Abstract
Free full text

Mutations associated with base excision repair deficiency and methylation-induced genotoxic stress
Abstract
The long-term effect of exposure to DNA alkylating agents is entwined with the cell's genetic capacity for DNA repair and appropriate DNA damage responses. A unique combination of environmental exposure and deficiency in these responses can lead to genomic instability; this “gene–environment interaction” paradigm is a theme for research on chronic disease etiology. In the present study, we used mouse embryonic fibroblasts with a gene deletion in the base excision repair (BER) enzymes DNA β-polymerase (β-pol) and alkyladenine DNA glycosylase (AAG), along with exposure to methyl methanesulfonate (MMS) to study mutagenesis as a function of a particular gene–environment interaction. The β-pol null cells, defective in BER, exhibit a modest increase in spontaneous mutagenesis compared with wild-type cells. MMS exposure increases mutant frequency in β-pol null cells, but not in isogenic wild-type cells; UV light exposure or N-methyl-N′-nitro-N-nitrosoguanidine exposure increases mutant frequency similarly in both cell lines. The MMS-induced increase in mutant frequency in β-pol null cells appears to be caused by DNA lesions that are AAG substrates, because overexpression of AAG in β-pol null cells eliminates the effect. In contrast, β-pol/AAG double null cells are slightly more mutable than the β-pol null cells after MMS exposure. These results illustrate that BER plays a role in protecting mouse embryonic fibroblast cells against methylation-induced mutations and characterize the effect of a particular combination of BER gene defect and environmental exposure.
Base excision repair (BER) (1) is considered the predominant DNA repair system in mammalian cells for eliminating small DNA lesions generated either exogenously or endogenously at DNA bases (1–3). Such DNA damage can be caused by exposure to environmental agents or by normal cellular metabolic processes that produce reactive oxygen species, alkylating molecules, and other reactive metabolites capable of modifying DNA. The first two steps of the mammalian BER pathway are removal of a damaged base residue by a DNA glycosylase and subsequent action of apurinic/apyrimidinic (AP) endonuclease. The product of these reactions is single-nucleotide gapped DNA with 5′ deoxyribosephosphate (dRP) and 3′ OH at the margins of the gap. A DNA β-polymerase (β-pol)-mediated DNA synthesis step fills the single-nucleotide gap (4–6), and the 5′ dRP group is removed by the dRP lyase activity of β-pol (7–11). DNA ligase I or III conducts the final, nick-sealing step in the pathway. Many laboratories have presented evidence in support of the roles of the enzymes noted here in single-nucleotide BER, mediated either by crude cell extracts or purified enzymes (3, 4, 12–16). Another important feature of single-nucleotide BER is that some of the enzymes in the pathway can interact with each other, forming a series of “subassemblies” that may pass along the substrates and products like a baton in a relay (for review, see ref. 17).
It is well known that β-pol null mouse cells are hypersensitive to monofunctional DNA alkylating agents, such as methyl methanesulfonate (MMS) (6, 15, 18). This finding indicates that other DNA polymerases or repair mechanisms do not completely substitute for the β-pol role(s) in protecting these cells against MMS-induced lesions. However, the status of DNA damage and DNA repair in β-pol null cells has not yet been thoroughly examined. In addition, any relationship between a deficiency in β-pol-mediated BER and mutagenesis has not been evaluated. In this study, isogenic mouse embryonic fibroblast (MEF) cell lines, wild type and β-pol null, were characterized to further define the importance of β-pol in the cellular response to MMS. We have examined DNA damage and repair and evaluated the question of whether a deficiency in a BER enzyme acting downstream of the DNA glycosylase and AP endonuclease steps can lead to an accumulation of genomic mutations. Quantitative PCR (QPCR) analysis, the comet assay, and AP site analysis were used to assess MMS-induced DNA damage. These analyses documented that β-pol null cells have a deficiency in repair of methylation-induced DNA lesions. Mutagenesis in the cells was examined by using a transgenic, chromosome-borne λ cII gene mutation assay (19). The β-pol null cells exhibited a weak spontaneous mutator phenotype and a stronger increase in mutant frequency after MMS treatment than observed with wild-type cells. Further, the β-pol-dependent increase in mutant frequency was specific for MMS, as both UV light exposure and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) exposure induced a similar increase in cII mutant frequency in both wild-type and β-pol null cells. We provided two lines of evidence that these mutations arise from AAG substrates by evaluating: (i) the influence of overexpression of AAG on MMS-induced mutations in β-pol null cells, and (ii) β-pol/AAG double null cells for the level of mutant frequency after MMS exposure. These results are discussed in the context of the significance of a specific combination of genetic deficiency in BER and MMS-induced DNA damage leading to cellular mutations.
