Abstract
Free full text

Widespread and Persistent Populations of a Major New Marine Actinomycete Taxon in Ocean Sediments
Abstract
A major taxon of obligate marine bacteria within the order Actinomycetales has been discovered from ocean sediments. Populations of these bacteria (designated MAR 1) are persistent and widespread, spanning at least three distinct ocean systems. In this study, 212 actinomycete isolates possessing MAR 1 morphologies were examined and all but two displayed an obligate requirement of seawater for growth. Forty-five of these isolates, representing all observed seawater-requiring morphotypes, were partially sequenced and found to share characteristic small-subunit rRNA signature nucleotides between positions 207 and 468 (Escherichia coli numbering). Phylogenetic characterization of seven representative isolates based on almost complete sequences of genes encoding 16S rRNA (16S ribosomal DNA) yielded a monophyletic clade within the family Micromonosporaceae and suggests novelty at the genus level. This is the first evidence for the existence of widespread populations of obligate marine actinomycetes. Organic extracts from cultured members of this new group exhibit remarkable biological activity, suggesting that they represent a prolific resource for biotechnological applications.
The recently proposed class Actinobacteria (20) is comprised of high-G+C-content gram-positive bacteria and includes the actinomycetes (order Actinomycetales), whose members have an unparalleled ability to produce diverse secondary metabolites. These bacteria are primarily saprophytic and are best known from soils where they contribute significantly to the turnover of complex biopolymers, such as lignocellulose, hemicellulose, pectin, keratin, and chitin (27). Additionally, nitrogen-fixing actinomycetes of the genus Frankia have one of the broadest host ranges known, forming root nodule symbioses in more than 200 species of flowering plants (7).
Despite their importance in soil ecology, actinomycetes are best known as a source of antibiotics. This became apparent in 1940, following Selman Waksman's seminal discovery of actinomycin (24), and was fully realized by the 1980s, when actinomycetes accounted for almost 70% of the world's naturally occurring antibiotics (15). In the past two decades, however, there has been a decline in the discovery of new lead compounds from common soil-derived actinomycetes as culture extracts yield unacceptably high numbers of previously described metabolites. For this reason, the cultivation of rare or novel actinomycete taxa has become a major focus in the search for the next generation of pharmaceutical agents (2).
It is interesting that the world's oceans, which cover 70% of the earth's surface and include some of the most biodiverse ecosystems on the planet, have not been widely recognized as an important resource for novel actinomycetes. In fact, the distributions of actinomycetes in the sea remain largely undescribed, and even today, conclusive evidence that these bacteria play important ecological roles in the marine environment has remained elusive. Speculation regarding the existence of indigenous populations of marine actinomycetes arises because these bacteria produce resistant spores that are known to be transported from land into the sea where they can remain viable but dormant for many years (2, 3, 5). Thus, it has been frequently assumed that actinomycetes isolated from marine samples are merely of terrestrial origin. This assumption has persisted despite evidence that actinomycetes can be recovered from deep-ocean sediments (25) and that marine-derived actinomycetes can be metabolically active (13) and physiologically adapted to growth in seawater (8). Despite evidence supporting the growth of actinomycetes in the sea, only one marine species, Rhodococcus marinonascens, has been described (6) and the inclusion of the actinomycetes within the autochthonous marine microbiota has not been widely accepted (2).
We report here the isolation and phylogenetic characterization of members of a new actinomycete taxon that we refer to as MAR 1. These bacteria represent the first major actinomycete taxon to be reported exclusively from the ocean. MAR 1 strains can be consistently isolated from marine sediments and are distinguished by morphological characteristics, small-subunit rRNA signature nucleotides, and an obligate requirement of seawater for growth. MAR 1 strains have been isolated on five separate occasions from both tropical and subtropical near-shore sediments collected from the Atlantic Ocean, the Red Sea, and the Sea of Cortez, suggesting a worldwide distribution.
MATERIALS AND METHODS
Sample collection and bacterial isolation.
