Abstract
Free full text

Structural insight into the complex formation of latent matrix metalloproteinase 2 with tissue inhibitor of metalloproteinase 2
Abstract
Matrix metalloproteinases (MMPs) are a family of multidomain enzymes involved in the physiological degradation of connective tissue, as well as in pathological states such as tumor invasion and arthritis. Apart from transcriptional regulation, MMPs are controlled by proenzyme activation and a class of specific tissue inhibitors of metalloproteinases (TIMPs) that bind to the catalytic site. TIMP-2 is a potent inhibitor of MMPs, but it has also been implicated in a unique cell surface activation mechanism of latent MMP-2/gelatinase A/type IV collagenase (proMMP-2), through its binding to the hemopexin domain of proMMP-2 on the one hand and to a membrane-type MMP activator on the other. The present crystal structure of the human proMMP-2/TIMP-2 complex reveals an interaction between the hemopexin domain of proMMP-2 and the C-terminal domain of TIMP-2, leaving the catalytic site of MMP-2 and the inhibitory site of TIMP-2 distant and spatially isolated. The interfacial contact of these two proteins is characterized by two distinct binding regions composed of alternating hydrophobic and hydrophilic interactions. This unique structure provides information for how specificity for noninhibitory MMP/TIMP complex formation is achieved.
Matrix metalloproteinases 2 and 9 (MMP-2 and MMP-9) (also termed gelatinase A and B or 72-kDa and 92-kDa type IV collagenases, respectively) distinguish themselves from other secreted MMPs in that their latent proenzyme form can make a complex with tissue inhibitor of MMP (TIMP) (1–4). This complex has been proposed to facilitate a unique activation mechanism of the gelatinase A on the cell surface. According to the current central paradigm, which has been studied mainly for latent MMP-2/gelatinase A/type IV collagenase (proMMP-2), TIMP-2 first forms a complex with proMMP-2 by binding to its hemopexin domain, after which the complex localizes to the cell surface where it binds to the active site of a membrane-type MMP 1 (MT1-MMP) molecule (5–8). This ternary proMMP-2/TIMP-2/MT1-MMP complex then facilitates the activation of its proMMP-2 by another MT1-MMP molecule. A large body of data shows that this complex is entirely different from the inhibitory complex of TIMP-2/active MMP-2. It is formed between the C-terminal domain of the inhibitor and the C-terminal hemopexin of MMP-2, so that both molecules maintain their proteolytic and inhibitory properties, respectively (9, 10). These noninhibitory complexes between progelatinases and TIMPs are restricted to proMMP-2 and TIMP-2, TIMP-3, or TIMP-4 on the one hand and to MMP-9 and TIMP-1 on the other. This specificity has been addressed in several earlier studies, and sequence elements on both inhibitor and proteinase critical for the specific interaction have been identified. TIMP-2 has a negatively charged C terminus, which differs from that of TIMP-1. This C terminus has been suggested to mediate specificity and kinetics of the complex formation (11–13). In contrast, the hemopexin domain of MMP-2 features a characteristic pattern of positive side chains, which by site-directed mutagenesis experiments have been shown to be involved in the interaction with TIMP-2 (14).
In recent years, structural information has been compiled on many of the key features in MMP biochemistry. Crystal structures for isolated domains of several MMPs have revealed the architecture of the catalytic site and also the structure of the C-terminal hemopexin domain (15–21). Three-dimensional structures containing the prodomain are solved, revealing how MMPs are kept in latent form and how activation may occur (19, 22). Two structures of full-length MMPs showing the topology of the multidomain arrangement have been determined (22, 23). The inhibition of MMPs by TIMPs is addressed in crystal structures of free TIMPs, as well as in complex with catalytic domains of different MMPs (24–27). To shed light on the interaction of proMMP-2 with TIMP-2 and how their complex could possibly interact with an MT-MMP at the cell surface, we have determined the crystal structure of the proMMP-2/TIMP-2 complex.
Methods
Crystallization and Data Collection.