Materials and Methods
Cell Lines.
Primary MEFs (wild type, β-pol null, and β-pol null/AAG null) were isolated and transformed essentially as described (15). The wild-type cell lines MB16tsA and MB36.3 and the β-pol null cell lines MB19tsA and MB38Δ4 have been described (15, 20).
DNA Damage and Repair Methods.
AP sites were measured by the aldehyde reactive probe-slot-blot method as described (21, 22). Heat labile sites were determined as follows: 10 μg DNA extracted from control cells or cells exposed to MMS was incubated at 100°C for 20 min in PBS. The AP sites induced by this heat incubation were measured by the aldehyde reactive probe-slot-blot method. For QPCR, high molecular weight DNA was isolated, and quantitative amplification of the β-globin target sequence was performed as described (23). Single-cell gel analysis (comet assay) was conducted as described (24).
Determination of Mutant Frequency at λ cII.
Cells (40,000) were added to each of six wells in a 6-well dish. After 24 h, the media were replaced with growth media alone or media supplemented with MMS or MNNG, and the cells were incubated at 34°C for 1 h. Alternatively, the media were removed and the cells were washed twice with PBS. After removal of the PBS, the cells were exposed to UV (15 J/m2) essentially as described (15). After genotoxicant exposure (MMS, MNNG, or UV), cells were washed once with growth media, refed with growth media, and incubated until the cells reached confluence. The cells were then seeded into 150-mm dishes and grown to confluence again. Normal culture time after exposure was 5–8 days. Genomic DNA was extracted from the cells (5–10 × 106 cells per sample) with Recoverease DNA Isolation Kits (Stratagene) and stored at 4°C. λ-LIZ shuttle vectors were rescued from genomic DNA with Transpack packaging extract according to the manufacturer's protocol (Stratagene). Plaques with mutations in the cII gene were determined essentially as described (19). Detailed procedures are described in additional Materials and Methods, which are published as supporting information on the PNAS web site, www.pnas.org. Briefly, mutant frequencies were determined by using genomic DNA isolated from at least three separate cell samples. For each DNA sample, a minimum of 300,000 plaques were analyzed from at least three packaging reactions to ensure an accurate mutant frequency determination.
Spontaneous and MMS-Induced λ cII Mutation Spectra.
The spontaneous and MMS-induced mutation spectra were determined at the cII locus in wild-type (MB36.3) or β-pol null (MB38Δ4) embryonic fibroblasts with a modified fluctuation test. Forty-eight individual cell populations (24 from each cell line) were established by plating 1,000 cells into each well of two 24-well dishes. Cell cultures were then expanded and DNA was isolated as described above. If the same mutation was observed more than once in each of the cell populations, then that mutation was counted only once in the spectra.
Overexpression of AAG in β-Pol Null Cells.
The full-length human AAG cDNA was cloned into pIRESneo (CLONTECH), a bicistronic vector expressing both AAG and neor gene products. β-Pol null cells were transfected with either pIRESneo or pIRESneo/AAG, and stable populations of cells were selected with G418 (600 μg/ml). G418-resistant cultures were analyzed by Western blotting as described (20) with an AAG-specific polyclonal antibody kindly provided by Timothy R. O'Connor, City of Hope National Medical Center, Duarte, CA (25). Functional glycosylase assays were as described (26) by using a hypoxanthine containing double-stranded oligonucleotide (Trevigen, Gaithersburg, MD).
Results
A DNA Repair Deficiency in β-Pol Null Cells.
β-Pol null cells are hypersensitive to the cytotoxic effect of MMS exposure (15, 16) and therefore more MMS-induced lesions may persist in genomic DNA of β-pol null cells than wild-type cells. To test this idea, QPCR was used to measure MMS-induced DNA polymerase blocking lesions (e.g., N3-methyl adenine, AP sites, and single-strand breaks) in a segment of genomic DNA (23, 27–35). Wild-type and β-pol null cells were examined with and without a repair incubation after termination of MMS exposure. At the end of the exposure period, after treatment with 1 mM MMS, an equivalent amount of DNA damage was found in both cell lines; an average of 0.1 MMS-induced lesions/kb was present in the genomic DNA (Fig. (Fig.11a). After a “repair incubation,” wild-type cells had fully repaired the MMS-induced lesions measured in this assay (Fig. (Fig.11a). In contrast, only a portion of the MMS-induced lesions were repaired in β-pol null cells (Fig. (Fig.11a). To further evaluate the capacity of β-pol null cells to repair MMS-induced lesions, DNA damage in genomic DNA was measured with the comet assay (Fig. (Fig.11b); this assay detects single-strand breaks plus alkali-labile sites (e.g., AP sites) (36). MMS induced an increase in comet assay tail moment, indicating an increase in DNA lesions in both cell lines. When measured after a repair incubation interval, DNA damage was substantially reduced in wild-type cells, but no reduction in DNA damage was detected in β-pol null cells (Fig. (Fig.11b). These results taken together indicate that the β-pol null cells are deficient in repair of MMS-induced lesions.
