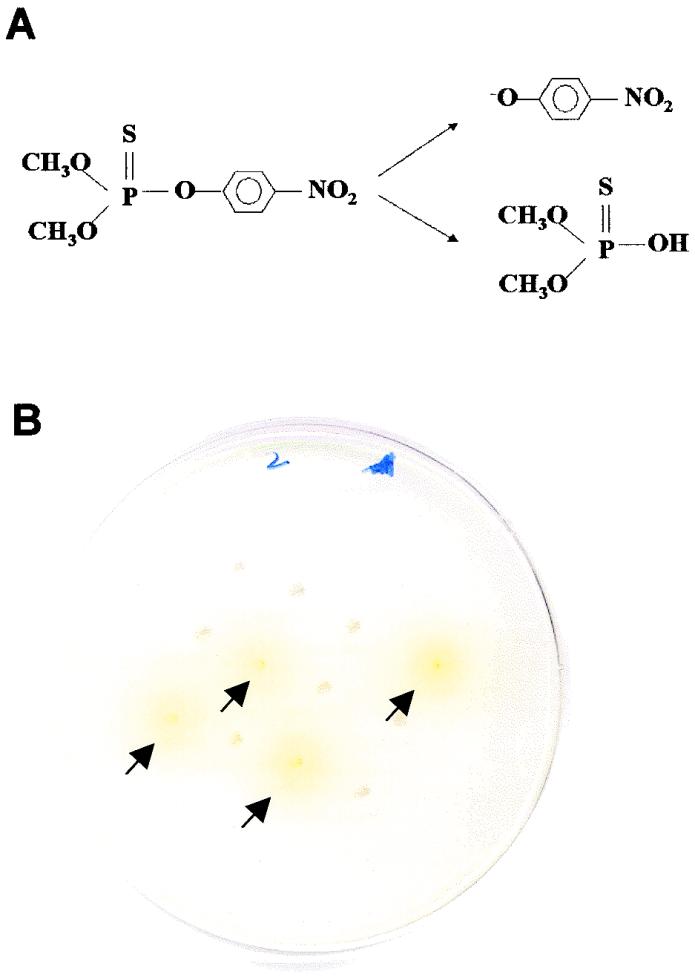
Principles of top agar prescreening assays for E. coli colonies displaying OPH variants. (A) Hydrolysis of methyl parathion by OPH into p-nitrophenol (a yellow product) and dimethylthiophosphoric acid. (B) Selection of potential variants by comparing the rates of p-nitrophenol (yellow) formation within 1 h. Colonies with high intensities of yellow color are indicated by arrows.
DNA shuffling and selection.
Following random mutagenesis of a 1.1-kb opd fragment by DNA shuffling, mutated DNA fragments were subcloned into the INPNC surface display vector, pINCOP. The mutant library was transformed into E. coli XL1-Blue and subjected to the top agar prescreening assay. Of the 1,800 colonies screened on supplemented M9 plates in the first generation of directed evolution, 700 potential variants showed higher levels of yellow color than the wild-type OPH. All 700 clones were rescreened in a 96-well microplate assay using citrate-phosphate buffer (pH 8) containing 0.5 mM methyl parathion. The turbidity of the cell suspension was used to estimate cell concentration. The rate of product appearance (yellow color) was normalized to cell concentration to provide a measure of specific activity. Those colonies with at least twofold-higher specific activity were tested again with degradation of methyl parathion in liquid assays. Four variants in particular (2H2, 2F6, 5A6, and 6D4) demonstrated a two- to sevenfold improvement in their abilities to hydrolyze methyl parathion, as indicated by an increase in whole-cell activity (Fig. (Fig.2A2A).
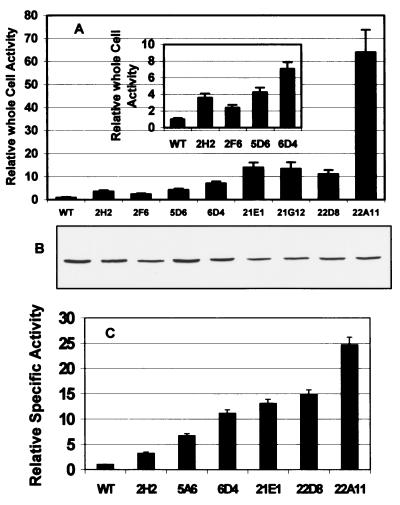
Comparisons of OPH activities and expression of improved variants and the wild-type (WT) OPH. (A) Relative whole-cell activities of the surface-displayed variants. (B) Western blot of INPNC-OPH fusion of each variant. (C) Specific activities of selected purified variants. Rates are normalized to that of the wild-type OPH.