ProMMP-2 (Glu-385 → Ala mutant) and TIMP-2 were prepared essentially as described for crystallization of the free proteins (22, 27). The complex was formed with a stoichiometric excess of TIMP-2 and purified by anion exchange HPLC where it elutes at a higher salt concentration than free TIMP-2 or MMP-2. The proMMP-2/TIMP-2 complex was crystallized in hanging drops at a concentration of 11 mg/ml at 4°C in 0.2 M Li2SO4, 0.2 mM reduced/0.2 mM oxidized glutathione, 4% polyethylene glycol 4000, and 0.1 M ammonium acetate buffer, pH 6.25. Poorly shaped thin sticks appeared in 3–4 weeks and they were transferred into cryobuffer (reservoir solution with 30% polyethylene glycol 550 monomethyl ether) and incubated for 5 min before data collection. A data set to 3.1-Å resolution (Table (Table1)1) was collected from a single crystal at 100 K at the wiggler beamline BW7B at DORIS (European Molecular Biology Laboratory outstation, Deutsches Elektronen Synchrotron, Hamburg). The data set was evaluated and scaled by using denzo/scalepack (28). The anisotropic correction was performed by sfcheck (29).
Table 1
Crystallographic statistics
statistics
| Data collection | |
 Resolution range, Å Resolution range, Å | 12.94–3.1 |
 Total number of reflections Total number of reflections | 46,286 (475,848) (475,848) |
 Unique reflections Unique reflections | 46,264 |
 Completeness, % Completeness, % | 94.1 |
 Highest resolution shell (3.2–3.1) Highest resolution shell (3.2–3.1) | 70.8 |
 Rmerge, %* Rmerge, %* | 11 |
 I/σ I/σ | 10.26 |
| Refinement statistics | |
 Number of reflections used in refinement (working set) Number of reflections used in refinement (working set) | 43,921 |
 Number of reflections used in refinement (test set) Number of reflections used in refinement (test set) | 2,314 (5%) (5%) |
 Number of nonhydrogen atoms Number of nonhydrogen atoms | 12,934 |
 R factor, %† R factor, %† | 28.2 |
 Rfree factor, %‡ Rfree factor, %‡ | 33.5 |
 Overall B factor Å2 Overall B factor Å2 | 37.8 |
 rms bond length, Å rms bond length, Å | 0.009 |
 rms bond angle, ° rms bond angle, ° | 1.639 |
 I(h)
I(h) |/ΣhΣiI(h)i, where I(h)i is the ith measurement.
|/ΣhΣiI(h)i, where I(h)i is the ith measurement.  FO| − |FC
FO| − |FC /ΣFO for all reflections.
/ΣFO for all reflections.  FO| − |FC
FO| − |FC /ΣFO, calculated on the 5% of data excluded from refinement.
/ΣFO, calculated on the 5% of data excluded from refinement. Structure Determination and Refinement.
The crystals belonged to the orthogonal space group C2221 with cell dimensions a = 75.7 Å, b = 374.6 Å, c = 191.0 Å and contained two complexes per asymmetric unit. Position and orientation of the two proMMP-2 molecules per asymmetric unit was determined by molecular replacement with coordinates of the full-length proMMP-2 as a search model by using blanc (29). Four Zn2+ ions, excluded from the search model, appeared in the Fourier difference map to confirm the solution found. The TIMP-2 molecules were traced with fffear (version 0.9) (30) by using the structure of the human TIMP-2 as a search model. Two independent positions near the C-terminal domains of proMMP-2 molecules were found. Rigid body refinement with cns (31) shifted both molecules to their final positions.
Several cycles of manual rebuilding with o (32) and refinement with refmac (33) helped to resolve the AB loops for both molecules of TIMP-2 and to trace some missing residues in the hinge region of proMMP-2 but nevertheless the density for TIMP-2 molecules was not so well defined as for MMPs. For this reason the refinement was continued with REFMAC 5 by using the tls option (33) to take account for the anisotropic displacement of domains in the subunits of both proteins. In this procedure, molecules were divided in two domains to calculate T, L, and S displacement tensors for each domain. The refinement with eight TLS groups was performed in a cyclic manner, together with the remodeling step with o. This process provided a better defined electron density map for both TIMP-2 molecules and allowed finalization of the structures for both complexes.
Current Model.
