Abstract
Free full text

Fast chromatin immunoprecipitation assay
Abstract
Chromatin immunoprecipitation (ChIP) is a widely used method to explore in vivo interactions between proteins and DNA. The ChIP assay takes several days to complete, involves several tube transfers and uses either phenol–chlorophorm or spin columns to purify DNA. The traditional ChIP method becomes a challenge when handling multiple samples. We have developed an efficient and rapid Chelex resin-based ChIP procedure that dramatically reduces time of the assay and uses only a single tube to isolate PCR-ready DNA. This method greatly facilitates the probing of chromatin changes over many time points with several antibodies in one experiment.
INTRODUCTION
Chromatin is composed of DNA, proteins and RNA (1–3). Chromatin structure is dynamic, responds to extracellular signals, and controls gene expression, cell division and DNA repair (3–5). Chromatin is one of the most intensely studied structures in biology and ChIP assays have proven to be a powerful means to investigate a host of DNA-dependent processes (3,6,7). Along with other techniques (8,9), Chromatin immunoprecipitation reveals an extraordinarily rich and dynamic chromatin environment (3,9).
The traditional method has several limitations, it takes several days to complete, requires DNA precipitations and involves multiple tube transfers (6). It is becoming increasingly clear that chromatin is a very dynamic structure (1,3,8,10–13). Thus, the traditional method may not be sufficient enough to explore the rich environment of chromatin. In the traditional ChIP protocol (6), DNA extractions and precipitations, are not only time consuming and tedious but also introduce steps for potential sample loss and contamination. This becomes a bigger problem in experiments involving multiple chromatin samples. We set out to simplify the standard ChIP protocol by eliminating the multiple tube transfers to purify DNA.
MATERIALS AND METHODS
Mammalian cells
Rat mesangial cells were grown in 150 mm plastic cell culture dishes in RPMI 1640 media supplemented with 10% FBS, 2 mM glutamine, penicillin (100 U/ml), streptomycin (0.01%) and humidified with 7/93% of CO2/air gas mixture (14).
Yeast strains
Media used for the growth of Saccharomyces cerevisiae were previously described (15); cells were grown at 30°C. The strain used in this study was MATα HMLα HMRa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1Δ-1 ura3-52 (16).
Reagents
Antibodies and blocking peptides
The antibody to the C-terminal peptide of K protein was raised in rabbits as described before (17). Anti-Sir2p serum was a gift from Jasper Rine, University of California, Berkeley (18). The other antibodies used, anti-RNA polymerase II, anti-histone H3 and rabbit IgG were from Santa Cruz (cat# sc-899), Abcam (cat# 1791), and Vector Laboratories (cat# I-1000), respectively. The blocking peptide to K protein antibody was previously described (17) and the blocking peptide to the RNA polymerase II antibody was obtained from Santa Cruz (cat# sc-899p).
Chelex-100 was purchased from BioRad (cat# 142–1253) and proteinase K from Invitrogen (cat# 25530–015).
In vivo cross-linking and immunoprecipitations
Cell cross-linking was done by adding 0.8 ml of 37% formaldehyde to 20 ml of overlaying media for 15 min at RT, followed by the addition of glycine to a final concentration of 125 mM (6). After cross-linking, cells were harvested and then washed twice with 10 ml phosphate-buffered saline (PBS). Cells from one dish were lysed with 1.0 ml IP buffer [150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% NP-40, 50 mM Tris–HCl (pH 7.5) and 0.5 mM DTT] containing the following inhibitors; 10 µg/ml leupeptin, 0.5 mM phenylmethlysulfonyl fluoride (PMSF), 30 mM p-nitrophenyl phosphate, 10 mM NaF, 0.1 mM Na3VO4, 0.1 mM Na2MoO4 and 10 mM β-glycerophosphate. After one wash with 1.0 ml IP buffer the pellet was resuspended in 1 ml IP buffer (containing all inhibitors) and sheared with a sonicator microprobe for 4 rounds of 15, 1 s pulses at output level 7 (Misonix 3000). Sheared chromatin was cleared by centrifugation (10 min at 17 000 g, Eppendorf 5403), split into two 0.5 ml fractions, and was used immediately or stored at −70°C. After adding antibody pre-incubated (30 min at room temp) with or without blocking peptide, 0.5 ml of the sheared chromatin fraction was incubated in an ultrasonic water bath (15 min, 4°C) (Bronson 3510). Tubes were centrifuged (10 min at 17 000 g, Eppendorf 5403) and the supernatant was transferred to fresh tubes containing 20 µl of washed protein A beads (Pharmacia) (19). The slurry was rotated for 45 min (4°C) and then the beads were washed five times with 1 ml cold IP buffer containing no inhibitors. Yeast chromatin was prepared as previously described (6).
