Abstract
Free full text

Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins
Abstract
Now that it is likely that all yeast nucleoporins are known, one of the ultimate goals is the in vitro assembly of the entire nuclear pore complex from its ~30 individual components. Here, we report the reconstitution of seven proteins (Nup133p, Nup145p-C, Nup120p, Nup85p, Nup84p, Seh1p and Sec13p) into a heptameric 0.5 MDa nuclear pore subcomplex. We found that double plasmid transformation combined with bi-cistronic mRNA translation allow the expression and assembly of distinct subcomplexes of up to five nucleoporins in a single Escherichia coli cell. During the sequential reconstitution of the Nup84p complex, smaller assembly intermediates can be isolated, which exhibit modular structures determined by electron microscopy that finally make up the whole Y-shaped Nup84p complex. Importantly, a seventh subunit, Nup133p, was incorporated into the complex through its interaction with Nup84p, thereby elongating one arm of the Y-shaped assembly to an ~40 nm long stalk. Taken together, our data document that the Nup84p–Nup133p complex self-assembles in a modular concept from distinct smaller nucleoporin construction sets.
Introduction
The nuclear pore complex (NPC) is an intriguing nanostructure in the eukaryotic cell, which is functionally and structurally conserved throughout evolution and acts as the only gateway for transport of cargo between the nucleus and the cytoplasm (Stoffler et al., 1999; Bagley et al., 2000; Vasu and Forbes, 2001). In yeast, the NPC is ~60 MDa in size and consists of ~30 different subunits (nucleoporins), which assemble into a highly symmetrical supramolecular structure having octagonal symmetry in the plane of the nuclear envelope (NE) (Rout et al., 2000). Distinct NPC substructures such as the central spoke complex, nuclear and cytoplasmic rings, the nuclear basket and short cytoplasmic fibrils can be visualized by electron microscopy (EM) (Stoffler et al., 1999; Allen et al., 2000).
It is well established that different nucleoporins have distinct roles in NPC structure and function. Nucleoporins with repeat sequences of the FG, GLFG and FXFG type provide the ‘stationary phase’ for nuclear transport by being localized at the entry and exit and probably along the nuclear pore channel exposing their repeat domains, which act as stepping stones for karyopherin-mediated transport reactions (Rexach and Blobel, 1995; Bayliss et al., 2000; Rout et al., 2000; Allen et al., 2001). Only a few nucleoporins are integral membrane proteins (Pom152p, Pom121p and gp210) that are thought to anchor the pores within the nuclear membrane (Gerace et al., 1982; Hallberg et al., 1993; Wozniak et al., 1994). Finally, several of the nucleoporins are used as structural building blocks, which assemble into distinct subcomplexes to constitute the core as well as peripheral structures of the NPC. At least some of these structural nucleoporins lack FXFG, GLFG and FG repeat sequences (for a review see Vasu and Forbes, 2001).
To date, very little is known about the pathway in which the NPC assembles and of which nucleoporins finally constitute the certain substructures. Furthermore, in most cases, it is not understood how direct physical contacts are generated among the 30 nuclear pore proteins (Vasu and Forbes, 2001). Therefore, only a ‘crude’ topological map exists, in which nucleoporins have been localized by immuno-EM to distinct sites of the structural NPC framework. Accordingly, nucleoporins have been localized to the central pore channel, the cytoplasmic fibrils or the nuclear basket (Stoffler et al., 1999; Rout et al., 2000). For example, the yeast Nsp1p–Nup49p–Nup57p complex and its homologous vertebrate counterpart, the p62 complex, line the entrance and exit of the central pore channel (Panté et al., 1994). In contrast, the Nup82p– Nup159p–Nsp1p complex is located asymetrically at the cytoplasmic side of the NPC (Kraemer et al., 1995), while Nup1p, Nup2p and Nup60p are located exclusively at the nuclear side of the NPC (Rout et al., 2000). In vertebrates, Nup358p and Nup214p/CAN are constituents of the cytoplasmic fibrils, whereas Nup153p is thought to be part of the nuclear basket and Tpr/Mlp1/2 is thought to constitute long intranuclear pore filaments (for a recent review see Vasu and Forbes, 2001). Recent work, however, has indicated that Nup153 is localized by immunogold EM to a position on the nuclear ring of the NPC (Walther et al., 2001).
A paradigm for studied NPC subcomplexes is the Nup84p complex, which consists of six subunits; five of them are bona fide nucleoporins (Nup145p-C, Nup120p, Nup85p, Nup84p and Seh1p) and one of them, Sec13p, is also part of the COPII complex, which facilitates vesicular transport from the endoplasmic reticulum (ER) to the Golgi (Siniossoglou et al., 1996). When nucleoporins of the Nup84p complex are mutated, strong defects in the export of mRNA, the distribution of NPCs and the organization of the nuclear membrane are observed (Aitchison et al., 1995; Heath et al., 1995; Goldstein et al., 1996; Siniossoglou et al., 1996). Furthermore, Nup84p genetically interacts with two novel nuclear/ER membrane proteins, Spo7p and Nem1p, which are required for formation of a spherical nucleus (Siniossoglou et al., 1998). Thus, the Nup84p complex was suggested to play (i) a structural role in NPC formation, and (ii) to be involved in nucleocytoplasmic transport (e.g. by interacting with other nucleoporins or shuttling transport receptors). The Nup84p complex appears to have a metazoan counterpart, the Nup160p complex (Fontoura et al., 1999; Vasu and Forbes, 2001), which has recently been characterized further and named the hNup107p complex (Belgareh et al., 2001).
Since the Nup84p complex is biochemically stable in vitro and therefore can be purified from yeast with a molecular weight of 375 kDa and a distinct 25 nm diameter Y-shaped structure (Siniossoglou et al., 2000), we sought to study this complex as a paradigm for a distinct structural module of the NPC. Our results document that the Nup84p complex can be fully reconstituted from its six recombinantly expressed components. During this self-assembly process, we were able to define which subunits interact directly with each other. Furthermore, we succeeded in the recombinant expression of another nucleoporin, Nup133p, and established its specific binding to the Nup84p complex via direct interaction with Nup84p. Thus, our work provides the first insight into the assembly pathway of a conserved NPC subcomplex and it documents how the Y-shaped structure is generated and elongated from the different nucleoporin building blocks in a modular fashion.