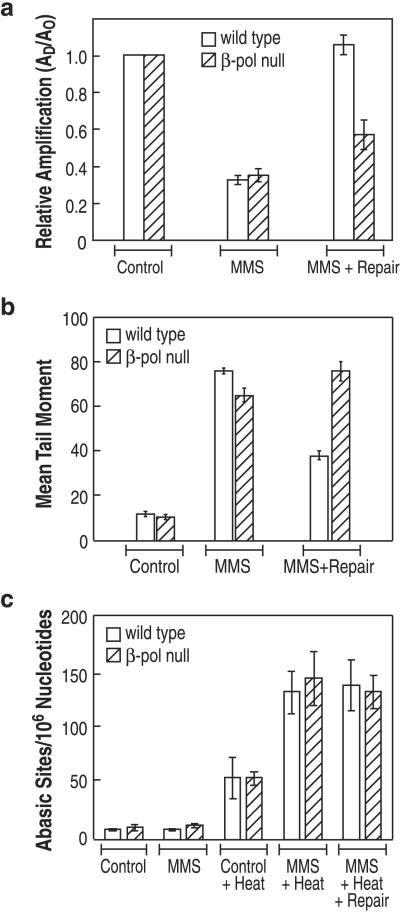
In vivo DNA repair deficiency in β-pol null cells. Wild-type cells (open bars) and β-pol null cells (hatched bars) were treated with control media or media supplemented with 1 mM MMS for 1 h. The MMS-supplemented media were then replaced with MMS-free growth media for 2–4 h. At the indicated times (either immediately after the treatment or after the repair period), cells were isolated for DNA preparation (a and c) or electrophoresis (b). (a) DNA damage analysis by QPCR: effect of MMS on the templating capacity of genomic DNA from wild-type and β-pol null cells. QPCR amplification of an 8.7-kb region of the β-globin gene was performed in triplicate on 15 ng genomic DNA. Control refers to cells without MMS treatment. The data are expressed as the mean, with bars indicating ± SD (n = 3 PCR analyses from each of three DNA samples). (b) Mean tail moment as determined in the comet assay. The data represent the tail moment of wild-type and β-pol null cells either untreated (control), treated with MMS for 1 h (MMS), or treated with MMS and allowed a repair interval of 2 h (MMS + Repair). The plot represents the mean of 100 scored cells from each of three separate experiments ± SD. (c) The number of AP sites and alkylated bases in genomic DNA from control and MMS treatment was determined for both wild-type and β-pol null cells. AP sites were measured directly whereas alkylated bases were determined from the number of AP sites after heat depurination of the genomic DNA, using the aldehyde reactive probe-slot-blot method as described in Materials and Methods. The values represent the number of AP sites per 106 nucleotides in control cells or MMS-treated cells as indicated. The plot represents the mean of three independent experiments from each of the cell lines, with bars indicating SD.
Status of AP Sites in Genomic DNA After Methylation.
MEFs deleted in the β-pol gene are known to be hypersensitive to the cytotoxic effects of MMS (5, 6, 15, 18, 20, 37), and this hypersensitivity is linked to a failure to remove the 5′ dRP group in the BER intermediate (20). However, it is not known whether a β-pol deficiency can impact levels of earlier intermediates in the repair mechanism, such as AP sites. We found that AP sites did not increase in DNA from either wild-type or β-pol null cells immediately after exposure to MMS (Fig. (Fig.11c). In addition, when assayed after a repair incubation, i.e., 4 h after exposure, there was still no difference in AP sites in genomic DNA from MMS-treated or untreated wild-type and β-pol null cells (not shown). As expected, MMS exposure in both cell lines gave rise to methylated bases, as revealed by a large increase in measured AP sites after heat treatment of the purified DNA (Fig. (Fig.11c). These results indicate that for both cell lines most of the MMS-induced methylated purine bases remained in genomic DNA (38) rather than being repaired or accumulated as AP sites.
Spontaneous Mutations at the λ cII Gene.
DNA lesions and the absence of DNA repair functions can result in mutations and/or unique mutation spectra. To evaluate these possibilities in wild-type and β-pol null cells, mutations in the chromosome-borne λ cII transgene were quantified, and independent mutants were selected and characterized by sequencing. Wild-type and β-pol null cells had spontaneous mutant frequencies of 1.4 ± 0.2 × 10−4 and 2.3 ± 0.6 × 10−4, respectively (Fig. (Fig.22a). A total of 130 and 102 independent spontaneous mutations from wild-type and β-pol null cells, respectively, were sequenced. The frequencies of mutations observed in these spontaneous mutant spectra are summarized in Table Table1.1. The relative spontaneous mutation frequencies of G:C → A:T and A:T → G:C transitions were comparable in both cell lines. The frequencies of all other mutations were elevated in β-pol null cells by between 1.8- and 3-fold.