The final refined model consists of residues 1–422 and 429–631 for both proMMP-2 molecules and residues 1–192 for both TIMP-2 molecules. There are two Zn2+ ions and one Ca2+ ion per proMMP-2 molecule and two sulfate ions in the region where the two proMMP-2 molecules meet through their second fibronectin type II domains. The Ca2+ ion, usually found in the tunnel of the MMP-hemopexin domains, was absent in both molecules. Structural changes are not likely to be the reason for this finding because distances between the main-chain oxygens of Asp residues coordinating Ca2+ ion in other structures remained virtually unchanged. Figures were prepared by using molscript (34), bobscript (35), RASTER3D (36), and grasp (37).
Results and Discussion
Overall Complex Organization.
The structure of the proMMP-2/TIMP-2 complex was solved with the molecular replacement technique. The asymmetric unit contains two complex pairs AC and BD, where A and B are proMMP-2 molecules and C and D are TIMP-2 molecules. The proMMP-2 molecules are organized into layers with close contacts between their fibronectin domains whereas TIMP-2 molecules are excluded from this dense MMP net and directed into the space between where sterical restrictions do not allow perfect crystal packing. The comparison of complexed proMMP-2 and TIMP-2 with their uncomplexed counterparts does not reveal any significant shifts or perturbations within the domains of both proteins. Superposition of molecules A and B from the complex with free proMMP-2 (22) gives rms values of 1.34 Å for 454 Cα atoms and 1.17 Å for 564 Cα atoms, respectively. The difference in rms is caused by a slight lateral shift of the hemopexin domain in molecule A compared with B, which might be explained by crystal packing.
In the case of TIMP-2, comparison of molecules C and D with free TIMP-2 (27) shows rms values of 1.25 Å for 166 Cα atoms and 1.09 Å for Cα atoms, respectively, and rms 1.37 Å for 178 Cα atoms between each other. The main difference is observed in the conformation of the β−hairpin AB loop that is not involved in the complex formation. In subunit C, this loop suffers from a steric clash with the corresponding loop of the symmetry-related molecule. The electron density at this location is rather weak and partially diffuse. The conformation of this hardly traceable loop is similar to that found in bovine TIMP-2 complexed with the catalytic domain of MT1-MMP, whereas in the D subunit the AB loop is determined clearly and shows a conformation that resembles that in the free TIMP-2 molecule. The arrangement of C-terminal loops surrounding the complex formation area does not show essential differences in the C subunit when compared with free TIMP-2 or an MT1-MMP-bound one. In the D subunit loop, GH has close crystallographic contact with a symmetry-related molecule. This causes a slight change in its conformation, but does not influence the formation of the complex.
Interaction between proMMP-2 and TIMP-2 is channeled through two contact areas, one comprising the GH loop of TIMP-2 bound to the fourth blade of the hemopexin domain of MMP-2 and the other being the C-terminal tail of TIMP-2 inserted between the third and fourth β-propeller blades (Fig. (Fig.1).1). The estimated common buried surface area between the two proteins is 2,489 Å2. The active sites of the proteinase and inhibitor are clearly separated in the complex, and neither the catalytic domain of MMP-2 nor the N-terminal domains of TIMP-2 interact.
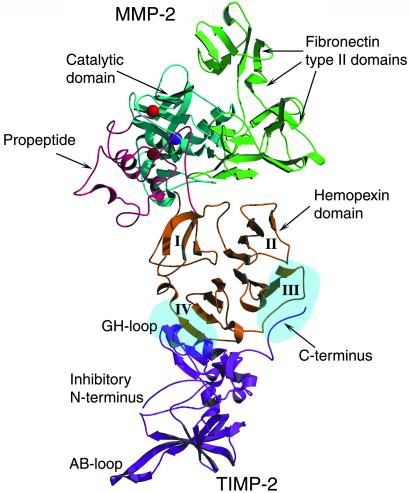
Structure of the proMMP-2/TIMP-2 complex. Overall conformation: the proteinase and inhibitor interact via their C-terminal domains. The catalytic site of MMP-2 and the inhibitory active site of TIMP-2 are turned away from each other. This topology excludes an inhibitory interaction between the proteinase and inhibitor and implies that both proteins remain fully functional in the complex. Catalytic and structural Zn2+ ions are colored red and Ca2+ ion purple. The β-propeller blades of the hemopexin domain are numbered from I to IV. Two light blue ellipsoids in blades III and IV indicate two areas of interaction between proMMP-2 and TIMP-2 molecules.