Results
Reconstitution of the Nup84p complex in E.coli
Here, we analyzed the yeast Nup84p complex, a paradigm for a structurally and functionally defined NPC subcomplex that constitutes ~15% of the mass of the NPC (assumed 16 copies per pore complex, see Rout et al., 2000), for its ability to self-assemble in vitro. We sought to study the requirements for its assembly in order to identify the subunits that are in direct contact with each other and also morphologically dissect how the Y-shaped structure of the complex is built up from its constituent subunits. If the six subunits of the Nup84p complex are able to self-assemble, it should be possible to reconstitute the entire complex in vitro. In a first attempt at reconstitution, the open reading frames (ORFs) encoding the six nucleoporins were cloned individually under the control of the T7 promoter, fused with a tag to facilitate affinity purification (either GST or His6) and expressed individually in Escherichia coli. This revealed that Seh1p and Sec13p were strongly expressed (Figure 1A), whereas the large nucleoporins (Nup84p, Nup85p, Nup145p-C and Nup120p) were obtained in lower amounts (Figure 1A). Nup120p and Nup145p-C were not, and Nup85p and Nup84p were only poorly soluble (data not shown). We therefore sought to increase the solubility of these individual nucleoporins by co-expressing them with their cognate partner nucleoporins.

Fig. 1. Expression of nucleoporins of the Nup84p complex in E.coli. BL21 cells were transformed with plasmids harboring the indicated nucleoporin genes. Cells were grown under non-induced (–) and IPTG-induced (+) conditions. Whole-cell lysates were prepared and analyzed by SDS–PAGE and Coomassie Blue staining. The positions of the induced nucleoporins are indicated by asterisks. In (A), single nucleoporins were expressed, whereas in (B) pairs of nucleoporins were expressed from bi-cistronic constructs. Also shown is a 10 kDa molecular weight ladder.
Co-expression was first performed with Nup85p and Seh1p, since they form a complex in vivo (Siniossoglou et al., 1996), which indeed led to a partially soluble complex of these two nucleoporins. We noticed that complex formation and solubility of Nup85p and Seh1p were strongly increased when a GST-TEV-NUP85 and an SEH1 ORF were expressed from a single bi-cistronic mRNA (Figure 1B; see also below). Accordingly, when an E.coli lysate containing soluble GST–Nup85p and untagged Seh1p (Figure 2A, lane 1) was applied to a glutathione–Sepharose column, a GST–Nup85p–Seh1p complex was affinity purified efficiently and eluted by tobacco etch virus (TEV) proteolytic cleavage (Figure 2A, lane 3). Finally, the eluted Nup85p–Seh1p complex was purified to >95% homogeneity by FPLC gel filtration. The elution profile and analysis by ultracentrifugation suggest a heterodimeric complex of ~120 kDa (Figure 2A, right panel).
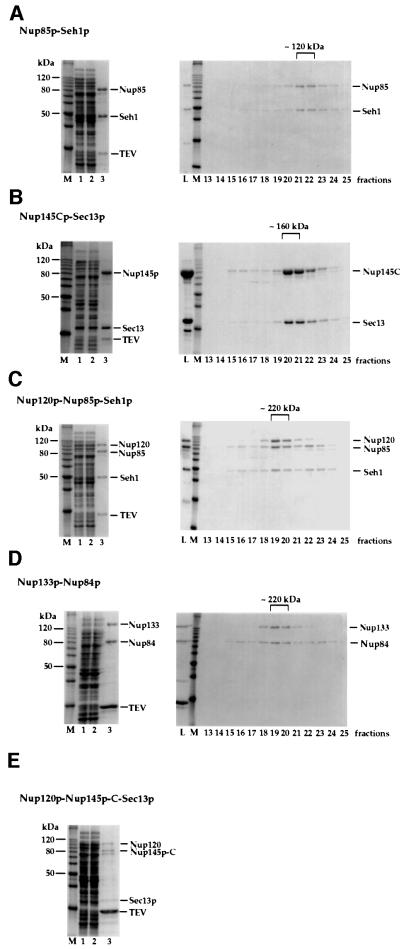
Fig. 2. Isolation of heterodimeric and heterotrimeric nucleoporin complexes. Purification of (A) the Nup85p–Seh1p complex, (B) the Nup145p-C–Sec13p complex, (C) the Nup120p–Nup85p–Seh1p complex, (D) the Nup133p–Nup84p complex and (E) the Nup120p–Nup145p-C–Sec13p complex. Note that the band below Nup145p-C is a degradation product of this protein as shown by western blot analysis (data not shown). Left panels (A–E): Coomassie-stained SDS–PAGE with a protein standard (M), the soluble E.coli lysate after high speed centrifugation (lane 1), the flow through after incubation with glutathione beads (lane 2) and the TEV eluate (lane 3). Right panels (A–D): the TEV eluates containing the indicated complexes were applied on an FPLC Superdex 200 column. Fractions 13–25 from the column were analyzed by SDS–PAGE and Coomassie Blue staining. Also shown is the TEV eluate input fraction (L) and a protein standard (M). The Superdex 200 column was calibrated with several molecular weight marker proteins of 670, 158, 44 and 17 kDa (Bio-Rad).
By further co-expression experiments, we found that Nup145p-C (which alone is insoluble) and Sec13p also form a complex. In this case, a GST-TEV-NUP145C and a SEC13 ORF were expressed from a bi-cistronic mRNA in E.coli (see Figure 1B) and the now soluble Nup145p-C–Sec13p complex was affinity purified by glutathione–Sepharose chromatography and TEV cleavage as described above (Figure 2B). The highly purified Nup145p-C–Sec13p complex eluted in a sharp peak from a Superdex 200 gel filtration column, corresponding to an apparent mol. wt of 160 kDa (Figure 2B). By comparison of the intensities of the Coomassie Blue-stained protein bands after SDS–PAGE, we conclude that Nup145p-C and Sec13p form a heterodimeric complex, which apparently runs slightly abnormally by gel filtration due to its elongated shape (see below).