A total of 112 sediment samples were collected during four separate research expeditions. Samples of the top 1 cm of sediment were collected in sterile 50-ml plastic Whirl-Pak bags (NASCO, Modesto, Calif.) by divers using scuba gear when necessary. Sediment sample depth ranges, numbers, locations, and dates were as follows: 0 to 30 m, 45 sediment samples (Bahamas, July 1999); 0 to 30 m, 20 sediment samples (Bahamas, August 2000); 10 to 30 m, 42 sediment samples (Red Sea, Egypt, February, 2000); and 0 to 30 m, 5 sediment samples (Sea of Cortez, Mexico, 2000). These samples were processed as soon as possible after collection (generally, <4 h) using the selective methods of (i) stamping and (ii) dilution and heat shock or both (as described below) and were inoculated onto isolation media (M1 to M5). The dilution-and-heat-shock method was carried out as follows: 1 ml of wet sediment was added to 4 ml of sterile seawater, heated for 6 min at 55°C, vigorously shaken, and further diluted (1:4) in sterile seawater, and 50 μl of each dilution was inoculated by spreading with a sterile glass rod onto agar-based isolation media. The stamping method was carried out as follows: 10 ml of wet sediment was aseptically placed into a sterile aluminum dish, dried (ca. 24 h) in a laminar flow hood, ground lightly with a pestle, pressed into a sterile foam plug (14 mm in diameter), and inoculated onto agar media by stamping eight or nine times in a circular fashion, giving a serial dilution effect. All media were prepared with 100% filtered natural seawater. Actinomycetes generally appeared after 2 to 6 weeks of incubation at 25 to 28°C and were considered to be any colony with a tough leathery texture, dry or folded appearance, and branching filaments with or without aerial mycelia. Hence, this study was focused on culturable, filamentous actinomycetes. Isolation media consisted of the following: M1, 10 g of starch, 4 g of yeast extract, 2 g of peptone, 18 g of agar, and 1 liter of natural seawater; M2, 6 ml of 100% glycerol, 1 g of arginine, 1 g of K2HPO4, 0.5 g of MgSO4, 18 g of agar, and 1 liter of natural seawater; M3, 6 g of glucose, 2 g of chitin (United States Biochemical, Cleveland, Ohio), 18 g of agar, and 1 liter of natural seawater; M4, 2 g of chitin, 18 g of agar, and 1 liter of natural seawater; and M5, 18 g of agar and 1 liter of natural seawater. All isolation media were amended with filtered (0.2-μm pore size) cycloheximide (100 μg/ml) and rifampin (5 μg/ml), after autoclaving.
Effects of seawater and sodium chloride on growth.
Isolates were screened for seawater and sodium growth requirements and sodium chloride tolerance. All media were modified from M1 (no antibiotics added) and prepared with either natural seawater (M1/natural seawater), deionized water (M1/DI water), artificial seawater (M1/ASW Na+, prepared using the recipe of Sieburth) (19), artificial seawater in which all sodium ion components were replaced with equimolar amounts of potassium ions (M1/ASW K+), or M1 prepared with deionized water and 1 M sodium chloride added (M1/DI water + 1 M Na+). Macerated vegetative mycelia were inoculated onto the analytical media using a sterile cotton swab, and the plate contents were incubated at 25 to 28°C for 6 to 8 weeks. Growth was monitored at up to ×64 magnification by using a Leica stereoscope (Leica Microscopy Systems Ltd., Heerbrugg, Switzerland).
Nucleic acid extraction.
Genomic DNA was prepared as follows: 10 mg of vegetative mycelia grown on M1 agar for 2 to 4 weeks at 25 to 28°C was harvested and macerated, and an aqueous cleared lysate was prepared using a method modified from the one described by Marmur (12). Genomic DNA within this cleared lysate was precipitated using 0.7 volumes of isopropanol, and the resultant DNA pellet was washed with 70% ethanol and resuspended in 10 mM Tris buffer (pH 8.5) to a final concentration of 100 μg/ml.
16S rRNA gene (16S rDNA) amplification and sequencing.
16S ribosomal DNA (rDNA) sequencing templates were amplified from 10 to 50 ng of genomic DNA by PCR using primers FC27 (5′-to-3′ AGAGTTTGATCCTGGCTCAG) and RC1492 (5′-to-3′ TACGGCTACCTTGTTACGACTT). PCR products were purified with a Qiagen QIAquick PCR cleanup kit using the manufacturer's protocols (Qiagen Inc., Chatsworth, Calif.). Partial sequences of morphologically diverse strains were obtained from nucleotides 80 to 480 (Escherichia coli numbering) by using the FC27 primer. Select 16S rDNA amplicons were sequenced almost in their entirety on both top and bottom strands by using a total of 10 primers: FC27 and RC1492 (used in template amplification), F357 (5′-to-3′ TACGGGAGGCAGCAG), FM536 (5′-to-3′ CAGCAGCCGCGGTAAGAC), F803 (5′-to-3′ ATTAGATACCCTGGTAG), F1114 (5′-to-3′ GCAACGAGCGCAACCC), R343 (5′-to-3′ CTGCTGCCTCCCGTA), RM519 (5′-to-3′ GTCTTACCGCGGCTGCTG), RM907 (5′-to-3′ CCGTCAATTCCTTTGAGTTT), and RM1378 (5′-to-3′ CGGTGTGTACAAGGCCCGGGAACG). The above sequencing primers were from the method described by Rainey et al. (17) except for FC27, RC1492, and those primers denoted with an “M” that were created specifically for this study. The resultant 10 individual sequences were then assembled, yielding 1,479 to 1,483 nucleotides.