Surprisingly, the lack of any ions and particularly Ca2+ ion in the channel of hemopexin domain has not been accompanied with any dramatic changes in the structure of this domain. Putatively Ca2+ ion has been suggested to participate in the binding of various ligands, such as heparin and fibronectin, and also in the stabilization of the structure (18, 38), but it is not necessary for TIMP-2 binding to this domain (39). However, the stabilization function in the complex appears to be carried out by hydrophobic residues of each blade directed into the interblade area and by a disulfide bridge formed between two cysteines, Cys-440–Cys-631, connecting the first and fourth blades together and closing the ring. Also, structurally homologous hemopexin does not have any Ca2+ ions in the channel (40), suggesting that divalent cations are not crucial in maintaining the β-propeller structure.
ProMMP-2/TIMP-2 Interface.
Analysis of the contact area in the complex between TIMP-2 and proMMP-2 reveals an association through two areas of hydrophobic interactions surrounded by electrostatic contacts (Figs. (Figs.11 and and2).2). The hydrophobic interactions appear to contribute significantly to the stability of the complex. The main cluster of hydrophobic interactions is focused around Met-149 of TIMP-2, involving additionally Ile-136, Leu-147, Val-152, Phe-165, Trp-177, and Cys-133 and Cys-138, which form a disulfide bridge (Fig. (Fig.22 a and b). In the proMMP-2 molecule, this cluster is recognized by Ala-583, Tyr-607, Leu-609, Val-619, and, most significantly, Phe-621, which is in close proximity to the Met-149 residue of TIMP-2. The second hydrophobic contact is made by Phe-188 of TIMP-2 that is inserted into a pocket-like structure on the surface of proMMP-2 formed by Tyr-552, Phe-559 and Phe-573, Ala-580, and Trp-581 (Fig. (Fig.22 c and d).
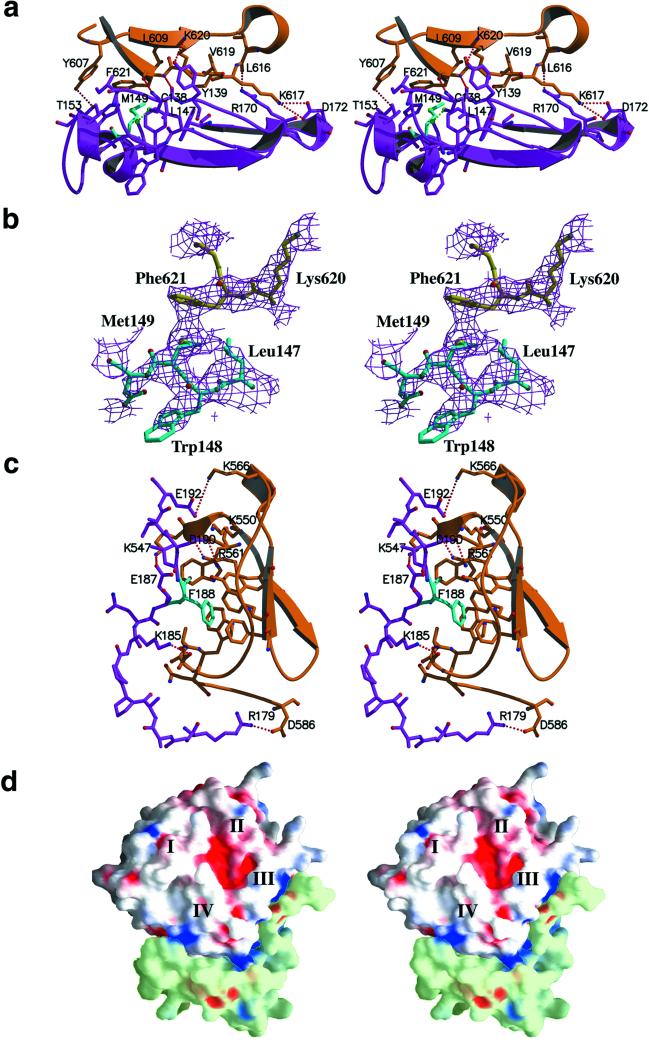
Contact areas in the proMMP-2/TIMP-2 complex. To clarify the details of binding areas, the orientation of the complex in a–c was turned on 180o compared with the so-called standard orientation presented in Fig. Fig.1.1. (a) Stereo view of the GH loop of TIMP-2 (magenta) in contact with the fourth blade of the hemopexin β-propeller of proMMP-2 (orange). Met-149 of TIMP-2, a central residue in the hydrophobic interaction, colored cyan, forms a close contact with Phe-621 of proMMP-2. Red dashed lines illustrate hydrogen bonds and salt bridges, and the green dashed line represents an S-S bridge formed by Cys-133–Cys-138. (b) Stereo view of the 2 Fo − Fc electron density map (contoured at 1.0 σ) with the model structure shows the hydrophobic contact between Met-149 (blue) molecules and Phe-621 (yellow). (c) The C terminus of TIMP-2 (magenta) is inserted between the third and fourth blades of the hemopexin domain (orange). Phe-188 (cyan) is buried in a hydrophobic groove formed by aromatic side chains on the surface of the hemopexin domain. Red dashed lines illustrate hydrogen bonds and salt bridges. (d) Stereo view of the grasp (37) representation of the contact areas between hemopexin domain of proMMP-2 (white) and C-terminal domain of TIMP-2 (light green). Molecular surfaces corresponding to positive potential are colored blue and areas with negative potential are shown in red. The negatively charged C tail of TIMP-2 is positioned in a positively charged cluster formed by residues of blade III of the hemopexin domain.