Isolation of DNA using conventional phenol–chlorophorm method
This procedure is based on previously described methods (6,20). Briefly, DNA is eluted twice from the protein A beads with 250 µl of elution buffer (1% SDS and 0.1 M NaHCO3) for 15 min with periodic vortexing at room temperature. Cross-linking is reversed by adding 20 µl of 5 M NaCl and incubating the eluate overnight at 65°C. After adding 5 µg linear acrylamide, as a carrier (21), DNA is precipitated with 1.0 ml 100% EtOH. The pellet is washed with 1 ml 70% EtOH, and then dissolved in 100 µl TE buffer (pH 8.0). Proteins are digested by adding 11 µl 10× proteinase K buffer [0.1 M Tris (pH 7.8), 50 mM EDTA and 5% SDS] and 1 µl of 20 µg/µl proteinase K at 50°C for 30 min. DNA is extracted using phenol/chlorophorm and then chloroform, precipitated with ethanol and the final DNA pellet is dissolved in 200 µl TE buffer.
Isolation of PCR-ready DNA using the new method
A total of 100 µl of 10% Chelex (10 g/100 ml H2O) is added directly to the washed protein A beads and vortexed. After 10 min boiling, the Chelex/protein A bead suspension is allowed to cool to room temperature. Proteinase K (100 µg/ml) is then added and beads are incubated for 30 min at 55°C while shaking, followed by another round of boiling for 10 min. Suspension is centrifuged and supernatant is collected. The Chelex/protein A beads fraction is vortexed with another 100 µl water, centrifuged again, and the first and the second supernatants are combined. Eluate is used directly as a template in PCR and makes up to 25% of the final reaction volume.
Real-time PCR
The reaction mixture contained 5 µl 2× SYBR Green PCR Master Mix (Applied Biosystems), 2.5 µl DNA template and 0.3 µM primers (10 µl final volume) in 384-Well Optical Reaction Plate (Applied Biosystems). Amplification (three step, 40 cycles), data acquisition and analysis were done using the 7900HT Real-Time PCR system and SDS Enterprise Database (Applied Biosystems).
Calculations
Factor density from ChIP assays are expressed as a signal ratio, R, using the following formula, R = exp2(CTmock − CTspecific), where CTmock and CTspecific are mean threshold cycles of PCR done in triplicates on DNA samples from specific and mock immunoprecipitations.
RESULTS AND DISCUSSION
Method development
Chelex-100 resin has been used previously for DNA extraction from forensic specimens (22). We reasoned that addition of Chelex resin to immunoprecipitated chromatin samples could facilitate DNA extraction. To test if the Chelex-based method can efficiently extract DNA from immunoprecipitated chromatin we used antibodies to hnRNP K (K protein). K protein is a conserved DNA/RNA-binding protein involved in gene expression including transcription (23–27). Using the traditional ChIP assay, we have previously shown the binding of K protein to multiple gene loci (20). Recruitment of this factor to DNA was estimated by comparing the DNA signal obtained with the specific antibody to that obtained in mock immunoprecipitation where the antibody is blocked with a specific peptide (20). Control experiment (western blot, Figure 1A) shows that K protein immunoprecipitation is blocked with the specific peptide. Sheared chromatin from 3H-thymidine-labeled cells was pulled down with protein A beads and anti-K protein antibody pre-incubated with or without blocking peptide. Chelex-100 suspension was added to the washed protein A immunoprecipitates and after boiling, the Chelex/protein A bead suspensions were treated with proteinase K. 3H counts in the bead and supernatant fractions were measured by liquid scintillation. As shown in Figure 1B most of the 3H counts were recovered in the supernatants indicating that the DNA is efficiently extracted from the immunoprecipitated chromatin. In agreement with previous studies (20), these results also show that a fraction of chromatin binds to the antibody-loaded protein A beads non-specifically.