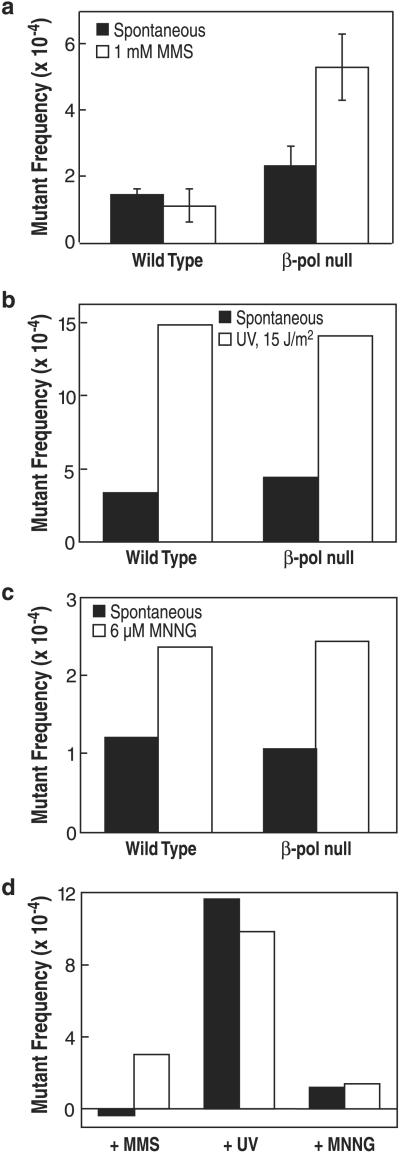
MMS-induced, UV-induced, and MNNG-induced mutations at the λ cII gene in β-pol null cells. (a) Spontaneous (filled bar) and MMS-induced (open bar) mutant frequencies at λ cII in wild-type or β-pol null MEFs. Bars represent the SD from three experiments. Mutant frequencies were calculated after analysis of 2.2 × 106 plaque-forming units (pfu) (wild type; spontaneous), 1.6 × 106 pfu (β-pol null; spontaneous), 1.1 × 106 pfu (wild type; +1 mM MMS), and 1.0 × 106 pfu (β-pol null; +1 mM MMS), respectively. (b) Spontaneous (filled bar) and UV-induced (open bar) mutant frequencies at λ cII in wild-type or β-pol null MEFs. Mutant frequencies were calculated after analysis of 4.7 × 105 pfu (wild type; spontaneous), 3.9 × 105 pfu (β-pol null; spontaneous), 4.5 × 105 pfu (wild type; +UV), and 4.2 × 105 pfu (β-pol null; +UV), respectively. (c) Spontaneous (filled bar) and MNNG-induced (open bar) mutant frequencies at λ cII in wild-type or β-pol null MEFs. Mutant frequencies were calculated after analysis of 4.9 × 105 pfu (wild type; spontaneous), 6.1 × 105 pfu (β-pol null; spontaneous), 5.3 × 105 pfu (wild type; +6 μM MNNG), and 6.7 × 105 pfu (β-pol null; +6 μM MNNG), respectively. (d) Induced mutant frequencies at λ cII in wild-type (filled bar) or β-pol null (open bar) MEFs. The induced mutant frequency value was determined by subtracting the value for the spontaneous mutant frequency from the value for the indicated genotoxicant-induced mutant frequency as shown in a–c.
Table 1
Comparison of spontaneous mutation frequency as a function of β-pol status
status
| Mutation type | Wild type
| β-pol null
| Fold β-pol-dependent change | ||||
|---|---|---|---|---|---|---|---|
| No. | % | Mutation frequency, × 10−5 | No. | % | Mutation frequency, × 10−5 | ||
| G:C to A:T | 39 | 30 | 4.2 | 24 | 24.5 | 5.6 | 1.3 |
| A:T to G:C | 24 | 18.4 | 2.6 | 8 | 7.8 | 1.8 | 0.7 |
| G:C to T:A | 23 | 17.7 | 2.5 | 21 | 20.6 | 4.7 | 1.9 |
| G:C to C:G | 17 | 13 | 1.8 | 24 | 23.5 | 5.4 | 3.0 |
| A:T to T:A | 9 | 6.9 | 1.0 | 10 | 9.8 | 2.3 | 2.4 |
| A:T to C:G | 18 | 13.8 | 1.9 | 15 | 14.7 | 3.4 | 1.8 |
| Total | 130 | 102 | |||||
The mutant frequencies used in the calculation of spontaneous mutation frequencies were 1.4 × 10−4 and 2.3 × 10−4 for wild-type and β-pol null cells, respectively. These values are graphically represented in Fig. Fig.2.2. Mutation frequencies were calculated by multiplying the fraction of each class of mutations observed by the average spontaneous mutant frequency. For example, the frequency of spontaneous G:C to C:G mutations in wild-type cells was calculated as follows: (0.13)(1.4 × 10−4) = 1.8 × 10−5. Fold change is calculated by dividing the mutation frequency of a given type of mutation in β-pol null cells by the frequency of the same type of mutation in wild-type cells.