Phylogenetic analyses.
All phylogenetic analyses were performed on nearly complete 16S rDNA sequences (nine strains). 16S rDNA sequences were compared to sequences in available databases by use of the Basic Local Alignment Search Tool online service to determine approximate phylogenetic positions (1). 16S rDNA similarity values were calculated by the Ribosomal Data Project (RDP) similarity matrix online analysis and compared to the three nearest neighbors in the RDP database (11). Hypervariable regions in the 16S rDNA sequences were excluded, yielding a total of 1,408 aligned nucleotides. Sequences were aligned to the secondary structure of members of the family Micromonosporaceae in the RDP (11) by using BioEdit software (4). Phylogenetic analyses were performed using the neighbor-joining and parsimony-based algorithms in Clustal W (23) and PHYLIP software packages (5), respectively. The dendrogram (Fig. (Fig.1)1) was drawn using TreeView 1.6.1 (16).
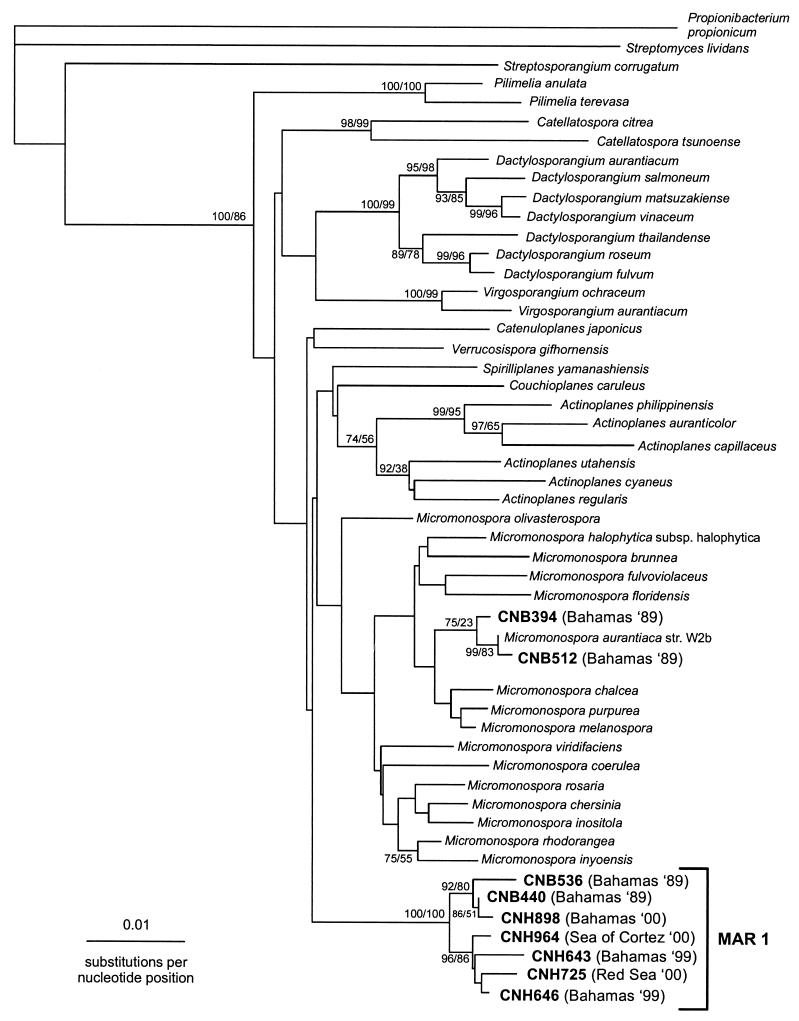
Phylogenetic relationships determined from almost complete 16S rDNA sequences of select MAR 1 isolates, CNB512 and CNB394 (Micromonospora spp.) and representatives of all 10 presently accepted genera within the Micromonosporaceae (9, 18, 22). Bootstrap values (in percent) calculated from 1,000 resamplings using neighbor-joining (value on left) and parsimony methods (value on right) are shown at their respective nodes when the values calculated using at least one method were 70% or greater. Propionibacterium propionicum, Streptosporangium corrugatum, and Streptomyces lividans were used as outgroups.