A Chelex-100-based method to isolate PCR-ready DNA from immunoprecipitated chromatin. (A) Anti-K protein antibody (10 µg) was pre-incubated with (+) or without (−) blocking peptide (4 µg) (30 min, RT) (17). Lysates were incubated with the antibodies, complexes were pulled down with protein A beads (IP), and after washing, proteins were eluted by boiling in SDS–PAGE loading buffer. Proteins were resolved by SDS–PAGE and, after transfer to PVDF membranes, immunostained (IS) with anti-K protein antibody. (B) Cells were labeled with 3H-thymidine overnight (10 µCi in 10 ml media). After cross-linking with formaldehyde, cells were lysed and sonicated. Half of the sonicated chromatin was incubated with antibody blocked with the peptide [(+) blocking peptide] and the other half with antibody that was not blocked [(−) blocking peptide]. After five washes with 1 ml of IP buffer, Chelex was added and the mixture was boiled. After cooling, proteinase K (200 µg/ml) was added and the tubes were incubated at 55°C for 60 min, mix was again boiled and beads were centrifuged. 3H counts in the supernatant and the beads were measured using a liquid scintillation counter. (C) Purified rat genomic DNA (Total DNA) was boiled with the Chelex/protein A beads suspension. After cooling to room temperature the suspension was treated with [(+) proteinase K] or without [(−) proteinase K] proteinase K and then the mix was boiled again for 10 min. The suspension was centrifuged and the supernatant was used as a template in real-time PCR using primers to the egr-1 and β-globin genes. Three step real-time PCR was run for 40 cycles. Results are expressed as 40-CT, (Threshold Cycle, Applied Biosystems, ABI7900 manual), which directly reflects levels of amplicons. (D) Sonicated chromatin was incubated with anti-K protein antibody as before. After five washes with 1 ml of IP buffer, Chelex was added and the mixture was boiled. After cooling the mix was treated without or with proteinase K (100 or 200 µg) for 60 min (55°C), suspension was boiled again and the released DNA was used as a template in real-time PCR. Plots show values mean ± SD n = 3.
The DNA eluted from protein A beads is typically treated with proteinase K to remove associated proteins. After proteolysis DNA is purified to remove the proteinase and peptide fragments (6). To avoid this DNA purification step, after treatment with proteinase K, we boiled the Chelex/protein A bead suspension containing genomic DNA. After centrifugation, the supernatant was used as template in real-time PCR with primers to the egr-1 and β-globin genes. Figure 1C illustrates that this procedure eliminated the potential inhibitory effects of proteinase K on PCR.
We next estimated the concentration of proteinase K needed to isolate DNA from the chromatin precipitated with antibodies to hnRNP K (Figure 1D). We found that DNA can be extracted even without proteinase K, however, addition of the enzyme increases DNA yield. The enhancing effect of proteinase K digestion was greater for the silent β-globin locus (Figure 1D, lower panel) compared with the active egr-1 locus (Figure 1D, upper panel), with a 2.4- compared with 1.5-fold increase in specific DNA yield, respectively. In subsequent experiments, we used 100 µg/ml concentration of proteinase K. In addition, we found that 30 min incubation with proteinase K is as effective as 4 h (data not shown). In subsequent experiments we used 30 min proteinase K treatment.
We also introduced an improvement in the immunoprecipitation step itself. In the ChIP assay, immunoprecipitation is typically carried out over several hours (6). We found that pre-incubating sheared chromatin with the antibody in an ultrasonic water bath (15 min at 4°C) first and then binding to protein A beads (45 min at 4°C) is sufficient to immunoprecipitate chromatin. This faster method for chromatin IP works well with different antibodies (20,28).
Verification of the fast ChIP assay in mammalian cells
To verify the new method, we examined kinetics of the recruitment of RNA polymerase II and K protein to the PMA-inducible loci. Using the traditional ChIP assay, we have previously demonstrated transient recruitment of hnRNP K to the inducibly transcribed egr-1 locus (20). The recruitment of RNA polymerase II is a highly regulated key step controlling the rate of mRNA synthesis from target gene loci (4,29). Treatment of rat mesangial cells with mitogens potently activates the immediate-early egr-1 gene (20). We compared kinetics of PMA-induced recruitment of RNA polymerase II and K protein to the egr-1 locus to the induction of egr-1 mRNA measured by real-time RT–PCR. This comparison revealed that the kinetics of RNA polymerase II recruitment to the egr-1 gene parallels the PMA-induced changes in the transcript (Figure 2A). The nearly identical changes in mRNA levels (Figure 2A, top panel, blue lines) and recruitment of RNA polymerase II (Figure 2, middle panel, blue lines) validate our ChIP protocol. Note reproducibility of the results of two independent ChIP is similar to that of two separate RT–PCR experiments. Comparison of the new (Figure 2A, middle panel, solid blue line) and the conventional (Figure 2A, dotted blue line) ChIP protocols done on the same extracts reveals similar PMA-induced kinetics of RNA polymerase II recruitment to the egr-1 locus. Both methods also revealed similar kinetics for K protein recruitment to this locus (Figure 2A, lower panel). Thus, results obtained with the new and the traditional methods were very similar.