Both hydrophobic clusters are accompanied by polar and electrostatic contacts. Met-149 and Phe-621, together with the other hydrophobic residues, are tightly packed in a large extended cavity lined at one end by a salt bridge between Lys-617 and Asp-172 and a hydrogen bond between the main-chain oxygen of Leu-616 and the side chain of Arg-170.
The opposite end of this cavity is fixed by two H-bonded pairs: Tyr-607–Thr-153 and Lys-620–Tyr-139. The side chain of Lys-610 is directed to form a double positive cluster with Lys-620. The main-chain nitrogen and oxygen atoms of Phe-621 and Cys-138 interact and form two hydrogen bonds (Fig. (Fig.22a). This positively charged surrounding is in good agreement with the previous results of single and double mutations of Lys-617 and Lys-617/Lys-610 to alanine described to be important in the TIMP-2 interaction with the hemopexin domain of MMP-2 (14). Similar arrangement of interwoven electrostatic interactions accompanied by hydrogen bonding is located at the C terminus of TIMP-2. In previous structures of TIMP-2, both as free molecules and in an inhibitory complex, the 10 or 12 last C-terminal amino acid residues of the molecule were disordered and could not be traced in the electron density maps (26, 27). The structure of the complex with proMMP-2 shows a more fixed peptide chain (Figs. (Figs.11 and and22 c and d) that, although not perfectly resolved, allows an estimation of the contribution of the C terminus of TIMP-2 to the formation of the complex. Two salt bridges, Arg-179–Asp-586 and Lys-185–Asp-579, line the hydrophobically fixed Phe-188 from one side. Additionally, the side-chain nitrogen of Asn-582 forms a hydrogen bond to the main-chain oxygen of Trp-177. At the other side of this area, the negatively charged C terminus (net charge −4) faces toward a positively charged environment formed by Lys-547, Lys-550, Arg-561, and Lys-566. The major role of Arg-561 in this cluster has been shown earlier based on the results of site-directed mutagenesis (14). In the structure this region is characterized by at least three salt bridges: Lys-566 forms a salt bridge to Glu-192, Arg-561 to Asp-190, and Lys-547 to Glu-187 (Fig. (Fig.22c). A putative function for this region as a recognition motif for negatively charged C tail of TIMP-2 molecule has been suggested earlier and recently from the results of yeast two-hybrid analysis (39). This recognition mechanism greatly resembles the mechanism by which clathrin recognizes ligands (41). Clathrin is a multidomain protein containing a β-propeller domain, and ligands of clathrin are bound at the interface between two blades, very similar to the way the C terminus of TIMP-2 is bound into the hemopexin domain. Clathrin ligands have a 5-aa sequence motif, the Clathrin box, which contains two or three charged residues and hydrophobic side chains. This motif resembles the C terminus of TIMP-2 that may control the affinity of different TIMPs to latent gelatinases.
Specificity of the Interactions.