Mutation Spectral Analysis for the λ cII Gene After Methylation.
MMS was mutagenic in β-pol null cells, inducing an increase in mutant frequency when comparing MMS-exposed wild-type and β-pol null cells (Fig. (Fig.22a). This increase was observed for MMS treatment at 1 mM, as well as for a MMS dose of equivalent (75%) cytotoxicity (not shown). A β-pol-dependent increase in mutant frequency was not observed when cells were exposed to either UV or MNNG; wild-type and β-pol null cells demonstrated identical UV-induced λ cII mutant frequency (Fig. (Fig.22b) and similar results were obtained for MNNG-induced mutant frequency (Fig. (Fig.22c).
The spectra for the MMS-induced mutations in wild-type and β-pol null cells are summarized in Table Table2.2. Wild-type cells showed little or no increase in cII mutation frequency as a function of MMS treatment (see Table 3, which is published as supporting information on the PNAS web site). In contrast, MMS-treated β-pol null cells had higher mutation frequencies for all base substitution mutations than MMS-treated wild-type cells (Table (Table2).2). There are several options for analysis of these mutational data, e.g., plus versus minus MMS, plus versus minus β-pol, changes within class of mutation, and changes as a function of DNA sequence in the cII gene (see Tables 3 and 4, which are published as supporting information on the PNAS web site). As seen in Table Table2,2, β-pol null cells showed greater MMS-induced mutation frequency than wild-type cells for both transitions and transversions. Yet, some of the increase in transversions in MMS-treated β-pol null cells (Table (Table2)2) could have been caused by the modest increase (≈2-fold) in spontaneous transversions seen in these cells. In contrast, none of the increase in A:T to G:C transitions seen in MMS-treated β-pol null cells (Table (Table2)2) appears to be explained by any increase in spontaneous mutations of this type (see Table 3). Finally, the mutation spectra contained six mutational hotspots, defined as sites where five or more mutations were detected in each cell type, but these mutations did not appear to depend on β-pol expression or MMS treatment (data not shown; see Fig. 5, which is published as supporting information on the PNAS web site).
Table 2
MMS-induced λ cII mutation frequency in wild-type and β-pol null cells
cells
| Mutation type | Mutation frequency, × 10−5
| Fold β-pol-dependent change | |
|---|---|---|---|
| Wild type | β-pol null | ||
| G:C to A:T | 3.2 | 10.4 | +3.2 |
| A:T to G:C | 0.8 | 7.2 | +9.0 |
| G:C to T:A | 3.1 | 14 | +4.5 |
| G:C to C:G | 1.2 | 8.6 | +7.1 |
| A:T to T:A | 1.5 | 7.5 | +5.0 |
| A:T to C:G | 1.1 | 5.4 | +4.9 |
The mutant frequencies used in the calculation of MMS-induced mutation frequencies were 1.1 × 10−4 and 5.3 × 10−4 for wild-type and β-pol nulls cells, respectively. These values are graphically represented in Fig. Fig.2.2. Mutation frequencies were calculated by multiplying the fraction of each class of mutations observed by the average spontaneous mutant frequency. For example, the frequency of MMS-induced A:T to G:C mutations in wild-type cells was calculated as follows: (0.076)(1.1 × 10−4) = 0.8 × 10−5. Fold change is calculated by dividing the mutation frequency of a given type of mutation in β-pol null cells by the frequency of the same type of mutation in wild-type cells.
AAG Dependence and the MMS-Induced Mutation Effect in β-Pol Null Cells.
The increase in methylation-induced mutations in β-pol null cells indicates that the β-pol deficiency is linked to alkylation damage-induced mutagenesis. To further evaluate this, we modified β-pol null cells to overexpress AAG (39). These cells expressed higher levels of AAG protein (Fig. (Fig.33a), and the crude cell extract had more AAG enzymatic activity than the control β-pol null(Neo) cell extract (Fig. (Fig.33b). These β-pol null(AAG) cells were compared with β-pol null(Neo) cells for MMS-induced mutagenesis at the λ cII gene. As shown in Fig. Fig.33c, overexpression of AAG eliminated the MMS-induced increase in mutant frequency seen in β-pol null cells or β-pol null(Neo) cells (Fig. (Fig.33c). These results are consistent with the idea that methylated purine bases are linked to the MMS-induced mutant frequency effect seen in β-pol null cells.