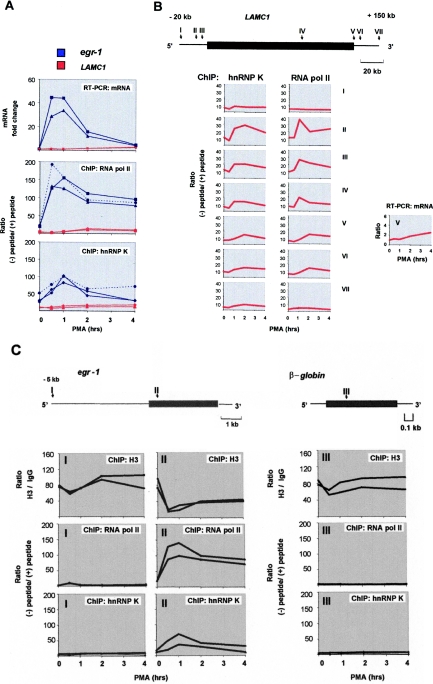
Verification of the new ChIP protocol. (A) Serum-deprived rat mesangial cells were treated with PMA (10−7 M) for indicated time points. Whole cell RNA was used in RT with random hexamer primers. Real-time PCR was carried out with primers to either egr-1 (exon 1) or LAMC1 (exon 28) genes. Results normalized to β-actin mRNA are shown as fold induction, mean ± SD, two experiments, PCR done in triplicates (top panel). Serum-deprived mesangial cells were treated with PMA as above (top panel). After cross-linking with formaldehyde, cells were lysed, pelleted and sonicated. Chromatin IPs were prepared with either RNA polymerase II (4 µg) (Middle panel) or K protein (10 µg) (Bottom panel) antibody with or without blocking peptides (4 µg). Equal amounts of chromatin fraction were used in the IPs. DNA purified with either the new (solid blue line) or the conventional (dotted blue line) ChIP protocols was used as a template in real-time PCR. The results are expressed as a ratio of the level of PCR products obtained without (−) and with (+) blocking peptide. The graphs show results from two independent IPs done with the new ChIP protocol and results of one representative experiment is shown for the traditional method. PCR was done in triplicates (middle and bottom panel). (B) New ChIP protocol was used to assesses PMA-induced kinetics of RNA polymerase II and K protein recruitment to the different regions (I–VII) along the LAMC1 gene in rat mesangial cells. The graph with results of RT–PCR analysis of mRNA is shown in the right panel (V). Results are are shown as mean values of two independent experiments. Diagram above the graphs represents LAMC1 transcribed (rectangle) and flanking regions (lines). The arrows point at the sites of the respective pair of primers (I and II are 20 and 5 kb 5′ to the start of transcription, respectively, III is the promoter region, IV is exon 2, V is exon 28 and VI and VII are 5 and 20 kb 3′ to the end of the last exon, exon 28). (C) Comparison of the density of RNA polymerase II, K protein and histone H3 in the 5′ flanking (I) and transcribed (II) regions of egr-1, and at the silent β-globin (III) locus. Equal aliquots of sheared chromatin were used in the new ChIP assay with either anti-H3 (4 µg), anti-RNA polymerase II or K antibodies. For H3 ChIP, purified rabbit IgG fraction (4 µg) was used as a mock IP control. Diagram above the graphs represents egr-1 and β-globin genes (rectangle) and flanking regions (lines). The arrows represent the sites of the respective pair of primers (I–III).
In comparison to the rapid and robust activation of the short egr-1 gene (3.8 kb), induction of the long laminin γ1 (LAMC1) (128 kb) gene is slow and of small magnitude (Figure 2A, upper panel, red lines); 2 to 3-fold induction for LAMC1 mRNA levels compared with 30–50-fold induction for egr-1 transcript. Results of ChIP analysis show that recruitment of RNA polymerase II and K protein to the last exon of LAMC1 (exon 28, primer V, diagram in Figure 2B) is much lower than to the egr-1 locus and parallels low levels of mRNA induction (Figure 2A, compare the red and blue lines).
