Abstract
Free full text

Hypervariable Sites in the mtDNA Control Region Are Mutational Hotspots
Abstract
Hypervariable sites in human mtDNA are readily identified in evolutionary studies and are usually assumed to represent mutational hotspots. Recently, an alternative hypothesis was proposed that holds that hypervariable sites may instead reflect ancient mtDNA mutations that have been “shuffled” among different lineages via recombination. These hypotheses can be tested by examining the evolutionary rates for sites at which new mtDNA mutations are observed; if hypervariable sites are mutational hotspots, then newly arisen mtDNA mutations should occur preferentially at hypervariable sites. Results of this study show that both germline and somatic mtDNA mutations occur preferentially at hypervariable sites, which supports the view that hypervariable sites are indeed mutational hotspots.
The existence of hypervariable sites (sites that evolve at a rate much faster than average) in the noncoding human mtDNA control region has been well documented via various analyses of human mtDNA variation (Hasegawa et al. 1993; Wakeley 1993; Excoffier and Yang 1999; Meyer et al. 1999); however, it is not known why such hypervariability exists. In the absence of any evidence, it has been generally assumed that hypervariable sites represent mutational hotspots, but recently it was suggested that hypervariable sites may actually reflect ancient mutations that have been redistributed among mtDNA lineages via recombination (Eyre-Walker et al. 1999; Hagelberg et al. 1999). Although recent claims of recombination in human mtDNA (Awadalla et al. 1999; Eyre-Walker et al. 1999; Hagelberg et al. 1999) have not been substantiated in some instances (Macaulay et al. 1999; Hagelberg et al. 2000) and remain controversial (Jorde and Bamshad 2000; Kivisild and Villems 2000; Kumar et al. 2000; Parsons and Irwin 2000), the suggestion that hypervariable sites are not mutational hotspots deserves consideration.
One way to test this hypothesis is to examine the evolutionary rate for sites at which new mutations have been observed. If hypervariable sites are indeed mutational hotspots, then new mutations should occur preferentially at hypervariable sites. However, if hypervariable sites are not mutational hotspots but, instead, reflect recombination (or some other process), then new mutations would not be expected to occur preferentially at hypervariable sites. In this report, I analyze the evolutionary rate for sites at which new mutations are observed, to test the hypothesis that hypervariable sites are mutational hotspots.
The evolutionary rate for each site in the two hypervariable segments (HV1 and HV2; positions 16024–16383 and 57–372, respectively, numbered according to the reference sequence [Anderson et al. 1981]) of the human mtDNA control region has been estimated elsewhere (Meyer et al. 1999), from patterns of variation in worldwide populations; the overall average evolutionary rate has been found to be .60 for HV1 alone and .54 for HV1 and HV2. Two large family studies reported a total of 12 germline mtDNA mutations for HV1 and HV2 (Parsons et al. 1997; Sigurdardottir et al. 2000), and the average evolutionary rate for the sites at which these 12 mutations occurred is 2.82 (table 1), which is 5.2 times greater than the overall average evolutionary rate for HV1 and HV2. Data are also available for 12 somatic mtDNA mutations in HV1 and HV2 that were observed in tumors but not in control normal tissues from the same individuals (Fliss et al. 2000). The average evolutionary rate for the sites at which these 12 tumor mutations occurred is 2.86 (table 1), which is 5.3 times greater than the overall average evolutionary rate for HV1 and HV2. Finally, a number of studies have documented the existence of heteroplasmy (i.e., the existence of more than one mtDNA type) for sites in the mtDNA control region (Comas et al. 1995; Bendall et al. 1996, 1997; Wilson et al. 1997; Hühne et al. 1998; Sigurdardottir et al. 2000; Tully et al. 2000). In principle, the heteroplasmic state of an individual could have arisen either via transmission from the mother (i.e., through the germline) or via a new somatic mutation occurring within the individual; most of the studies that have reported heteroplasmy have not attempted to determine which mechanism might be responsible. Therefore, for this analysis, all heteroplasmic mutations were grouped together, resulting in data on 51 mutations (table 1) in HV1 alone. The average evolutionary rate for the sites at which these 51 heteroplasmic mutations were observed is 2.87 (table 1), which is 4.8 times greater than the overall evolutionary rate for HV1.