Electron microscopy.
Cells were grown for 5 to 10 days in M1 broth shaken at 250 rpm at 25 to 28°C. Cells were then fixed by adding formalin to a final concentration of 3.7% and washed three times with 2 volumes of deionized water. Resuspended cells were spotted on a glass slide, flash frozen in a bath of 2-methylbutane in liquid nitrogen, freeze-dried, and then sputter coated with gold-palladium alloy under vacuum. Samples were visualized at a magnification of ×4,000 by using a Cambridge S-360 scanning electron microscope.
Cultivation, extraction, and bioactivity testing.
One hundred and five strains were cultured in shake flasks at 230 rpm for 7 to 14 days in 100-ml volumes of seawater-based media. Whole cultures were extracted with equal volumes of ethyl acetate, and the ethyl acetate layers were removed and dried with anhydrous sodium sulfate. The ethyl acetate fractions were concentrated by rotary evaporation, and the resulting extracts were weighed and brought up to 25 mg/ml in dimethyl sulfoxide and stored at −20°C in 96-well microtiter plates. Extracts were tested at a single dose for biological activity against the human colon tumor cell line HCT-116 (25 μg/ml), vancomycin-resistant Enterococcus faecium (VREF) (50 μg/ml), and amphotericin-resistant Candida albicans (ARCA) (150 μg/ml) by using standard microtiter plate methods. Extracts inhibiting cell growth by ≥50% (HCT-116) or ≥95% (VREF and ARCA) were considered active, serially diluted, and retested to generate 50% inhibitory concentrations (HCT-116) and MICs (VREF and ARCA).
Nucleotide sequence accession numbers.
The 16S rDNA sequences of the nine strains used in this study were submitted to GenBank and assigned the following accession numbers: CNB440 (AY040617), CNB536 (AY040618), CNH643 (AY040619), CNH646 (AY040620), CNH725 (AY040621), CNH898 (AY040622), CNH964 (AY040623), CNB512 (AY040624), and CNB394 (AY040625).
RESULTS
MAR 1 isolation and morphology.
Overall, 53 out of 112 sediment samples (47%) yielded colonies with MAR 1 morphologies. In general, these colonies first appeared after 3 weeks of incubation but sometimes took as long as 6 weeks to appear, depending upon the medium and isolation method. MAR 1 colonies were recognized by their lack of aerial hyphae and pigmentation that ranged from bright to pale orange to black on M1 medium and the frequent production of spreading substrate mycelia when grown on the low-nutrient media M4 and M5. Dark brown to black, bright orange, or pink diffusible pigments are frequently produced. Colonies can become darkened during sporulation with spores produced singly or in clusters on the colony surface or within substrate mycelia. Vegetative hyphae are finely branched and do not fragment. Neither sporangia nor spore motility has been observed for any isolate, suggesting that, within the Micromonosporaceae, the morphology of MAR 1 strains resembles those of genera such as Micromonospora and Catellatospora (9). Scanning electron microscopy (magnification, ×4,000) showed that spore diameter ranged from 0.8 to 3.8 μm and vegetative hyphae diameter ranged from 0.25 to 0.5 μm.
Seawater and sodium ion requirements.
All strains tested grew equally well on media prepared with either natural or artificial seawater (Table (Table1).1). No detectable growth was observed for any of the MAR 1 isolates on M1/DI water. The two Micromonospora isolates, CNB394 and CNB512, grew better on M1/DI water than on M1/natural seawater. MAR 1 isolates did not tolerate a sodium chloride level of 1 M (ca. twice the NaCl content of natural seawater), whereas growth was clearly evident for the two Micromonospora strains CNB394 and CNB512 on the medium M1/DI + 1 M Na+.
TABLE 1.
Seawater and sodium growth requirements of selected isolatesa
| Mediumb | Results for isolates tested
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CNB394 | CNB512 | CNB440 | CNB536 | CNH643 | CNH646 | CNH721 | CNH898 | CNH964 | |
| M1/natural seawaterc | + | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| M1/DI water | ++ | ++ | − | − | − | − | − | − | − |
| M1/ASW Na+d | + | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| M1/ASW K+d | + | + | − | − | − | − | − | − | − |
| M1/DI + 1 M Na+e | +/− | +/− | − | − | − | − | − | − | − |
Phylogenetic analyses.