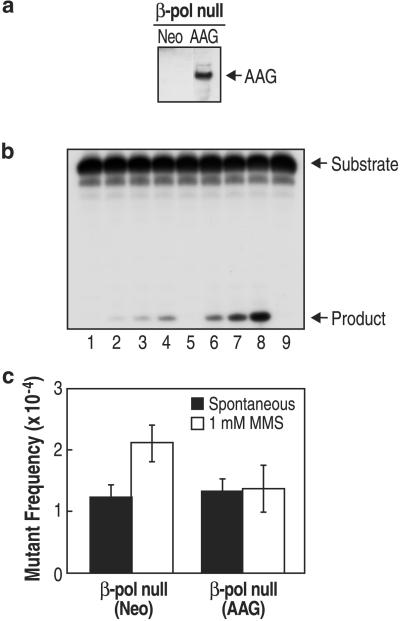
Overexpression of AAG in β-pol null cells. (a) Detection of AAG protein by immunoblotting of whole-cell extracts from 20 μg β-pol null(Neo) cells (Left) and 20 μg β-pol null(AAG) cells (Right). The arrow indicates the AAG band. (b) AAG activity in whole-cell extracts. The relative functional expression of AAG in the β-pol null(Neo) and β-pol null(AAG) cells was determined by release of hypoxanthine from a single lesion-containing 24-mer double-stranded oligonucleotide, as described in Materials and Methods. Lanes 1–4: β-pol null(Neo) cell extract incubated for 0, 2, 4, and 6 min, respectively. Lanes 5–8: β-pol null(AAG) cell extract incubated for 0, 2, 4, and 6 min, respectively. Lane 9: no extract control incubated for 6 min. (c) Methylation-induced increase in mutant frequency at λ cII. Spontaneous (filled bar) and MMS-induced (open bar) mutant frequencies at λ cII in β-pol null(Neo) cells or β-pol null(AAG) cells. Mutant frequencies were calculated after analysis of 3.2 × 106 pfu [β-pol null (Neo); spontaneous], 2.9 × 106 pfu [β-pol null (Neo); +1 mM MMS], 3.1 × 106 pfu [β-pol null (AAG); spontaneous], and 3.2 × 106 pfu [β-pol null (AAG); +1 mM MMS], respectively.
Because the increases in mutations seen in β-pol null cells may be caused by AAG substrates, a genetic knockout of AAG in the β-pol null background may augment the mutagenic effect of MMS exposure. To further delineate lesions responsible for the mutagenic effect, we developed β-pol and AAG double null fibroblast cell lines. As shown in Fig. Fig.4,4, the β-pol null/AAG null cells exhibited a 2.45-fold increase in MMS-induced mutant frequency at the λ cII locus. Thus, the β-pol null/AAG null fibroblasts exhibited a slightly higher increase in MMS-induced mutant frequency as compared with β-pol null cells. This finding indicated that in the β-pol null background the effect of AAG elimination on mutation accumulation was noticeable, but minimal.
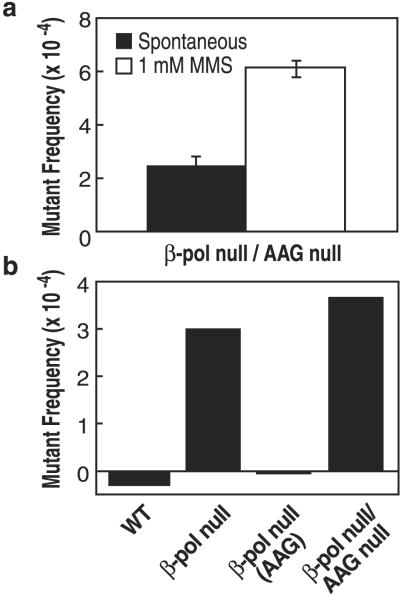
MMS-induced increase in cII mutations in β-pol null/AAG null cells. (a) Spontaneous (filled bar) and MMS-induced (open bar) mutant frequencies at λ cII in β-pol null/AAG null MEF cells. Mutant frequencies were calculated after analysis of 3.2 × 106 pfu (β-pol null/AAG null; spontaneous) and 3.0 × 106 pfu (β-pol null/AAG null; +1 mM MMS), respectively. (b) Induced mutant frequencies at λ cII in wild-type, β-pol null, β-pol null(AAG), and β-pol null/AAG null MEFs. The induced mutant frequency value was determined by subtracting the value for the spontaneous mutant frequency from the value for the MMS-induced mutant frequency as shown in a and Figs. Figs.22a and 3c.