We next examined the inducible recruitment of hnRNP K and RNA polymerase II to the long and weakly induced LAMC1 gene in greater details (Figure 2B). Results of the ChIP analysis revealed that there was PMA-inducible increase in hnRNP K and RNA polymerase II recruitment to the LAMC1 gene and 5′ and 3′ flanking regions. The highest levels were observed in the promoter region with the density not only decreasing along the transcribed region but also exhibiting different kinetics (Figure 2B). Likewise, in the case of the egr-1 locus (Figure 2A), at most of the examined LAMC1 sites the recruitment of K protein resembled but was not identical to that of RNA polymerase II (II–VI). At the intergenic sites 20 kb 5′ and 3′ from the gene there were low constitutive and PMA-inducible levels of K protein with little or no RNA polymerase II detected. The finding here that K protein binding differs from that of RNA polymerase II is consistent with the notion that hnRNP K is involved not only in transcription but also in chromatin remodeling (24). The higher density of hnRNP K at regions that encode laminin γ1 protein (IV and V) than at the intergenic sites (I and VII) indicates preferential K protein recruitment to domains that include LAMC1 open reading frame (ORF). The analysis of the LAMC1 gene illustrates that the new method has a sufficient signal to noise ratio to study genes that exhibit low levels of induction.
The ability to simultaneously monitor DNA-binding of several factors enhances chromatin studies. We used equal aliquots of chromatin from the same time course experiment to compare density of RNA polymerase II, histone H3 and K protein at 5′-flanking and transcribed region of egr-1 gene and the silenced β-globin locus (Figure 2C). These experiments revealed that at zero time point the histone H3 density was comparable at transcribed (Figure 2C, upper panels, II) and non-transcribed (I) regions of egr-1 and at the silent β-globin locus (III). In the transcribed region of egr-1 (II) there was large PMA-inducible loss of H3-DNA contact associated with RNA polymerase II recruitment to this site (Figure 2C, center top and middle panel). The kinetics of K protein recruitment (Figure 2C, bottom panel) is similar but not identical to that of RNA polymerase II. In the intergenic region 5 kb 5′ to the egr-1 gene and at the silent β-globin locus RNA polymerase II was not detected (Figure 2C, I and III) and the level of H3 decreased slightly or remained the same after PMA treatment (Figure 2C, top panels I and III). As in the case of the regions flanking the LAMC1 gene (Figure 2B, I and VII) there were low levels of K protein (Figure 2C, bottom panels I and III). Finding K protein in the intergenic regions is consistent with the previous suggestion that its binding is genome-wide (20). These data confirm previous observations of the inverse relationship between the density of RNA polymerase II and histones (30–36). These results also further validate the new ChIP method and show the ability to simultaneously monitor DNA-binding of several factors.
The presence of RNA polymerase II in the transcribed and promoter regions but not in the intergenic (Figure 2B and C) domains and the differential loss of H3-DNA contact (Figure 2C, top panels, I–III) confirms the specificity of the fast ChIP assay.
The high reproducibility of the results obtained with the new ChIP method is illustrated by close kinetics of changes in the density of RNA polymerase II, histone H3 and hnRNP K protein at egr-1 locus observed in two separate experiments (Figure 2C). Also, note that in these experiments (Figure 2C) the kinetics of RNA polymerase II and hnRNP K protein recruitment to the transcribed region of egr-1 is similar to the set of two other experiments shown in Figure 2A. These experiments illustrate that the new method yields reproducible results and allows probing of multiple factors in chromatin preps from one experiment.
We use one 15 cm (~107 cells) dish to prepare chromatin sufficient to IP with one antibody, with and without blocking peptide. DNA purified with this method from one chromatin IP is sufficient for 80–100 PCR.
Verification of the fast ChIP assay in yeast
S.cerevisiae has proven to be a valuable system to study chromatin processes (6) and many heterochromatin factors were initially characterized in yeast [reviewed in (37)]. Among these factors, Sir2p is a very conserved protein that binds to all major silenced domains in the yeast nucleus, including HML and HMR mating type loci (37). We estimated the density of Sir2 at HMR and at a control locus that does not bind this protein using the new and traditional ChIP methods. Equal amounts of extracts and antibodies were used in IPs. The results of real-time PCR analysis demonstrate that both methods reveal Sir2p recruitment to the silenced HMR region but not to the control locus (Figure 3). These results demonstrate that the fast ChIP method can be also used to study chromatin processes in yeast.