Phylogenetic analyses of the nearly complete 16S rDNA sequences of seven MAR 1 strains (Table (Table2)2) indicate that they form a coherent clade within the Micromonosporaceae (Fig. (Fig.1).1). Signature nucleotides unify this clade (Table (Table3),3), and a high bootstrap value supports clear separation from the 10 presently described genera within the family. The three species to which members of this clade show closest homology are Micromonospora olivasterospora (97.1 to 97.7% similarity), Verrucosispora gifhornensis (96.8 to 97.4% similarity), and Catenuloplanes japonicus (96.3 to 97.0% similarity). The MAR 1 clade shares 9 of the 12 previously published Micromonospora-specific signature nucleotide positions with the most deeply rooted member of that genus, M. olivasterospora. This is more dissimilar than the recently described species V. gifhornensis, which shows 98.0% similarity to M. olivasterospora and shares 11 of 12 Micromonospora-specific signature nucleotides (18).
TABLE 2.
Strains used for phylogenetic and physiological studies
| Straina | Source or reference | Yr (location) | Habitat description and depth | Taxonomic designation |
|---|---|---|---|---|
| CNH643 | This study | 1999 (Bahamas, Sweetings Cay) | Coarse sand, 1 m | MAR 1 |
| CNH646 | This study | 1999 (Bahamas, Andros Island) | Spur and groove, 10 m | MAR 1 |
| CNH725 | This study | 2000 (Red Sea, Sha'b el utal) | Coarse sand, 20 m | MAR 1 |
| CNH898 | This study | 2000 (Bahamas, Little San Salvador) | Coarse sand, 30 m | MAR 1 |
| CNH964 | This study | 2000 (Sea of Cortez, Caleta Partida) | Coarse sand, 30 m | MAR 1 |
| CNB440 | 8 | 1989 (Bahamas, Chub Cay) | Spur and groove, 20 m | MAR 1 |
| CNB536 | 8 | 1989 (Bahamas, Acklins Island) | Coarse sand and sea grass, 10 m | MAR 1 |
| CNB394 | 8 | 1989 (Bahamas, Chub Cay) | Coarse sand and sea grass, 1 m | Micromonospora |
| CNB512 | 8 | 1989 (Bahamas, San Salvador Island) | Spur and groove, 30 m | Micromonospora |
TABLE 3.
16S rRNA signature nucleotides distinguishing the MAR1 group from all other genera within the Micromonosporaceaea
| Nucleotide position (16S rRNA)b | Signature for:
| |
|---|---|---|
| Micromonosporaceaec | MAR 1d | |
| 207 | (U/C) | A |
| 366 | (A/G) | C |
| 467 | (A/G) | U |
| 468 | A | U |
| 1456 | A | G |
Initial MAR 1 identification.
The MAR 1 group was initially recognized after phylogenetic characterization of sediment-derived actinomycetes isolated during a 1999 expedition to the Bahamas. Of the 45 actinomycetes obtained, 40 possessed MAR 1 morphological characteristics, indicating that this group was the numerically dominant filamentous actinomycete cultured. All of these MAR 1 morphotypes failed to grow on an agar medium when seawater was replaced with deionized water. Two strains (CNH643 and CNH646 [Table [Table2])2]) were further tested for the requirement of sodium for growth, a physiological characteristic commonly associated with obligate marine bacteria, and both required this cation (Table (Table1).1). Partial 16S rDNA gene sequences were obtained for eight morphologically diverse strains, and all eight were found to possess four signature nucleotides between positions 207 and 468 (E. coli numbering [Table [Table3]).3]). Of these, two strains showing the highest phylogenetic diversity (CNH643 and CNH646) were sequenced nearly in their entirety and were found to form a distinct clade within the Micromonosporaceae (Fig. (Fig.11).
Persistence and abundance.