Analyses of the contact areas and sequences (Fig. (Fig.3)3) clearly indicate why TIMP-2, and, with lower affinity TIMP-3 and TIMP-4, but not TIMP-1 can bind to proMMP-2. This result emphasizes the pivotal role of the C terminus for the complex formation. TIMP-1 is C-terminally truncated and lacks not only the electrostatic interactions, but also the second area of hydrophobic interactions around Phe-188. An overlay of TIMP-2 in the complex structure with TIMP-1 (not shown) suggests that the first area of hydrophobic interactions is also compromised, because the central Met-149 is replaced by a threonine in TIMP-1. TIMP-4 (42) and TIMP-3 (43) are known to complex with proMMP-2, and our results suggest this is caused mainly by the C terminus, where TIMP-4 has three and TIMP-3 two of the four crucial residues conserved. TIMP-4 has a phenylalanine in position 189, but the last Asp-193 is replaced by a glutamine residue. TIMP-3 has that Asp conserved, but the phenylalanine is replaced with isoleucine, and the first aspartate is changed to asparagine. The hydrophobic contacts with the fourth blade of the hemopexin domain probably do not exist for TIMP-3 or TIMP-4, because Met-149 is also replaced in those by threonine.

Sequence alignment of human TIMPs and MMPs. Hydrophobic residues stabilizing the complex between TIMP-2 and MMP-2 are colored blue and green, respectively, representing the two separate contact areas. The polar and charged residues of the two areas are colored yellow and red. Residues that are predicted to participate in hydrophilic interactions, but not traceable in our model, are boxed.
Sequence comparison reveals the specific contribution of MMP-2 for formation of the complex. Some key residues are unique for MMP-2 and, most importantly, they are not found in MMP-9 (Fig. (Fig.3).3). Leu-609, a part of the above-mentioned hydrophobic pocket, is replaced with Trp-678 in MMP-9, and by Phe or Tyr in nearly all of the other MMPs. Their aromatic side chains are too bulky for the contact area and they would cause steric problems in the complex. For residue Phe-621, the situation is the opposite. The aromatic Phe-621 in MMP-2 is replaced by the smaller Val-694 in MMP-9 (and MMP-11), whereas all other MMPs have polar side chains in that position. Ala-583 is unique for MMP-2, as other MMPs have Gly at that site. The electrostatic interactions (Fig. (Fig.2)2) are likewise specific for MMP-2. This applies mainly to the Lys-566–568 triplet and Lys-547, which are the backbone of the charged surface into which the C-terminal tail of TIMP-2 is bound. Asn-582, involved in a hydrogen bond, has no counterpart in many other MMPs, and Lys-617 is replaced by Leu in MMP-9.
Application for Activation Mechanism of ProMMP-2.
The orientation of the two molecules in the complex excludes an intramolecular interaction between the catalytic site and inhibitor. Thus, the proposed function (12) of the complex as an initial, rate-enhancing step for proteinase inhibition appears unlikely. Instead, the inhibitor probably has a docking or bridging function to connect MMP-2 to other MMPs by forming a ternary complex (Fig. (Fig.4).4). This hypothesis is central to our current understanding of physiological activation of MMP-2 where the proMMP-2/TIMP-2 complex is believed to inhibit MT-MMP and localize MMP-2 to the cell membrane where it is activated by membrane proteinases. However, in the case of the analogous proMMP-9/TIMP-1 complex such involvement in cell surface activation has not been found, but proMMP-9/TIMP-1/Stromelysin-1 soluble ternary complex has been observed in vitro (44), proving that the inhibitory N terminus of TIMP-1 is functional in the complex. A major unsolved question in this context is how activated MMP-2 is released from the ternary complex.
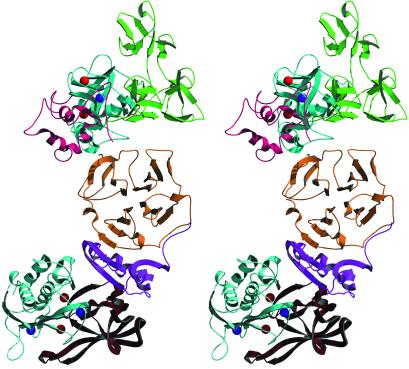
Stereo ribbon diagram of the hypothetical model of complex formed between proMMP-2, TIMP-2, and the catalytic domain of MT1-MMP. In the model shown, TIMP-2 is a hybrid with its C-terminal domain taken from the presented proMMP-2/TIMP-2 complex structure (magenta) and the N-terminal TIMP-2 half (red) is obtained from the model of the MT1-MMP/TIMP-2 inhibitory complex (Protein Data Bank code 1BUV) (26). Such combining was done because of the structural differences between complexed and uncomplexed forms of TIMP-2 molecules. Coloring for proMMP-2: the propeptide is pink, catalytic domain is blue, three fibronectin-like domains are green, and the hemopexin domain is yellow; for MT1-MMP: the catalytic domain is light blue.