We next wanted to find out whether Nup120p associates with the Nup85p–Seh1p or the Nup145p-C–Sec13p complex. When untagged NUP120 was co-expressed with bi-cistronic GST-NUP85/SEH1, the expression and solubility of Nup120p increased significantly due to heterotrimeric complex formation. A Nup120p–Nup85p–Seh1p complex was purified to apparent homogeneity by glutathione– Sepharose affinity chromatography, TEV cleavage and Superdex gel filtration (Figure 2C). To investigate whether Nup120p also binds the Nup145p-C–Sec13p complex, Nup120p was co-expressed with bi-cistronic GST–Nup145p-C–Sec13p. Indeed, a trimeric complex of these three proteins could be purified by affinity chromatography and TEV cleavage (Figure 2E). Due to the relatively low yield of this heterotrimeric complex, no further purification step such as gel filtration was performed. The fact that Nup120p binds independently to both of the dimeric complexes, which however do not bind to each other (data not shown), demonstrates that Nup120p is the linking protein between the Nup85p–Seh1p complex and the Nup145p-C–Sec13p complex (see below).
Since Nup84p failed to bind to the Nup85p–Seh1p complex, we tested whether Nup84p associates with the Nup145p-C–Sec13p complex, which indeed was the case. Furthermore, since Nup84p does not bind to Sec13p alone, we conclude that Nup84p interacts directly with Nup145p-C (data not shown; see also below). However, the lack of an in vitro interaction does not exclude the possibility that both proteins interact in vivo.
The entire Nup84p complex consisting of six subunits was then reconstituted in vitro, using the following strategy: heterotrimeric Nup120p–Nup85p–Seh1p, heterodimeric Nup145p-C–Sec13p and monomeric Nup84p were purified to apparent homogeneity from E.coli as described above. The two complexes and Nup84p were then mixed in equimolar amounts, determined with a Bio-Rad assay, and incubated for 1.5 h at room temperature. This mixture was then applied to a Superose 6 gel filtration column to separate the reconstituted hexameric complex from the unassembled input fractions. The peak fractions, which appear to contain the six subunits in roughly stoichiometric amounts (as estimated from the intensity of the Coomassie Blue-stained bands), correspond to ~600–700 kDa apparent molecular weight, which was also found for the authentic Nup84p complex purified from yeast (Figure 3A; see also Siniossoglou et al., 2000). When the pooled peak fractions were loaded for a second time on the same gel filtration column, the Nup84p complex again eluted in the same fractions (Figure 3A), showing that the reconstituted complex is biochemically stable. The in vitro assembled Nup84p complex thus greatly resembles the native complex, in both its protein composition and its behavior during gel filtration (Siniossoglou et al., 2000). Hence, we conclude that the purified subcomplexes of the Nup84p complex are capable of self-assembly in vitro to form a hexameric complex.


Fig. 3. Purification of the reconstituted Nup84p complex with or without Nup133p. The upper gels show the in vitro assembled Nup84p complex without Nup133p (A) and with bound Nup133p (B) before Superose 6 gel filtration chromatography (input fraction = L), a protein standard (M) and fractions 16–28 from the first Superose 6 column run. The lower gels are the result of a second Superose gel filtration of the pooled fractions 21–24 (L). Note that the additional band at ~65 kDa is a degradation product of Nup145p-C (revealed by western blot; data not shown). The Superose 6 column was calibrated with molecular weight marker proteins of 670, 158, 44 and 17 kDa (Bio-Rad). A typical calibration profile is shown at the bottom of (A).
Finally, we succeeded in expressing and assembling five subunits of the Nup84p complex in a single E.coli cell. We constructed an amp-containing plasmid, which harbors bi-cistronically arranged GST-tagged NUP145C and SEC13 plus NUP120 under the control of a separate T7 promoter. This plasmid was co-transformed with a kan-containing plasmid harboring untagged bi-cistronical NUP85 and SEH1. Expression of all five ORFs could be induced by isopropyl-β-d-thiogalactopyranoside (IPTG). This in vivo-assembled pentameric complex was isolated finally as described above utilizing the GST tag attached to Nup145p-C (see below, data not shown).
The finding that the entire Nup84p complex could be reconstituted in vitro without the help of co-factors raised the question of whether additional recombinantly expressed nucleoporins could be incorporated into the Nup84p complex. To date, direct interaction partners of the Nup84p complex have not been identified. However, vertebrate Nup107, which is homologous to yeast Nup84p, interacts with vertebrate Nup133 within a conserved hNup107 complex (also designated the Nup160 complex) (Belgareh et al., 2001; Vasu et al., 2001). Furthermore, it was suggested that yeast Nup133p associates with the Nup84p complex (Allen et al., 2001). Hence, we wanted to find out whether recombinant Nup133p binds specifically to Nup84p and can be assembled into the recombinant Nup84p complex.
To test this possibility, we first performed bi-cistronic co-expression of Nup133p and Nup84p in E.coli. This revealed that Nup84p and Nup133p indeed form a complex in which they most probably were present in equimolar amounts (Figure 2D). Most importantly, Nup133p could then be incorporated into the hexameric Nup84p complex by mixing heterotrimeric Nup120p– Nup85p–Seh1p, heterodimeric Nup145p-C–Sec13p and heterodimeric Nup84p–Nup133p, all purified from E.coli as described above. This in vitro-reconstituted heptameric complex was then purified to homogeneity by two successive Superose 6 chromatography steps (Figure 3B). The heptameric Nup133p–Nup84p complex revealed equimolar stable association of Nup133p with Nup84p (Figure 3B, lower panel). Thus, we could identify Nup133p as a direct binding partner of the Nup84p complex. Evidently, Nup133p is linked via the Nup84p subunit to the Nup84p complex.
Electron microscopic analysis of the reconstituted Nup133p–Nup84p complex
Concomitantly with the biochemical reconstitution, we analyzed the structure of these in vitro assembled nucleoporin complexes, including the final Nup133p– Nup84p, complex by transmission electron microscopy (TEM). The negatively stained samples used for TEM analysis were purified by affinity chromatography and gel filtration (see above) and were essentially pure as assayed by the Coomassie Blue-stained SDS–polyacrylamide gels (Figures 4 and and55).