In August 2000, a follow-up study was undertaken in the Bahamas to determine the persistence of the MAR 1 group. Of the 111 actinomycete strains isolated from 20 sediment samples, 90% displayed characteristic MAR 1 morphologies, again supporting the observation that this group may be the numerically dominant filamentous actinomycete in marine sediments. Interestingly, 9 of the 11 non-MAR 1 actinomycetes obtained were isolated from shoreline samples, suggesting that the distribution of some actinomycete taxa may be restricted to intertidal regions. Over 50% of the MAR 1 isolates appeared on the low-nutrient medium M4, indicating the importance of using appropriate isolation techniques. The average abundance of MAR 1 strains calculated from triplicate platings of samples from five transects (0 to 30 m), processed using the heat shock method and plated on media M1 to M4, ranged from 1.2 × 103 to 2.3 × 103 CFU/ml, suggesting that these bacteria are abundant in marine sediments. The numbers of CFU obtained from any one sample, however, ranged from 0 to 104/ml, indicating a patchy distribution. Thirteen representatives of eight different MAR 1 colony morphotypes obtained during the Bahamas 2000 expedition were partially sequenced, and the phylogenetically diverse isolate CNH898 (Table (Table2)2) was sequenced nearly in its entirety and found to belong to the MAR 1 clade (Fig. (Fig.11).
An examination of 30 actinomycetes that possessed MAR 1 morphological characteristics and were isolated from Bahamian sediments collected in 1989 (8) revealed that all but two of these strains had an obligate requirement of seawater for growth. All 30 of these strains were previously recognized as Micromonospora-like; however, their phylogenetic relationship to genera within the Micromonosporaceae had not been determined (8). Ten seawater-requiring strains from the 1989 expedition representing six different morphotypes were partially sequenced and found to possess the five MAR 1 signature nucleotides between positions 207 and 468 (Table (Table3).3). Analysis of the nearly complete 16S rDNA sequences of two of these, CNB440 and CNB536 (Table (Table2),2), indicates that they are diverse members of the MAR 1 clade (Fig. (Fig.1).1). Thus, strains belonging to this new taxon have been isolated from near-shore Bahamian sediments on three separate occasions over an 11-year period, indicating that they are persistent members of the sediment bacterial community.
Distribution.
To determine if MAR 1 members had a broader distribution, marine sediments were collected from the Red Sea and the Sea of Cortez. From 42 Red Sea sediment samples, 22 isolates with MAR 1 morphologies were obtained and all of these displayed an obligate requirement of seawater for growth. Six isolates representing four major morphotypes were partially sequenced and found to contain the MAR 1 signature nucleotides between positions 207 and 468 (Table (Table3).3). The almost complete 16S rDNA sequence of one Red Sea strain, CNH725 (Table (Table2),2), is represented in Fig. Fig.11 and is clearly a member of the MAR 1 clade. From five sediments collected in the Sea of Cortez, 20 seawater-requiring actinomycetes with MAR 1 morphologies were isolated. Eight strains representing five morphotypes were partially sequenced, and all eight possessed the MAR 1 signature nucleotides (Table (Table3).3). The phylogenetically diverse isolate CNH964 (Table (Table2)2) was sequenced almost in its entirety (Fig. (Fig.1)1) and shares membership within the MAR 1 clade. These data clearly indicate that MAR 1 members are widely distributed in tropical and subtropical marine sediments.
Occurrence of Micromonospora in marine sediments.
Two strains, CNB394 and CNB512 (Table (Table2),2), with colony morphologies similar to that of MAR 1, were isolated in 1989, but these strains did not require seawater for growth and were found to lack all of the MAR 1 signatures between positions 207 and 468. Analyses of the almost complete 16S rDNA sequences of these strains showed all previously published Micromonospora signatures (9), 99.6 to 99.9% similarity to Micromonospora aurantiaca strain W2b, and clear phylogenetic placement in the Micromonospora clade (Fig. (Fig.1).1). Micromonospora isolates have been reported from marine sediments (21), including deep-sea samples (26); however, unlike MAR 1 strains, this genus is well known from terrestrial soils and there is no evidence that marine isolates require seawater for growth. From six independent soil samples collected above the high-tide level (supralittoral) during the Bahamas 2000 expedition, we observed over 200 actinomycete colonies, including strains with Micromonospora-like morphologies (ca. 10%); however, none of these required seawater for growth. Our inability to recover MAR 1 strains from supralittoral samples supports our observation that these bacteria are restricted to the marine environment.
Biological activities of organic extracts.
MAR 1 isolates appear to represent a remarkable source of biologically active secondary metabolites. Of the 105 strains examined, 86% yielded culture extracts with significant cancer cell cytotoxicities (50% inhibitory concentrations ranging from 0.004 to 16.4 μg/ml against the HCT-116 cell line). Liquid chromatography-mass spectrometry analyses of these extracts indicate considerable strain-to-strain chemical diversity, with active peaks corresponding to metabolites with different retention times and molecular weights (data not shown). Significant antifungal and antibiotic activities were also observed, with 30% of the crude extracts yielding MICs of 19.5 μg/ml or less for ARCA and 35% yielding extract MICs of 25 μg/ml or less for VREF. Chemical studies of active extracts have thus far led to the isolation of a novel series of cytotoxic β-lactones (to be published elsewhere) and other diverse chemical classes of biologically active secondary metabolites.