MMP-2 is a drug target because its activity is often associated with excessive extracellular matrix turnover that e.g., is a prerequisite for tumor invasion and formation of metastasis or in inflammatory reactions. The design of specific antagonists as drug candidates has, however, proved difficult as all MMPs share a highly homologous catalytic domain. However, the interactions between TIMPs and proMMP-2 are highly unique and specific and, therefore, the structural data presented here may provide the basis for a novel approach to inhibitor design aimed at preventing the activation of proMMP-2 by blocking its interaction with TIMP-2.
Acknowledgments
We thank Margareta Andersson and Tiina Berg for technical assistance. We also thank Professors Ylva Lindqvist and Gunter Schneider for helpful discussions. We gratefully acknowledge access to synchrotron radiation at the European Molecular Biology Laboratory outstation, Deutsches Elektronen Synchrotron, Hamburg and thank the staff for help at the beamline. This work was supported by the Swedish Cancer Foundation, Medical Research Council, and European Community Research Contract QLG1-CT-2000-01131.
Abbreviations
| MMP | matrix metalloproteinase |
| proMMP-2 | latent MMP-2/gelatinase A/type IV collagenase |
| TIMP | tissue inhibitor of MMPs |
| MT-MMP | membrane-type MMP |
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1GXD).
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.102185399
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc124245?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The Role of Biomarkers in Predicting Outcomes of Anterior Cruciate Ligament Reconstruction: A Systematic Review.
Orthop J Sports Med, 12(10):23259671241275072, 07 Oct 2024
Cited by: 0 articles | PMID: 39380669 | PMCID: PMC11460236
Review Free full text in Europe PMC
Mapping Extracellular Protein-Protein Interactions Using Extracellular Proximity Labeling (ePL).
J Proteome Res, 23(10):4715-4728, 05 Sep 2024
Cited by: 1 article | PMID: 39238192
Extracellular proteolysis in cancer: Proteases, substrates, and mechanisms in tumor progression and metastasis.
J Biol Chem, 300(6):107347, 06 May 2024
Cited by: 4 articles | PMID: 38718867
Review
Matrix Metalloproteinases in the Periodontium-Vital in Tissue Turnover and Unfortunate in Periodontitis.
Int J Mol Sci, 25(5):2763, 27 Feb 2024
Cited by: 1 article | PMID: 38474009 | PMCID: PMC10932118
Review Free full text in Europe PMC
Structure and computation-guided yeast surface display for the evolution of TIMP-based matrix metalloproteinase inhibitors.
Front Mol Biosci, 10:1321956, 23 Nov 2023
Cited by: 2 articles | PMID: 38074088 | PMCID: PMC10702220
Review Free full text in Europe PMC
Go to all (116) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (2)
-
(1 citation)
PDBe - 1BUVView structure
-
(1 citation)
PDBe - 1GXDView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor.
EMBO J, 17(17):5238-5248, 01 Sep 1998
Cited by: 187 articles | PMID: 9724659 | PMCID: PMC1170851
E. coli expression of TIMP-4 and comparative kinetic studies with TIMP-1 and TIMP-2: insights into the interactions of TIMPs and matrix metalloproteinase 2 (gelatinase A).
Biochemistry, 41(50):15025-15035, 01 Dec 2002
Cited by: 42 articles | PMID: 12475252
Peptide from the C-terminal domain of tissue inhibitor of matrix metalloproteinases-2 (TIMP-2) inhibits membrane activation of matrix metalloproteinase-2 (MMP-2).
Matrix Biol, 30(7-8):404-412, 04 Aug 2011
Cited by: 8 articles | PMID: 21839835 | PMCID: PMC3191288
Insights into MMP-TIMP interactions.
Ann N Y Acad Sci, 878:73-91, 01 Jun 1999
Cited by: 107 articles | PMID: 10415721
Review