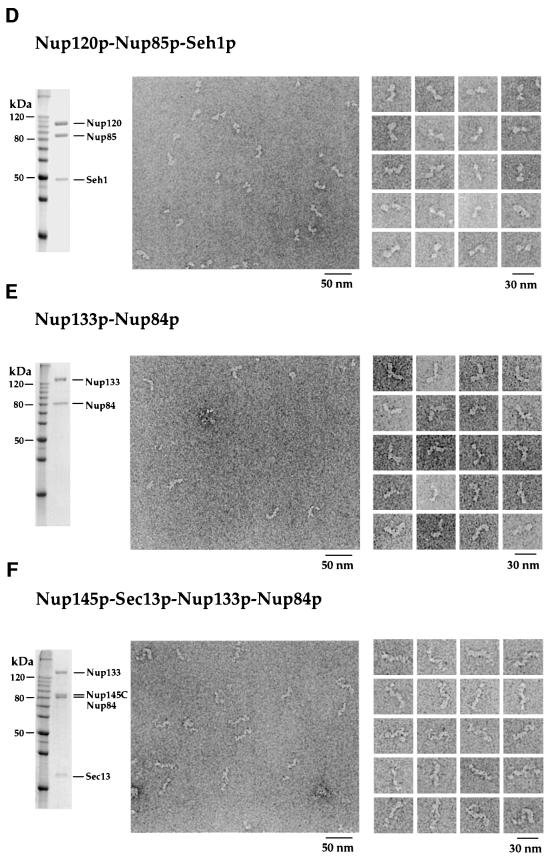
Fig. 4. The reconstituted intermediate heteromeric complexes display modular structures visualized by EM analysis after purification and negative staining. Shown is a Coomassie-stained SDS–polyacrylamide gel of the purified protein or complex (left panels), an overview electron micrograph of the negatively stained preparation (middle panels) and a gallery of selected particles (right panels). (A) Nup84p; (B) Nup85p–Seh1p complex; (C) Nup145p-C–Sec13p complex; (D) Nup120p–Nup85p–Seh1p complex; (E) Nup133p–Nup84p complex; (F) Nup133p–Nup84p–Nup145p-C–Sec13p complex.

Fig. 5. The reconstituted Nup84p complex displays a Y-shaped structure, in which one arm, the ‘stalk’ of the ‘Y’, is extended by Nup133p. Shown is the complex purified by SDS–PAGE with Coomassie Blue staining (left panels), an overview electron micrograph of the negatively stained preparation (middle panels) and a gallery of selected particles (right panels). (A) In vitro reconstituted hexameric Nup84p complex. (B) In vitro reconstituted pentameric Nup84p complex lacking the Nup84p subunit. (C) In vitro reconstituted heptameric Nup84p complex with bound Nup133p.
Purified Nup84p has a globular, donut-like shape with an apparent diameter of ~10 nm (Figure 4A). The Nup85p–Seh1p complex appears slightly elongated with a length of ~14 nm (Figure 4B). The Nup145p-C–Sec13p complex is elongated further, with a length of ~17 nm (Figure 4C). Addition of Nup120p to the Nup85p–Seh1p complex results in an elongated heterotrimeric complex with an apparent length of ~23 nm (Figure 4D) consisting of a larger (Nup120p) and a smaller lobe (Nup85p–Seh1p) joined in a kinked way. Evidently, the Nup120p– Nup85p–Seh1p complex exhibits a ‘two-winged’ morphology (Figure 4D).
Very much like the yeast purified Nup84p complex (Siniossoglou et al., 2000), the reconstituted Nup84p complex has a pronounced Y-shaped appearance with an overall diameter of ~25 nm in the electron microscope (Figure 5A). One arm (i.e. the ‘stalk’) of the Y-shaped complex is slightly longer than the other two arms (see below). The morphology of the complex varies somewhat, indicating flexible links between the ‘arms’ and the ‘stalk’. Moreover, the reconstituted complex appears to be heterogenous to a certain extent under the electron microscope, very much like the authentic complex, possibly due to partial denaturation during specimen preparation. We conclude that the morphology of the reconstituted complex is very similar to that of the authentic yeast complex (Siniossoglou et al., 2000).
Interestingly, a Nup84p complex that lacks the Nup84p subunit could be assembled in E.coli as well as in vitro. After negative staining, this penta meric complex consisting of Nup120p–Nup85p–Seh1p–Nup145p-C–Sec13p also exhibits the typical Y-shaped structure (Figure 5B). However, its stalk is significantly shorter that that of the entire reconstituted Nup84p complex (compare Figure 5B with A). Hence, stalk elongation correlates with binding of Nup84p to the Nup145p-C–Sec13p arm. These data clearly document that (i) the Nup84p subunit is dispensable for the formation of a core complex, and (ii) the Y-shaped morphology does not depend on the presence of Nup84p within the core complex (see Discussion).
Finally, we determined the structural contribution of Nup133p upon binding to the Nup84p complex. Since Nup133p alone cannot be purified in significant amounts (see above), the simplest soluble complex that contains Nup133p is the Nup133p–Nup84p heterodimer, which is found in an elongated and often boomerang-like shape (Figure 4E). This suggests that Nup133p is an extended molecule and that its joining with Nup84p at one tip generates a curved structure. When the intermediate tetrameric complex Nup145p-C–Sec13p–Nup84p– Nup133p is visualized by EM, it reveals an ~36 nm long stalk that sometimes appears twisted (Figure 4F). Finally, the Nup84p complex with bound Nup133p exhibits a Y-shaped, or sometimes somewhat T-shaped, structure with a pronounced 36 nm long stalk (Figure 5C) that is significantly extended compared with that of the Nup84p complex (compare Figure 5C with A). Taken together, our data demonstrate that binding of Nup133p causes elongation of the Nup145p-C–Sec13p–Nup84p stalk of the Y-shaped Nup84p complex (see Discussion).
Discussion
To date, it is not known how the huge NPC assembles and which nucleoporins constitute its various substructures, such as the spoke complex, the nuclear and cytoplasmic rings, the nuclear basket, the distal ring or the pore filaments, which protrude into the pore environment. Our work here provides a first step towards the in vitro reconstitution of the yeast NPC and reveals how distinct pore structures may be generated from individual nucleoporins. By combining double plasmid transformation with bi-cistronic mRNA translation, it was possible to express and purify all members of the yeast Nup84p complex from E.coli and to assemble them in vitro into a distinct multimeric NPC subcomplex. Although we do not have a functional assay for the reconstituted Nup84p complex, all published data support our observation that the in vitro assembled complex does not differ from the native complex in molecular weight, subunit composition or structural appearance under the electron microscope.