DISCUSSION
The data presented here provide the first conclusive evidence for the widespread and persistent occurrence of indigenous actinomycete populations in marine sediments. Phylogenetic analyses indicate that the MAR 1 group of actinomycetes comprises a new taxon within the Micromonosporaceae. MAR 1 strains possess characteristic signature nucleotides and a requirement of sodium for growth, a hallmark of obligate marine bacteria. Sodium requirements have been studied extensively in gram-negative marine bacteria and are indicative of highly evolved marine adaptations, such as a respiration-dependent sodium ion pump and/or a sodium-dependent membrane transport mechanism (10, 14). Sodium requirements are uncommon in gram-positive bacteria, and the discovery of the MAR 1 group appears to represent the first genus-level gram-positive taxon to reside exclusively in the ocean.
It is important to note that over 2,000 MAR 1 isolates have been added to our culture collection in addition to the 212 strains reported in this study. These strains were obtained during recent expeditions to the U.S. Virgin Islands and Guam and a second expedition to the Sea of Cortez. Thus, MAR 1 strains have been recovered from all five tropical and/or subtropical locations sampled to date. During the Guam expedition, strains with MAR 1 characteristics were recovered from sediments collected as deep as 600 m, suggesting that these bacteria are widely distributed in deep as well as shallow sediments (data to be reported in a future publication). The relative ease with which this new group can be cultured when appropriate isolation methods are employed suggests that traditional techniques need to be reevaluated and that further investment in new isolation methods will add significantly to studies of microbial diversity. Improved methodologies coupled with future culture-intensive studies may ultimately dispel the long-standing belief that the vast majority of marine bacteria cannot be cultivated.
It is interesting that Micromonospora isolates CNB394 and CNB512 did not require seawater for growth yet were tolerant to a higher concentration of sodium chloride than the MAR 1 strains (Table (Table1).1). This characteristic suggests that certain marine-derived Micromonospora isolates may be adapted to osmotically variable intertidal or supralittoral environments, whereas MAR 1 strains are adapted to the relatively constant salinities associated with deeper marine sediments.
Phylogenetic analyses of the nearly complete 16S rDNA sequences of the seven MAR 1 strains indicate that they form a robust and coherent clade within the Micromonosporaceae (Fig. (Fig.1).1). A high bootstrap value supports clear separation from the 10 presently described genera within the family. These data together with the signature nucleotides, similarity values, and physiological requirements for seawater suggest that the MAR 1 group comprises a new genus. Although it is unlikely that the diversity within the MAR 1 clade has been revealed in the present study, intragroup 16S rDNA sequence similarity (98.6%) and a robust clade topology suggest that it is composed of multiple species (Fig. (Fig.1).1). We are currently collaborating on a formal, polyphasic taxonomic description of this new group for which the generic epithet “Salinospora” will be proposed.
Placement of the MAR 1 group within the Micromonosporaceae is supported by the presence of a complete set of family-specific 16S rRNA signature nucleotides (20). MAR 1 strains share greater than 94% similarity to other genera within the Micromonosporaceae, and genera within this family form phylogenetically coherent entities using multiple treeing methods. However, the low bootstrap values calculated at the supra-generic nodes indicate uncertainty about the order in which various genera within the Micromonosporaceae emerged during evolution. Radiations such as these have been proposed to represent rapid, episodic evolutionary events possibly brought about by severe environmental stress (28). This radiation appears to have occurred about 38 to 75 million years ago (based on a calculation of 1 to 2% base change per 50 million years) (4) during a period in the earth's history when sea level dropped over 300 m. It is interesting to hypothesize that this change in sea level acted as a selective pressure leading to the emergence of species within the Micromonosporaceae that occur only in the ocean, species that are restricted to land, and species, such as the Micromonospora strains studied here, that appear to be capable of surviving in both habitats. The widespread distribution, phylogenetic divergence, abundance, and physiological adaptations of the MAR 1 group suggest that it represents a highly evolved and ecologically significant member of the marine sediment bacterial community. These actinomycetes may play important ecological roles, similar to their saprophytic relatives in soils, perhaps substantially impacting the cycling of complex carbon substrates in benthic ocean habitats. Studies comparing MAR 1 strains with other members of the Micromonosporaceae may reveal more details of how marine and terrestrial adaptations differ and the evolutionary events that led to the diversification of the family. Future work with the MAR 1 group is aimed at providing a better understanding of their distributions, biotechnological potential, and ecological roles in the marine environment.