In the second generation, all four positive variants in the first round were used as parental genes for DNA shuffling. Over 3,300 clones were screened by the top agar assay, and 170 clones with higher levels of yellow color than that of the best variant from the first round, 6D4, were selected. Only four variants (21E1, 21G12, 22D8, and 22A11) showed at least 50% higher activity than 6D4 when screened with 0.5 mM methyl parathion using the 96-well microplate assay, while the majority of the colonies were either not active or had much lower activity than did 6D4 (Fig. (Fig.2A).2A). One clone in particular, 22A11,was 64 times more active against methyl parathion than the wild-type OPH.
The expression levels of different variants were probed by Western blot analysis with the INPNC antisera (Fig. (Fig.2B).2B). All positive variants showed essentially the same level of expression relative to that of the wild-type OPH, indicating that changes in the activities of the variants were not the result of changes in protein expression.
Selected variants from the first and second rounds (2H2, 5A6, 6D4, 21G12, 22D8, and 22A11) were excised from INPNC and expressed as a His-tagged fusion in the cytoplasm. Individual variants were purified using a Ni-resin column, and the catalytic properties against methyl parathion were determined. Similar to results with whole-cell assays, the specific activities of the purified OPH variants were all higher than that of the wild-type OPH (Fig. (Fig.2C).2C). Although the extent of improvement was different, the trend was well correlated with the whole-cell screening assay. The best variant, 22A11, was 25-fold more active than the wild type. Thus, surface display of enzymes presents a new strategy for the isolation of improved variants based on catalytic turnover without any limitation on substrate accessibility.
Sequence and structural analysis of evolved OPHs.
The molecular structure of OPH has been determined in the presence of a nonhydrolyzable inhibitor, diethyl 4-methylbenzyl phosphonate (21), and Fig. Fig.33 shows the side views of the wild-type OPH with this inhibitor (green). Residues in the small (red; Gly60, Ile106, Leu303, and Ser308), large (purple; His254, His257, Leu271, and Met317), and leaving group (yellow; Trp131, Phe132, Phe306, and Tyr309) binding pockets are highlighted (21). For most substrates with a nitrophenolate as the leaving group, the small and large binding pockets could accommodate a variety of substituents attached to the phosphorus center, including methyl, ethyl, isopropyl, and phenyl groups (7, 8). However, previous studies with the substrate specificity of OPH revealed that the activities for dimethyl-substituted substrates are significantly lower than those for diethyl-substituted substrates (2, 8). This observation suggests that the two ethoxy groups contribute greatly to the binding interactions with the substrates. Because methyl parathion is a smaller substrate containing two methoxy groups, mutations toward higher activity for methyl parathion are likely to favor the formation of a smaller binding pocket.
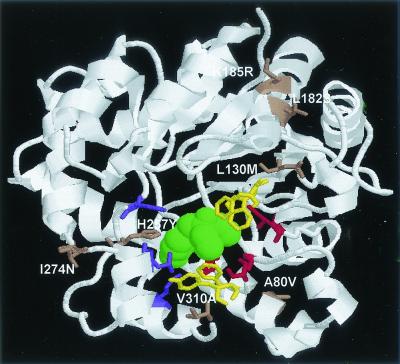
Structure of OPH in the presence of a nonhydrolyzable inhibitor, diethyl 4-methylbenzyl phosphonate (green). The residues for the large (purple), small (red), and leaving group (yellow) binding pockets are highlighted. Substitutions found in first- and second-round variants are indicated (brown).
Amino acid substitutions for each OPH variant are shown in Table Table1.1. In the first round of shuffling, the best variant, 6D4, contained a single mutation at histidine 257, which is situated in the larger binding pocket of OPH. The change of histidine into a bulkier tyrosine residue has been shown previously to reduce the size of the larger binding pocket and favor the binding of smaller substituents (23). The same observation is true in this case for methyl parathion. Other variants, 2F6 and 5A6, contained mutations such as I274N and V310A that are within 10 Å of, but not directly located in, the active site. It is difficult to comment on how this substitution may contribute to the improved activity against methyl parathion and the other substrates. However, it is clear that this important substitution will be impossible to predict solely based on structural information. This is precisely what makes directed evolution such a powerful strategy because of its ability to identify distal mutations that involve subtle, long-range interactions.