Also, to our knowledge, this is the first report demonstrating that as many as five different heterologous ORFs can be expressed simultaneously in a single E.coli cell. Hence, our work may be relevant for research on heterologous gene expression and reconstitution of multimeric complexes. The vast majority of proteins in a eukaryotic cell do not act by themselves, but rather function as multiprotein complexes that represent principal units of biological activity. One scientific as well as commercial goal in proteomic research is to develop and apply methodologies and technologies for the systematic, genome-wide functional analysis of multiprotein complexes and then to reconstitute these molecular machines as functional in vitro or in heterologous systems.
Unfortunately, a frequent problem in heterologous gene expression is that single recombinant proteins produced in E.coli or other hosts are insoluble and/or not properly folded because the correct binding partner is absent. Our work provides evidence that folding and complex formation are facilitated or enhanced when interacting partner proteins are co-expressed in the same E.coli cell. Expression, folding and assembly were improved even further when two cognate subunits were expressed from a bi-cistronic mRNA. Apparently, the status nascendi of partner proteins emerging from the ribosomes, which are bound to the same bi-cistronic mRNA, favors complex formation and thus increases their solubility. This strategy of heterologous gene expression may be useful for oligomeric complexes in general, which so far have been difficult or impossible to assemble in vitro. Finally, the E.coli assembled nucleoporin complexes may be well suited for crystallization trials to solve their structure at an atomic level.
Another important finding presented here is that nucleoporins exhibit an extremely high specificity for interaction with the co-expressed cognate partner nucleoporin in E.coli, since they heterodimerize in the presence of thousands of different E.coli proteins. Furthermore, the capability for self-assembly in E.coli points to a very high affinity between the various subunits within the Nup84p complex. This may suggest that in yeast the cognate partner nucleoporins assemble early in the cytoplasm or in other cell compartments (e.g. in the ER or at the NE) during or shortly after their synthesis rather than at the newly forming NPC.
Remarkable in this context is the fact, that some of the heteromeric interactions (e.g. the formation of the heterodimeric Nup85p–Seh1p and Nup145p-C–Sec13p complexes) within the Nup84p complex can only take place in E.coli (i.e. they require in vivo conditions for proper association). Evidently, Nup145p-C and most of the Nup85p without bound Sec13p and Seh1p, respectively, are insoluble. This suggests that in yeast too, Nup145p-C and Nup85p may require a co-translational assembly with Sec13p and Seh1p. On the other hand, once these heterodimeric complexes are formed, they are soluble and can also be used in vitro to build up larger multimeric complexes. As a consequence, in vivo, the Nup84p complex may be formed from heterodimeric or heterotrimeric modules, which assemble where they are needed to yield the hexameric complex that is finally incorporated into the newly forming NPC. Therefore, the need for an in vivo environment for some subunit associations may point to early steps in complex assembly.
Importantly, our reconstitution approach in E.coli was helpful in unravelling the direct interaction partners within the Nup84p complex. In this way, several distinct subcomplexes could be identified and were determined during sequential reconstitution: such as (i) dimeric Nup85p–Seh1p; (ii) trimeric Nup120p–Nup85p– Seh1p; (iii) dimeric Nup145p-C–Sec13p; (iv) trimeric Nup145p-C–Sec13p–Nup120p; (v) trimeric Nup145p-C– Sec13p–Nup84p; (vi) dimeric Nup133p–Nup84p; (vii) tetrameric Nup145p-C–Sec13p–Nup84p–Nup133p; and, finally, (viii) pentameric Nup84p complex lacking Nup84p. As sketched in Figure 6, taken together, this information now allows us to draw an assembly map charting the direct physical contacts between the six members within the Nup84p complex. Clearly, such a detailed assembly map could not be derived from cross-linking or in vivo depletion studies (Rappsilber et al., 2000; Siniossoglou et al., 2000). However, cross-linking studies revealed several binary and higher interactions, which are largely consistent with the studies presented in this work (Rappsilber et al., 2000).

Fig. 6. Structural model of the Nup84p–Nup133p complex. Representative single particles of the different subcomplexes and the whole Nup84p complex with or without Nup133p (see Figures 4 and 5) were taken from scanned electron micrographs using Adobe Photoshop. After magnification by the same factor, the pictures were blurred to overcome the pixelated appearance and arranged as depicted. The modular assembly of the Nup84p complex from the different subcomplexes and the elongation of its stalk by adding Nup133p to Nup84p are apparent. Note that the depicted EM pictures are processed extensively and thus should be considered only as models. The labels of the binary complexes (e.g. Nup145p-C–Sec13p, Nup85p–Seh1p) do not imply exactly where each subunit within the heterodimer is located.
Notably, a Nup84p complex which lacks the Nup84p subunit could be assembled in vivo and in vitro. Clearly, Nup84p is not required to form the Y-shaped complex. In yeast cells, NUP84 can be deleted without a significant growth defect at physiological temperatures, and a core complex lacking Nup84p can still be isolated, while deletion of NUP85, NUP120 or NUP145 is extremely deleterious to the cell and no core complexes can be isolated from such cells (Siniossoglou et al., 1996). Furthermore, yeast cells lacking NUP84 show, in contrast to the core subunit disrupted cells, only a very mild mRNA export defect, but, on the other hand, show strong abnormalities in NE organization (Siniossoglou et al., 1996). These facts, together with the finding that NUP84 genetically interacts with SPO7 and NEM1, two nuclear membrane proteins not associated with NPCs (Siniossoglou et al., 1998), suggest that Nup84p somehow links NPCs with other functions of the NE such as nuclear membrane biogenesis. These data are consistent with the in vitro findings described here that Nup85p, Nup145p-C and Nup120p are the core subunits, which constitute the Y-shaped Nup84p complex (see Figure 6). Nup84p, however, as discussed above, is a peripheral subunit, which localizes to the tip of the Nup145p-C–Sec13p arm and functions as a ‘joint’ to attach Nup133p to the complex (see below).