Acknowledgments
This research is a result of financial support from the National Science Foundation, Chemistry Division, under grant CHE 9807098; the National Institutes of Health, National Cancer Institute, under grant CA 44848; University of California BioSTAR project award no. 00-10102; and a fellowship granted to T.J.M. from the Khaled Bin Sultan Living Oceans Foundation. We thank His Royal Highness Prince Khaled Bin Sultan Bin Abdul-Aziz for his generous support of our Red Sea research program. The Molecular Pathology Shared Resource, University of California at San Diego Cancer Center, which is mentioned below, is funded in part by NCI Cancer Center Support Grant no. 5P0CA23100-16.
We also thank the officers and crew of the M/Y Golden Shadow and R/V Seward Johnson and J. Pawlik for his invitation to participate in the Bahamas 1999 and 2000 R/V Seward Johnson expeditions. DNA sequencing was performed by the Molecular Pathology Shared Resource, University of California at San Diego Cancer Center.
REFERENCES
Articles from Applied and Environmental Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aem.68.10.5005-5011.2002
Read article for free, from open access legal sources, via Unpaywall:
https://aem.asm.org/content/aem/68/10/5005.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101973102
Article citations
Endophytic Streptomyces: an underexplored source with potential for novel natural drug discovery and development.
Arch Microbiol, 206(11):442, 22 Oct 2024
Cited by: 0 articles | PMID: 39436470
Review
Anti-Melanogenic Activity of Undecylprodigiosin, a Red Pigment Isolated from a Marine Streptomyces sp. SNA-077.
Biomol Ther (Seoul), 32(4):492-498, 23 Apr 2024
Cited by: 0 articles | PMID: 38651201 | PMCID: PMC11214958
Dry Stamping Coral Powder: An Effective Method for Isolating Coral Symbiotic Actinobacteria.
Microorganisms, 11(12):2951, 10 Dec 2023
Cited by: 0 articles | PMID: 38138095 | PMCID: PMC10745815
Complete genome sequence analysis of plant growth-promoting bacterium, Isoptericola sp. AK164 isolated from the rhizosphere of Avicennia marina growing at the Red Sea coast.
Arch Microbiol, 205(9):307, 14 Aug 2023
Cited by: 2 articles | PMID: 37580455 | PMCID: PMC10425560
Discovery and biosynthetic assessment of 'Streptomyces ortus' sp. nov. isolated from a deep-sea sponge.
Microb Genom, 9(5), 01 May 2023
Cited by: 1 article | PMID: 37166955 | PMCID: PMC10272871
Go to all (251) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 9 of 9)
- (1 citation) ENA - AY040623
- (1 citation) ENA - AY040622
- (1 citation) ENA - AY040621
- (1 citation) ENA - AY040620
- (1 citation) ENA - AY040625
- (1 citation) ENA - AY040624
- (1 citation) ENA - AY040619
- (1 citation) ENA - AY040618
- (1 citation) ENA - AY040617
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Culturable marine actinomycete diversity from tropical Pacific Ocean sediments.
Environ Microbiol, 7(7):1039-1048, 01 Jul 2005
Cited by: 141 articles | PMID: 15946301
Prauserella marina sp. nov., isolated from ocean sediment of the South China Sea.
Int J Syst Evol Microbiol, 60(pt 4):985-989, 07 Aug 2009
Cited by: 17 articles | PMID: 19666793
Kineococcus marinus sp. nov., isolated from marine sediment of the coast of Jeju, Korea.
Int J Syst Evol Microbiol, 56(pt 6):1279-1283, 01 Jun 2006
Cited by: 21 articles | PMID: 16738104
Pseudonocardia antitumoralis sp. nov., a deoxynyboquinone-producing actinomycete isolated from a deep-sea sediment.
Int J Syst Evol Microbiol, 63(pt 3):893-899, 25 May 2012
Cited by: 18 articles | PMID: 22634702
Funding
Funders who supported this work.
NCI NIH HHS (5)
Grant ID: 5P01CA23100-16
Grant ID: CA 44848
Grant ID: R01 CA044848
Grant ID: R37 CA044848
Grant ID: P30 CA023100