Discussion
β-Pol Null Cells: A Genetic Defect in BER.
A genetic deficiency of β-pol in MEFs results in greater MMS sensitivity mainly because of failure to remove the 5′ dRP group in the BER intermediate in this mouse fibroblast system (15, 20). When characterized here for repair of MMS-induced lesions with QPCR, wild-type cells repaired more of the MMS-induced lesions than the β-pol null cells, confirming they are repair deficient; we obtained comparable results with the comet assay. These observations are consistent with the hypothesis that β-pol-dependent BER is important in repair of methylation-induced damage in this system. Yet, our measurements of AP sites and total methylated purines in genomic DNA (Fig. (Fig.11c) revealed that wild-type and β-pol null cells are quantitatively similar, both immediately after MMS treatment and after a period (4 h) of repair. Therefore, most of the MMS-induced lesions, i.e., N7-methylguanine, remained in genomic DNA during this repair period and did not accumulate as BER intermediates, blocked at a β-pol-mediated step.
Mutagenesis After MMS Exposure.
Our comparison of spontaneous mutant frequency in wild-type and β-pol null cells demonstrated a weak spontaneous mutator phenotype for the β-pol null cells (Table (Table11 and Fig. Fig.2).2). Mutant frequency in the MMS-treated β-pol null cells was higher than that in untreated β-pol null cells and much higher than in MMS-treated wild-type cells (Table (Table2).2). These results indicate that these BER-deficient MEF cells are hypermutable after MMS exposure. A similar observation had been made for the XRCC1-defective cell line EM-C11, in that mutations at the hprt locus increased severalfold after MMS treatment, as compared with a wild-type Chinese hamster ovary cell line (40). These XRCC1-defective cells, like the β-pol null cells, are hypersensitive to MMS and appear to be deficient in BER.
Sequencing of individual mutants revealed that the frequency of A:T → G:C mutations increased 9-fold and all other base substitution mutations were elevated from 3.2- to 7.1-fold in β-pol null cells after MMS treatment, as compared with wild-type cells (Table (Table2).2). The frequency of G:C → T:A and A:T → T:A mutations were 4.5- and 5-fold higher, respectively, in MMS-treated β-pol null cells (Table (Table2)2) than in wild-type cells. We note that these two base substitution mutations were also found to predominate in AAG null mice after exposure to MMS (41).
Methylation-Induced Mutagenesis as a Function of β-Pol Status.
Studies of several transgenic and endogenous genes in wild-type mice or cell lines have shown that MMS does not increase mutant frequency (40, 42–45). However, in repair-deficient rodent cells, MMS can be mutagenic (40, 42, 46, 47). This finding, along with the results reported here, suggested the use of the SN2 methylating agent MMS as the DNA damaging agent of choice for the mutagenesis studies described. Except as a reference, SN1 methylating agents such as MNNG, producing relatively more O6-methyl guanine (O6meG), were avoided in this study because of their strong tendency to produce G → A mutations and also because O6-meG is repaired by the methyl transferase mechanism, rather than by β-pol-dependent BER.
MMS exposure of cells carrying a genetic deficiency in AAG would be expected to give rise to methylation-induced DNA mutations resulting from replicative miscoding by MMS-induced lesions (41). We presented evidence here that a deficiency in β-pol, a downstream enzyme in the BER pathway, also imparts a methylation-induced increase in DNA mutations. Further, our evidence obtained with cells expressing different levels of AAG suggests that these mutations were caused by mutagenic AAG substrates. These results suggest a defect in processing methylated purine bases caused by the deficiency in the downstream BER enzyme β-pol.
Mechanism of Increase in MMS-Induced Mutants.
MMS is known to produce a number of methylated DNA lesions in addition to the predominant lesion N7-methylguanine, which is not miscoding during DNA replication. Several of these minor lesions are known to be capable of miscoding during DNA synthesis and could be repaired slower in β-pol null cells than in the wild-type cells, leading to replication-based mutations in the null cells, as noted above. To evaluate this idea, we examined the effect of overexpression of mammalian AAG. Such overexpression of AAG was found to suppress the increase in MMS-induced mutant frequency in the β-pol null cells (Fig. (Fig.33c), suggesting that substrates for this enzyme are responsible for the increased mutagenesis. We also found that MMS was mutagenic in β-pol null/AAG null fibroblasts (Fig. (Fig.4),4), again suggesting that AAG substrates left unrepaired are mutagenic in this system. Thus, it is likely that quantitatively minor methylated bases miscode with incoming dNTPs during DNA replication and that the β-pol deficiency somehow influences the processing of these mutagenic AAG substrates. In addition to miscoding, unrepaired methylpurines may undergo depurination, leading to AP sites that are mutagenic in the β-pol null cell. These mechanisms could explain an increase in mutant frequency, such as that observed in MMS-treated β-pol null cells.