Another novel finding derived from our in vitro reconstitution experiments is that Nup133p interacts directly via Nup84p with the Nup84p complex (see Figure 2D). This has not been detectable in vivo so far because the interaction may not be strong enough to persist during biochemical purification, or Nup133p may be proteolytically unstable. For the first time, we could also determine the structure of Nup133p using EM. Obviously, Nup133p is an elongated molecule, which associates with Nup84p through head to tail contact. Upon Nup133p binding to the Nup84p complex, the stalk (Nup145p-C–Sec13p– Nup84p) of the Nup84p complex becomes significantly extended (Figure 6, lower panel). It will be interesting to find out whether the Nup133p extension also constitutes an exposed and accessible stalk in vivo, to which other nucleoporins or shuttling transport factors can bind. Since a green fluorescent protein (GFP)-tagged version of Nup133p is still localized to the NPCs in a nup84 disrupted strain, Nup133p may interact with additional NPC constituents (M.Lutzman, unpublished results). In this context, it is worth mentioning that Nup133p plays a role in mRNA export (Doye et al., 1994; Li et al., 1995; Pemberton et al., 1995). mRNPs, during their transport through the pore may associate with the Nup133p stalk, which could act as a docking and/or release site. In addition, Nup133p was implicated in preventing NPCs from becoming clustered within the NE (Doye et al., 1994). It was speculated that Nup133p is exposed from the NPC so that it can interact with a nucleo- or cytoskeletal component to maintain the distribution of NPCs within the nuclear membrane (Doye et al., 1994). Such a role would be consistent with the elongated structure of the Nup133p molecule and its interaction with Nup84p, which is itself dispensable for maintenance of the core complex.
Recently, an in vivo interaction was seen between the corresponding vertebrate homologs Nup133p and Nup107p (which corresponds to yeast Nup84p) (Belgareh et al., 2001). Furthermore, an orthologous human hNup107p complex, which was also designated the Nup160 complex, was reported (Belgareh et al., 2001; Vasu et al., 2001). This metazoan complex, however, contains only four nucleoporins, hNup133p, hNup107p, hNup120p and hNup96p (which corresponds to yeast Nup145p-C) (Belgareh et al., 2001; Vasu et al., 2001). There is also evidence that homologs of Sec13p and Seh1p are present in this human complex (Fontoura et al., 1999; Vasu et al., 2001). However, no mammalian homolog of Nup85p has yet been found associated with the human hNup107p complex. We believe that a mammalian Nup85p also exists, but it may dissociate from the biochemically stable hNup107p core complex. According to our structural data, we predict that the isolated mammalian hNup107p complex exhibits an elongated, probably kinked, but not Y-shaped structure, because it lacks the hNup85p–hSeh1p heterodimer. Our data may also allow us to predict how the various subunits within the human complex are organized. Accordingly, hNup107p and hNup133p form a subcomplex, which probably binds via the hNup107p to a hNup96p– hNup120p subcomplex.
The work presented here demonstrates that nucleoporins within the Nup84p complex have distinct locations, interaction partners and structural appearance. Through assembly of structurally defined subcomplexes, the Y-shaped Nup84p complex is generated, and we were able to unravel how this is facilitated by in vitro reconstitution combined with EM analysis (see Figure 6). During this study, it became obvious that the larger nucleoporins in general have an elongated structure, and that the structures become even more elongated and specific by head to tail contacts between the heteromeric building units. The Y-shaped fork of the Nup84p complex finally is generated by two contacts with Nup120p, one with the Nup85p arm and the other with Nup145p-C. This interlocked interaction between Nup120p, Nup85p and Nup145p-C may stabilize the overall Y-shaped structure. Such an interlocking of nucleoporins at distinct positions by forming multiple contacts may be a general principle in the organization of the NPC structure to stabilize the overall three-dimensional network.
The longest elongated structure within the Nup84p– Nup133p complex is represented by a linear array of Nup133p–Nup84p–Nup145p-C–Sec13p, with a length of ~42 nm. This array already could constitute a thick filament or fibril. Whether such a stalk is exposed and can recruit other nucleoporins or nucleocytoplasmic transport factors remains to be determined. Therefore, we now aim to let the Nup84p complex grow in vitro by searching for further interaction partners and by co-expressing them together with already known subunits of the Nup84p complex.
Finally, an important goal for future research is to position and orient the Nup84 complex exactly into a three-dimensional mass density map of the entire NPC. From immuno-EM data, it was suggested that the subunits of the Nup84p–Nup133p complex are located on both faces of the NPC (Rout et al., 2000). A similar conclusion has been derived recently for the conserved human Nup106 complex (Belgareh et al., 2001). Furthermore, a member of the Nup84 complex, Nup145p-C, is involved in organizing the intranuclear pore filaments (Galy et al., 2000). Thus, it is likely that the Nup84p–Nup133p complex is a peripheral NPC structure that is part of either the nuclear and cytoplasmic rings or of the attached peripheral elements such as the nuclear basket or the cytoplasmic filaments.
Materials and methods
E.coli strains, microbiological techniques, plasmids and DNA manipulations
The BL21 codon plus RIL E.coli strain was used in this study for heterologous gene expression. Standard DNA manipulations (restriction analysis, ligation, PCR amplification) and microbiological techniques (growth and transformation of E.coli strains and induction of protein expression) were performed as described earlier (Santos-Rosa et al., 1998).