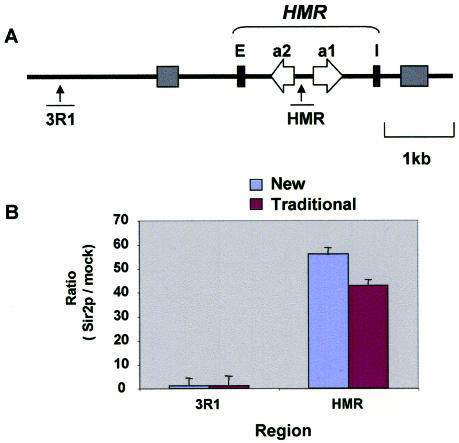
Verification of the new ChIP assay in yeast. The new and traditional ChIP methods were used to assess recruitment of the yeast Sir2p to HMR and an adjacent genomic locus. (A) Diagram of the yeast HMR locus (37). Primers for PCR analysis were designed to the indicated regions. (B) Results of ChIP analysis with either antibodies to Sir2p or no antibodies (mock IP). IPs and DNA purification using either the new (blue) or traditional (purple) methods were done in parallel with equal amounts of yeast chromatin. Purified DNA samples were analyzed as above with real-time PCR. Results are expressed as signal ratios of anti-Sir2p IP to mock IP. PCR were done in triplicates, data represented as mean ± SD.
In summary, we have developed a simple and efficient ChIP assay that is fast, and allows using of multiple antibodies in one experiment to simultaneously process many samples. The new ChIP assay will greatly facilitate experiments designed to study chromatin dynamics from yeast to mammals.
Acknowledgments
We thank Dr J. Rine for antibodies, and members of KB lab for valuable discussions of the method. This work was supported by NIH DK45978, GM45134 and Juvenile Diabetes Research Foundation (K.B.). Funding to pay the Open Access publication charges for this article was provided by NIH DK45978.
Conflict of interest statement. None declared.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gnj004
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/34/1/e2/7129312/gnj004.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Mechanisms of HIF1A-mediated immune evasion in gastric cancer and the impact on therapy resistance.
Cell Biol Toxicol, 40(1):87, 10 Oct 2024
Cited by: 0 articles | PMID: 39384651 | PMCID: PMC11464584
Deciphering the SOX4/MAPK1 regulatory axis: a phosphoproteomic insight into IQGAP1 phosphorylation and pancreatic Cancer progression.
J Transl Med, 22(1):602, 28 Jun 2024
Cited by: 0 articles | PMID: 38943117
Meta-analysis and transcriptomic analysis reveal that NKRF and ZBTB17 regulate the NF-κB signaling pathway, contributing to the shared molecular mechanisms of Alzheimer's disease and atherosclerosis.
CNS Neurosci Ther, 30(5):e14683, 01 May 2024
Cited by: 1 article | PMID: 38738952 | PMCID: PMC11090078
Implications of IFNγ SNP rs2069705 in primary Sjögren's syndrome: transcriptional activation and B cell infiltration.
Am J Physiol Cell Physiol, 326(5):C1494-C1504, 26 Feb 2024
Cited by: 2 articles | PMID: 38406824 | PMCID: PMC11371360
Amplification of poly(I:C)-induced interleukin-6 production in human bronchial epithelial cells by priming with interferon-γ.
Sci Rep, 13(1):21067, 29 Nov 2023
Cited by: 1 article | PMID: 38030681 | PMCID: PMC10687102
Go to all (150) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The fast chromatin immunoprecipitation method.
Methods Mol Biol, 567:45-57, 01 Jan 2009
Cited by: 18 articles | PMID: 19588084
Review
Protocol for the fast chromatin immunoprecipitation (ChIP) method.
Nat Protoc, 1(1):179-185, 01 Jan 2006
Cited by: 546 articles | PMID: 17406230
Microplate-based chromatin immunoprecipitation method, Matrix ChIP: a platform to study signaling of complex genomic events.
Nucleic Acids Res, 36(3):e17, 18 Jan 2008
Cited by: 57 articles | PMID: 18203739 | PMCID: PMC2241906
MicroChIP: chromatin immunoprecipitation for small cell numbers.
Methods Mol Biol, 567:59-74, 01 Jan 2009
Cited by: 18 articles | PMID: 19588085
Review
Funding
Funders who supported this work.
NIDDK NIH HHS (3)
Grant ID: R37 DK045978
Grant ID: DK45978
Grant ID: R01 DK045978
NIGMS NIH HHS (2)
Grant ID: R01 GM045134
Grant ID: GM45134