TABLE 1.
Amino acid substitutions and silent mutations of the variants
| Variant | Amino acid substitutions and silent mutations |
|---|---|
| First round | |
    2H2 2H2 | A80V, S365P |
    2F6 2F6 | L182S, V310A |
    5A6 5A6 | L87L(TTG→CTG), 1274N |
    6D4 6D4 | H257Y, A364A(GCG→GCA) |
| Second round | |
    21E1 21E1 | L87L, H257Y, I274N, S365P |
    21G12 21G12 | L87L, S218S(TCC→TCT), H257Y, I274N, S365P |
    22D8 22D8 | L87L, L130M, H257Y, I296I(ATC→ATT), I274N, A364A |
    22A11 22A11 | A14T, A80V, L87L, L185R, H257Y, I274N, T350T(ACT→ACC), A364A |
Variants from the second round were obtained by shuffling the first-round variants as the parental genes and were selected based on improved activity compared to the best variant identified in the first round, 6D4. Not surprisingly, these improved variants contained mutations accumulated from the first round as well as several new mutations. Specifically, all improved variants contained the I274N and H257Y double mutations, indicating their additive effects in improving the hydrolysis of methyl parathion. The additional substitution of L130M in 22D8 appears to be neutral, since no significant changes in specific activity were observed in comparison with those of variants containing only the I274N and H257Y double mutations (21E1). However, with the additional substitution of K185R, the best variant (22A11) was 1.7 times more active than other second-round variants and 25-fold more active against methyl parathion than the wild-type OPH, indicative of the importance of this key mutation.
With the success that we achieved with directed evolution of OPH for improved hydrolysis of methyl parathion, we believe that we can easily extend this method in creating OPH variants with improved activity against other commonly used but poorly degraded insecticides such as diazinon and chlorpyrifos and against chemical warfare agents such as sarin and soman.
Acknowledgments
This work was supported by a grant from the USDA (99-35102-8597).
We thank Mark Shimazu for constructing the plasmid pINCOP.
REFERENCES
Articles from Applied and Environmental Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aem.68.4.2026-2030.2002
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc123893?pdf=render
Citations & impact
Impact metrics
Article citations
Bacterial Membrane Vesicles for In Vitro Catalysis.
Bioengineering (Basel), 10(9):1099, 20 Sep 2023
Cited by: 1 article | PMID: 37760201 | PMCID: PMC10525882
Review Free full text in Europe PMC
Degradation strategies of pesticide residue: From chemicals to synthetic biology.
Synth Syst Biotechnol, 8(2):302-313, 23 Mar 2023
Cited by: 4 articles | PMID: 37122957 | PMCID: PMC10130697
Improving the specificity of organophosphorus hydrolase to acephate by mutagenesis at its binding site: a computational study.
J Mol Model, 27(6):164, 10 May 2021
Cited by: 0 articles | PMID: 33970322
Enzymatic Bioremediation of Organophosphate Compounds-Progress and Remaining Challenges.
Front Bioeng Biotechnol, 7:289, 08 Nov 2019
Cited by: 28 articles | PMID: 31781549 | PMCID: PMC6856225
Review Free full text in Europe PMC
Synthesis and application of magnetic molecularly imprinted polymers in sample preparation.
Anal Bioanal Chem, 410(17):3991-4014, 12 Apr 2018
Cited by: 16 articles | PMID: 29651522
Review
Go to all (72) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Altering the substrate specificity of organophosphorus hydrolase for enhanced hydrolysis of chlorpyrifos.
Appl Environ Microbiol, 70(8):4681-4685, 01 Aug 2004
Cited by: 53 articles | PMID: 15294802 | PMCID: PMC492387
Mutagenesis of organophosphorus hydrolase to enhance hydrolysis of the nerve agent VX.
Biochem Biophys Res Commun, 279(2):516-519, 01 Dec 2000
Cited by: 32 articles | PMID: 11118318
Functional analysis of organophosphorus hydrolase variants with high degradation activity towards organophosphate pesticides.
Protein Eng Des Sel, 19(3):99-105, 19 Jan 2006
Cited by: 24 articles | PMID: 16423845
Rational design of organophosphorus hydrolase for altered substrate specificities.
Chem Biol Interact, 119-120:211-223, 01 May 1999
Cited by: 29 articles | PMID: 10421455
Review