Insertion of yeast nucleoporin ORFs in E.coli expression plasmids
For expression of single nucleoporins in E.coli, all ORFs except NUP120 and NUP133 were cloned into a pET24d-derived vector containing a kanamycin (kan) resistance marker (pET24d-GST-TEV), which allowed expression of N-terminal tagged GST fusion proteins under the control of the T7/lac promoter. To create this vector, the coding sequence of the GST affinity tag followed by the recognition sequence for the TEV protease was inserted in front of the multiple cloning site of pET24d (Novagen) using standard PCR and cloning techniques. The ORFs of NUP120 and NUP133 were cloned into a pET8c vector, which contains an ampicillin (amp) resistance marker, allowing the expression of N-terminal His6-tagged fusion proteins under the control of the T7 plain promoter, which is not completely repressed in the non-induced state, but allows higher expression levels. For bi-cistronic expression of GST-NUP85 and SEH1, the NUP85 ORF was cloned initially into pET24d-GST-TEV. Then a second ribosomal-binding site was created in the 5′ region of the SEH1 ORF by PCR using a primer including the ribosomal-binding sequence GAAGGA in front of the start codon of SEH1. This 5′ primer sequence was 5′-GGGGGGGGATCCAATA ATTTTGTTTAACTTTAAGAAGGAGATATACATATGCAACAAT TTGATAGTGGG, with the ribosomal-binding site shown in italics and the beginning of the SEH1 ORF in bold. The PCR fragment of SEH1 containing the second ribosomal-binding site was inserted downstream of the stop codon of NUP85. This approach of introducing a second ribosomal-binding site via PCR for bi-cistronic mRNA expression was also used for the GST-NUP145C/SEC13 and GST-NUP84/NUP133 pairs. The bi-cistronic cloning of NUP145C/SEC13 was carried out in a pProEXHT-based vector, containing an ampicillin resistance marker, in which the GST–TEV sequence together with the multiple cloning site of the pET24d-GST-TEV vector was cloned 5′ in front of the His6 tag of pPROEXHT. This allowed for expression of GST-tagged proteins under the control of the TRC promoter, which in our experience is even stronger than the T7 promoter. The bi-cistronic NUP133/GST-NUP84 pair was expressed from a pET8c vector. In this case, NUP133 was expressed as an N-terminal His6 fusion and the 3′ adjacent NUP84 ORF was expressed as the GST–TEV fusion protein.
Expression of recombinant proteins in E.coli
Recombinant nucleoporins were expressed in E.coli BL21 codon plus RIL cells (Stratagene). Pre-cultures were grown in minimal medium containing the appropriate antibiotics (34 µg of chloramphenicol plus 50 µg/ml kanamycin, ampicillin or both) overnight at 37°C. Cultures to express the recombinant proteins were started with a 100-fold dilution of the pre-culture in minimal medium lacking the antibiotics and were grown at 37°C to an OD600 of 0.7. Cultures were then shifted to 23°C and induced with a final concentration of 0.8 mM IPTG. The induced cultures were grown up to 12 h before cells were pelleted by centrifugation, frozen in liquid nitrogen and stored at –20°C.
Affinity purification of GST-tagged proteins
Frozen E.coli cells containing the expressed nucleoporins were thawed and lysed by sonification in lysis buffer [300 mM NaCl, 150 mM KOAc, 20 mM Tris pH 7.9, 2 mM Mg(OAc)2 10% glycerol] containing 0.1% NP-40. After centrifugation (15 min, 27 000 r.p.m.), the supernatant was incubated with glutathione–Sepharose 4B beads (Amersham/Pharmacia) for 2 h at 4°C. After binding, beads were first washed with 20 vols of lysis buffer and finally with 5 vols of lysis buffer containing 1 mM dithiothreitol (DTT). TEV cleavage was performed in 1 vol. of lysis buffer + 1 mM DTT at 4°C for 8–12 h and the eluted proteins or protein complexes were concentrated by ultrafiltration. The concentrated eluates were used for further purification by FPLC gel filtration or used directly for binding assays with other purified complexes. For the in vitro reconstitution reactions, the different in vivo-assembled subcomplexes were mixed in approximately equimolar amounts and incubated for 90 min at 23°C on a turning wheel. Gel filtration analysis was performed in lysis buffer using an Äkta-Basic-System (Amersham/Pharmacia). Except for the hexameric Nup84p complex and the heptameric Nup84p–Nup133p complex, which were purified over a Superose 6 30/10 column (Amersham/Pharmacia), all the other complexes and single proteins were purified over a Superdex 200 HR 30/10 column (Amersham/Pharmacia). During gel filtration, 0.6 ml fractions were collected and analyzed by SDS–PAGE. Single fractions containing the highly purified proteins/complexes were used for EM.
Electron microscopy
Negative staining was performed as described previously (Siniossoglou et al., 2000). In brief, a 10 µl drop of sample was placed on a carbon–collodion-coated grid, which had been freshly glow-discharged in air. After washing with 100 µl of buffer, the sample was stained by washing with 100 µl of 1% (w/v) aqueous uranyl acetate and blotted dry. Micrographs were taken at 80 kV in a Philips 400T electron microscope at a magnification of ×50 000.
Acknowledgements
We thank Günther Stier (EMBL Heidelberg) for technical assistance during vector construction. The critical proofreading of Dr Matthew Grooves and Dr Tracy Nissan is also acknowledged. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB352; to E.C.H.), the Human Frontiers Science Program (HFSP; to E.C.H. and U.A.), the Swiss National Science Foundation (4036-044061 to N.P. and 3100-053034 to U.A.), and by the Kanton Basel-Stadt and the M.E.Müller Foundation of Switzerland.