Table 1
Sites in HV1 and HV2 at Which Germline, Tumor, and Heteroplasmic Mutations Were Observed and Their Estimated Evolutionary Rate
| No. of Mutations | ||||
| Sitea | Rateb | Germline | Tumor | Heteroplasmic |
| 16092 | .69 | 1 | 0 | 0 |
| 16093 | 2.98 | 2 | 0 | 13 |
| 16104 | .47 | 0 | 0 | 2 |
| 16111 | 2.57 | 1 | 0 | 1 |
| 16124 | .81 | 0 | 0 | 1 |
| 16129 | 4.78 | 0 | 0 | 6 |
| 16172 | 3.76 | 0 | 1 | 1 |
| 16183 | 3 | 0 | 1 | 0 |
| 16185 | .61 | 0 | 0 | 1 |
| 16187 | 3.67 | 0 | 1 | 0 |
| 16189 | 4.78 | 0 | 1 | 3 |
| 16192 | 3.6 | 0 | 0 | 3 |
| 16205 | 0 | 0 | 0 | 1 |
| 16222 | .41 | 0 | 0 | 1 |
| 16234 | .85 | 0 | 0 | 1 |
| 16239 | .79 | 0 | 0 | 2 |
| 16256 | 2.3 | 1 | 0 | 2 |
| 16257 | .44 | 0 | 0 | 1 |
| 16262 | .05 | 0 | 0 | 1 |
| 16265 | 2.18 | 0 | 1 | 0 |
| 16292 | 2.32 | 0 | 1 | 0 |
| 16293 | 3.9 | 0 | 0 | 3 |
| 16295 | .72 | 0 | 0 | 1 |
| 16300 | 2.1 | 0 | 1 | 0 |
| 16301 | .3 | 0 | 0 | 1 |
| 16309 | 4.03 | 0 | 0 | 2 |
| 16311 | 4.78 | 0 | 0 | 1 |
| 16355 | 2.16 | 0 | 0 | 1 |
| 16362 | 4.72 | 0 | 0 | 2 |
| 16380 | 0 | 0 | 1 | 0 |
| 75 | 0 | 0 | 1 | 0 |
| 94 | 0 | 1 | 0 | 0 |
| 114 | .17 | 0 | 1 | 0 |
| 150 | 6.12 | 0 | 1 | 0 |
| 153 | 3.99 | 1 | 0 | 0 |
| 185 | 4.86 | 1 | 0 | 0 |
| 189 | 6.22 | 1 | 0 | 0 |
| 195 | 6.22 | 0 | 1 | 0 |
| 207 | 3.29 | 2 | 0 | 0 |
| 234 | .69 | 1 | 0 | 0 |
 Totalc Totalc | 12 (2.82) | 12 (2.86) | 51 (2.87) | |
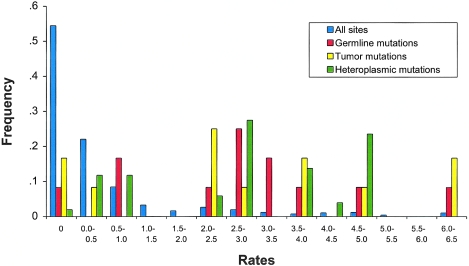
The distribution of inferred evolutionary rates for all sites in HV1 and HV2 is shown in figure 1, along with the distributions for the sites at which germline, tumor, and heteroplasmic mtDNA mutations were reported. A nonparametric (Mann-Whitney) test indicates that the distribution of evolutionary rates for sites at which germline, tumor, and heteroplasmic mutations were reported are all significantly different from the distribution of evolutionary rates for all sites in HV1 and HV2 (P<.001 for all comparisons). Thus, germline, tumor, and heteroplasmic mtDNA mutations all occur preferentially at hypervariable sites in the human mtDNA control region.
The average evolutionary rates for sites at which germline, tumor, and heteroplasmic mutations were observed were remarkably similar (table 1). Moreover, the distribution of evolutionary rates (fig. 1) does not differ among germline, tumor, or heteroplasmic mutations (pairwise Mann-Whitney tests; P>.05 for all comparisons). This overall similarity among germline, tumor, and heteroplasmic mutations may indicate that the underlying mutational processes for mtDNA in germ cells and in somatic cells are similar. However, it is also possible that some (or even all) of the supposed germline mutations inferred from family studies are actually somatic mutations that occurred early during the prenatal development of the individual and drifted to homoplasy and that heteroplasmic mutations may reflect an intermediate stage in this process. The observed close correspondence between the evolutionary rates for somatic mutations (both tumor and heteroplasmic) and for germline mutations would then arise because the supposed germline mutations are in fact somatic mutations. If not all mutations observed in family studies are true germline mutations, then this may also account for the fact that evolutionary rates that have been determined from family studies are higher than those inferred from evolutionary studies (Parsons et al. 1997; Jazin et al. 1998; Sigurdardottir et al. 2000). It would be of interest to know whether individuals who exhibit what appear to be newly arisen, germline mtDNA mutations actually transmit the new mutation to their offspring.