Susceptibility to Alkylation-Induced Mutagenesis Is a Model for Study of Gene–Environment Interactions.
Our results illustrate an example of a linkage between a genetic deficiency in BER and mutagenicity in a mammalian system and also illustrate a case where a specific combination of a genetic defect (in BER) and exposure to genotoxicant is required to observe the mutagenic effect. This particular “gene–environment interaction” appeared to be necessary, because we did not observe the β-pol-dependent hypermutable phenotype with alternative genotoxicant exposures: UV light, producing lesions repaired exclusively by nucleotide excision repair, or MNNG, producing the mutagenic O6-methyl guanine lesion repaired by the methyl transferase mechanism. These results should help us answer questions about study design for complex situations encountered in animal model and population-based research on gene–environment interactions. Our results suggest that even when a genetic defect in β-pol exists the phenotype of increased mutagenesis will not be appreciated unless environmental exposure to an appropriate (SN2 alkylating) agent is identified.
Acknowledgments
We thank Dr. Roel Schaaper (Laboratory of Molecular Genetics, National Institute of Environmental Health Sciences) and Dr. Barry Gold (University of Nebraska Medical Center, Omaha) for valuable discussions and critical evaluation of the manuscript. We also thank Drs. Raymond Tice and Barbara Shane (Integrated Laboratory Systems, Durham, NC) for assistance with the Comet and cII mutation assays, respectively. L.S. is the Ellison American Cancer Society Research Professor, and this research was supported by RO1s CA75576 and CA55042.
Abbreviations
| AP | apurinic/apyrimidinic |
| BER | base excision repair |
| MMS | methyl methanesulfonate |
| dRP | 5′ deoxyribosephosphate |
| β-pol | DNA β-polymerase |
| QPCR | quantitative PCR |
| AAG | alkyladenine DNA glycosylase |
| MEF | mouse embryonic fibroblast |
| MNNG | N-methyl-N′-nitro-N-nitrosoguanidine |
| pfu | plaque-forming units |
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.092662499
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc124494?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Mouse models to explore the biological and organismic role of DNA polymerase beta.
Environ Mol Mutagen, 65 Suppl 1:57-71, 01 Apr 2024
Cited by: 1 article | PMID: 38619421
Review
A Review of Numerical Models of Radiation Injury and Repair Considering Subcellular Targets and the Extracellular Microenvironment.
Int J Mol Sci, 25(2):1015, 13 Jan 2024
Cited by: 0 articles | PMID: 38256089 | PMCID: PMC10816679
Review Free full text in Europe PMC
Cooperative interaction between AAG and UV-DDB in the removal of modified bases.
Nucleic Acids Res, 50(22):12856-12871, 01 Dec 2022
Cited by: 4 articles | PMID: 36511855 | PMCID: PMC9825174
Base excision repair accessory factors in senescence avoidance and resistance to treatments.
Cancer Drug Resist, 5(3):703-720, 22 Jun 2022
Cited by: 4 articles | PMID: 36176767 | PMCID: PMC9511810
Review Free full text in Europe PMC
CometChip enables parallel analysis of multiple DNA repair activities.
DNA Repair (Amst), 106:103176, 10 Jul 2021
Cited by: 3 articles | PMID: 34365116 | PMCID: PMC8439179
Go to all (50) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mammalian DNA beta-polymerase in base excision repair of alkylation damage.
Prog Nucleic Acid Res Mol Biol, 68:57-74, 01 Jan 2001
Cited by: 60 articles | PMID: 11554313
Review
Protection against methylation-induced cytotoxicity by DNA polymerase beta-dependent long patch base excision repair.
J Biol Chem, 275(3):2211-2218, 01 Jan 2000
Cited by: 87 articles | PMID: 10636928
Defects in base excision repair combined with elevated intracellular dCTP levels dramatically reduce mutation induction in yeast by ethyl methanesulfonate and N-methyl-N'-nitro-N-nitrosoguanidine.
Environ Mol Mutagen, 32(2):173-178, 01 Jan 1998
Cited by: 5 articles | PMID: 9776180
BER, MGMT, and MMR in defense against alkylation-induced genotoxicity and apoptosis.
Prog Nucleic Acid Res Mol Biol, 68:41-54, 01 Jan 2001
Cited by: 57 articles | PMID: 11554312
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: R01 CA075576
Grant ID: R01 CA55042
Grant ID: R01 CA75576
Grant ID: R01 CA055042