References
- Aitchison J.D., Blobel,G. and Rout,M.P. (1995) Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J. Cell Biol., 131, 1659–1675. [Europe PMC free article] [Abstract] [Google Scholar]
- Allen N.P., Huang,L., Burlingame,A. and Rexach,M.F. (2001) Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem., 276, 29268–29274. [Abstract] [Google Scholar]
- Allen T.D., Cronshaw,J.M., Bagley,S., Kiseleva,E. and Goldberg,M.W. (2000) The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci., 113, 1651–1659. [Abstract] [Google Scholar]
- Bagley S., Goldberg,M.W., Cronshaw,J.M., Rutherford,S.A. and Allen,T.D. (2000) The nuclear pore complex. J. Cell Sci., 113, 3885–3886. [Abstract] [Google Scholar]
- Bayliss R., Littlewood,T. and Stewart,M. (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell, 102, 99–108. [Abstract] [Google Scholar]
- Belgareh N. et al. (2001) An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol., 154, 1147–1160. [Europe PMC free article] [Abstract] [Google Scholar]
- Doye V., Wepf,R. and Hurt,E.C. (1994) A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J., 13, 6062–6075. [Europe PMC free article] [Abstract] [Google Scholar]
- Fontoura B.M.A., Blobel,G. and Matunis,M.J. (1999) A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol., 144, 1097–1112. [Europe PMC free article] [Abstract] [Google Scholar]
- Galy V., Olivo-Martin,J., Scherthan,H., Doye,V., Rascalou,N. and Nehrbass,U. (2000) Nuclear pore complexes in the organization of silent telomeric chromatin. Nature, 403, 108–112. [Abstract] [Google Scholar]
- Gerace L., Ottaviano,Y. and Kondor-Koch,C. (1982) Identification of a major polypeptide of the nuclear pore complex. J. Cell Biol., 95, 826–837. [Europe PMC free article] [Abstract] [Google Scholar]
- Goldstein A.L., Snay,C.A., Heath,C.V. and Cole,C.N. (1996) Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol. Biol. Cell, 7, 917–934. [Europe PMC free article] [Abstract] [Google Scholar]
- Hallberg E., Wozniak,R.W. and Blobel,G. (1993) An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J. Cell Biol., 122, 513–521. [Europe PMC free article] [Abstract] [Google Scholar]
- Heath C.V., Copeland,C.S., Amberg,D.C., Del Priore,V., Snyder,M. and Cole,C.N. (1995) Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J. Cell Biol., 131, 1677–1697. [Europe PMC free article] [Abstract] [Google Scholar]
- Kraemer D.M., Strambio-de-Castillia,C., Blobel,G. and Rout,M.P. (1995) The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J. Biol. Chem., 270, 19017–19021. [Abstract] [Google Scholar]
- Li O., Heath,C.V., Amberg,D.C., Dockendorff,T.C., Copeland,C.S., Snyder,M. and Cole,C.N. (1995) Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol. Biol. Cell, 6, 401–417. [Europe PMC free article] [Abstract] [Google Scholar]
- Panté N., Bastos,R., McMorrow,I., Burke,B. and Aebi,U. (1994) Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol., 126, 603–617. [Europe PMC free article] [Abstract] [Google Scholar]
- Pemberton L.F., Rout,M.P. and Blobel,G. (1995) Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc. Natl Acad. Sci. USA, 92, 1187–1191. [Europe PMC free article] [Abstract] [Google Scholar]
- Rappsilber J., Siniossoglou,S., Hurt,E.C. and Mann,M. (2000) A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal. Chem., 72, 267–275. [Abstract] [Google Scholar]
- Rexach M. and Blobel,G. (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell, 83, 683–692. [Abstract] [Google Scholar]
- Rout M.P., Aitchison,J.D., Suprapto,A., Hjertaas,K., Zhao,Y. and Chait,B.T. (2000) The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol., 148, 635–651. [Europe PMC free article] [Abstract] [Google Scholar]
- Santos-Rosa H., Moreno,H., Simos,G., Segref,A., Fahrenkrog,B., Panté,N. and Hurt,E. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol., 18, 6826–6838. [Europe PMC free article] [Abstract] [Google Scholar]
- Siniossoglou S., Wimmer,C., Rieger,M., Doye,V., Tekotte,H., Weise,C., Emig,S., Segref,A. and Hurt,E.C. (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell, 84, 265–275. [Abstract] [Google Scholar]
- Siniossoglou S., Santos-Rosa,H., Rappsilber,J., Mann,M. and Hurt,E. (1998) A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J., 17, 6449–6464. [Europe PMC free article] [Abstract] [Google Scholar]
- Siniossoglou S., Lutzmann,M., Santos-Rosa,H., Leonard,K., Mueller,S., Aebi,U. and Hurt,E.C. (2000) Structure and assembly of the Nup84p complex. J. Cell Biol., 149, 41–53. [Europe PMC free article] [Abstract] [Google Scholar]
- Stoffler D., Fahrenkrog,B. and Aebi,U. (1999) The nuclear pore complex: from molecular architecture to functional dynamics. Curr. Opin. Cell Biol., 11, 391–401. [Abstract] [Google Scholar]
- Vasu S.K. and Forbes,D.J. (2001) Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol., 13, 363–375. [Abstract] [Google Scholar]
- Vasu S., Shah,S., Orjalo,A., Park,M., Fischer,W.H. and Forbes,D.J. (2001) Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol., 155, 3339–3354. [Europe PMC free article] [Abstract] [Google Scholar]
- Walther T.C., Fornerod,M., Pickersgill,H., Goldberg,M., Allen,T.D. and Mattaj,I.W. (2001) The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J., 20, 5703–5714. [Europe PMC free article] [Abstract] [Google Scholar]
- Wozniak R.W., Blobel,G. and Rout,M.P. (1994) POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J. Cell Biol., 125, 31–42. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1093/emboj/21.3.387
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc125826?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/emboj/21.3.387
Article citations
Docking a flexible basket onto the core of the nuclear pore complex.
Nat Cell Biol, 26(9):1504-1519, 13 Aug 2024
Cited by: 0 articles | PMID: 39138317 | PMCID: PMC11392808
Nuclear pore dysfunction and disease: a complex opportunity.
Nucleus, 15(1):2314297, 21 Feb 2024
Cited by: 2 articles | PMID: 38383349 | PMCID: PMC10883112
Review Free full text in Europe PMC
The scaffold nucleoporins SAR1 and SAR3 are essential for proper meiotic progression in Arabidopsis thaliana.
Front Cell Dev Biol, 11:1285695, 04 Dec 2023
Cited by: 0 articles | PMID: 38111849 | PMCID: PMC10725928
Implications of a multiscale structure of the yeast nuclear pore complex.
Mol Cell, 83(18):3283-3302.e5, 01 Sep 2023
Cited by: 6 articles | PMID: 37738963 | PMCID: PMC10630966
Distinct domains in Ndc1 mediate its interaction with the Nup84 complex and the nuclear membrane.
J Cell Biol, 222(6):e202210059, 08 May 2023
Cited by: 1 article | PMID: 37154843 | PMCID: PMC10165475
Go to all (162) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structure and assembly of the Nup84p complex.
J Cell Biol, 149(1):41-54, 01 Apr 2000
Cited by: 121 articles | PMID: 10747086 | PMCID: PMC2175103
A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores.
Cell, 84(2):265-275, 01 Jan 1996
Cited by: 230 articles | PMID: 8565072
Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins.
EMBO J, 16(16):5086-5097, 01 Aug 1997
Cited by: 67 articles | PMID: 9305650 | PMCID: PMC1170143
Structural analysis of the nuclear pore complex by integrated approaches.
Curr Opin Struct Biol, 19(2):226-232, 25 Mar 2009
Cited by: 40 articles | PMID: 19327984
Review
Funding
Funders who supported this work.
Swiss National Science Foundation (1)
Grant ID: 044061