In conclusion, both somatic and (supposed) germline mtDNA mutations occur preferentially at hypervariable sites in HV1 and HV2. This result is not consistent with the idea that such hypervariable sites may reflect ancient mutations that have spread to other mtDNA lineages via recombination (Eyre-Walker et al. 1999; Hagelberg et al. 1999). A possible counterargument would be that new mutations actually are rare recombination events between paternal and maternal mtDNA. For this to be true, however, an unusual recombination mechanism must be postulated, because observed mutant mtDNA types differ from the parental type almost exclusively by single mutations, whereas paternal and maternal mtDNA typically differ at several nucleotide positions in the control region. Thus, any supposed mtDNA recombination must instead operate more like gene conversion, involving only short stretches of mtDNA sequence, perhaps even single nucleotide positions. It is difficult to imagine that such a mechanism could be operating in both the germline and in somatic cells (especially tumor cells). Moreover, if recombination involves only single nucleotide positions, then there is no longer the expectation that linkage disequilibrium should be negatively correlated with the physical distance between pairs of sites, a position still held by some (Awadalla et al. 2000) despite criticisms concerning the measure of linkage disequilibrium used (Jorde and Bamshad 2000; Kumar et al. 2000).
Given these difficulties with a recombinational mechanism for new mutations, it does appear that hypervariable sites are hotspots for both germline and somatic mutations. The challenge now facing mitochondrial geneticists is to determine what features of these hypervariable sites and/or the mtDNA replication machinery cause them to be mutational hotspots.
Acknowledgments
I thank S. Meyer, for providing the evolutionary rates for HV1 and HV2, and S. Pääbo, for useful discussion.
References
 (2000) Questioning evidence for recombination in human mitochondrial DNA. Science 288:1931 [Abstract] [Google Scholar]
(2000) Questioning evidence for recombination in human mitochondrial DNA. Science 288:1931 [Abstract] [Google Scholar] (2000) Evidence for mitochondrial DNA recombination in a human population of island Melanesia [correction]. Proc R Soc Lond B Biol Sci 267:1595–1596 [Europe PMC free article] [Abstract] [Google Scholar]
(2000) Evidence for mitochondrial DNA recombination in a human population of island Melanesia [correction]. Proc R Soc Lond B Biol Sci 267:1595–1596 [Europe PMC free article] [Abstract] [Google Scholar]Articles from American Journal of Human Genetics are provided here courtesy of American Society of Human Genetics
Full text links
Read article at publisher's site: https://doi.org/10.1086/303092
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S000292970763300X/pdf
Citations & impact
Impact metrics
Article citations
Variant load of mitochondrial DNA in single human mesenchymal stem cells.
Sci Rep, 14(1):20989, 09 Sep 2024
Cited by: 0 articles | PMID: 39251776 | PMCID: PMC11385243
Mitochondrial DNA mosaicism in normal human somatic cells.
Nat Genet, 56(8):1665-1677, 22 Jul 2024
Cited by: 1 article | PMID: 39039280 | PMCID: PMC11319206
The effect of library preparation protocol on the efficiency of heteroplasmy detection in mitochondrial DNA using two massively parallel sequencing Illumina systems.
J Appl Genet, 65(3):559-563, 19 Dec 2023
Cited by: 0 articles | PMID: 38110828 | PMCID: PMC11310223
Mutational profiling of mitochondrial DNA reveals an epithelial ovarian cancer-specific evolutionary pattern contributing to high oxidative metabolism.
Clin Transl Med, 14(1):e1523, 01 Jan 2024
Cited by: 1 article | PMID: 38193640 | PMCID: PMC10775184
Investigating the role of mitochondrial DNA D-loop variants, haplotypes, and copy number in polycystic ovary syndrome: implications for clinical phenotypes in the Chinese population.
Front Endocrinol (Lausanne), 14:1206995, 06 Sep 2023
Cited by: 0 articles | PMID: 37745710 | PMCID: PMC10512090
Go to all (140) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Phylogenetic and familial estimates of mitochondrial substitution rates: study of control region mutations in deep-rooting pedigrees.
Am J Hum Genet, 69(5):1113-1126, 01 Oct 2001
Cited by: 61 articles | PMID: 11582570 | PMCID: PMC1274355
A sensitive denaturing gradient-Gel electrophoresis assay reveals a high frequency of heteroplasmy in hypervariable region 1 of the human mtDNA control region.
Am J Hum Genet, 67(2):432-443, 28 Jun 2000
Cited by: 68 articles | PMID: 10873789 | PMCID: PMC1287188
Analysis of phylogenetically reconstructed mutational spectra in human mitochondrial DNA control region.
Hum Genet, 111(1):46-53, 07 Jun 2002
Cited by: 32 articles | PMID: 12136235
Recombination or mutational hot spots in human mtDNA?
Mol Biol Evol, 19(7):1122-1127, 01 Jul 2002
Cited by: 35 articles | PMID: 12082131
Review