Abstract
Free full text

APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells
Abstract
While the ability of APOBEC3G to reduce the replication of a range of exogenous retroviruses is now well established, recent evidence has suggested that APOBEC3G can also inhibit the replication of endogenous retrotransposons that bear long terminal repeats. Here, we extend this earlier work by showing that two other members of the human APOBEC3 protein family, APOBEC3B and APOBEC3A, can reduce retrotransposition by the intracisternal A-particle (IAP) retrotransposon in human cells by 20-fold to up to 100-fold, respectively. This compares to an ~4-fold inhibition in IAP retrotransposition induced by APOBEC3G. While both APOBEC3G and APOBEC3B specifically interact with the IAP Gag protein in co-expressing cells, and induce extensive editing of IAP reverse transcripts, APOBEC3A fails to package detectably into IAP virus-like particles and does not edit IAP reverse transcripts. These data, which identify human APOBEC3A as a highly potent inhibitor of LTR-retrotransposon function, are the first to ascribe a biological activity to APOBEC3A. Moreover, these results argue that APOBEC3A inhibits IAP retrotransposition via a novel mechanism that is distinct from, and in this case more effective than, the DNA editing mechanism characteristic of APOBEC3G and APOBEC3B.
INTRODUCTION
The ability of members of the APOBEC3 protein family to confer intrinsic immunity to retroviral infection was first recognized in the case of APOBEC3G (hA3G), which can block the replication of human immunodeficiency virus type 1 (HIV-1) mutants lacking a functional copy of the vif gene (ΔVif) (1). Subsequently, APOBEC3F (hA3F) was found to also inhibit the replication of ΔVif, but not wild-type, HIV-1 variants (2–4). In contrast, APOBEC3B (hA3B) appears able to inhibit the replication of both wild-type and ΔVif HIV-1 (5,6). Two other human APOBEC3 proteins, APOBEC3C (hA3C) and APOBEC3A (hA3A), at most weakly inhibit either wild-type or ΔVif HIV-1 (2,5,7,8), although hA3C has been reported to block the replication of ΔVif, but not wild-type, forms of simian immunodeficiency virus (SIV) (9).
Inhibitory APOBEC3 proteins specifically interact with the nucleocapsid domain of the HIV-1 Gag protein and are packaged into progeny virions (10–16). During the subsequent infection of new target cells, the APOBEC3 proteins can interfere with reverse transcription by inducing extensive dC to dU editing of nascent proviral minus strands (15–19). This can induce degradation of the proviral intermediate, perhaps due to abortive efforts at DNA repair by cellular proteins, or can lead to fatal mutagenesis. While editing of the retroviral provirus is therefore a general characteristic of inhibitory APOBEC3 proteins, it has recently been reported that some hA3G mutants that are unable to edit can still inhibit HIV-1ΔVif replication (20).
While APOBEC3 proteins were first identified as inhibitors of HIV-1ΔVif replication, several human APOBEC3 proteins can also inhibit other retroviruses, including not only SIV (15,21) but also murine leukemia virus (MLV) (16,19) and primate foamy virus (PFV) (22,23). Moreover, APOBEC3 proteins have also been shown to act as inhibitors of LTR-retrotransposon function. Thus, hA3G, hA3F and hA3C can inhibit Ty1 retrotransposition in yeast cells by up to 100-fold, and this inhibition correlates with C to T hypermutation of the Ty1 genome (24,25). In the case of the murine LTR-retrotransposons intracisternal A-particle (IAP) and MusD, hA3G was found to reduce retrotransposition in HeLa cells by ~4-fold in the case of IAP and ~10-fold in the case of MusD (26). In both cases, this inhibition again correlated with C to T hypermutation.
In this manuscript, we have confirmed the ability of hA3G to modestly inhibit IAP retrotransposition and demonstrate that hA3B and hA3A are much more potent inhibitors, reducing productive IAP retrotransposition by up to 100-fold. Both hA3G and hA3B induce C to T hypermutation of the IAP genome and specifically interact with IAP Gag. In contrast, hA3A failed to detectably interact with IAP Gag in vivo and did not induce IAP hypermutation. These data are the first to ascribe a biological activity to hA3A and suggest that hA3A may have evolved a novel mechanism to block retrotransposon function.
MATERIALS AND METHODS
Molecular clones
We have previously described expression plasmids, based on pcDNA3, that express C-terminally HA-tagged versions of hA3A, hA3B, hA3C, hA3F, hA3G and β-arrestin (2,6,21). Because pcDNA3 contains the neo gene, the APOBEC3 and β-arrestin open reading frames were excised using HindIII and XhoI and cloned into the pK expression plasmid (27). The IAP retrotransposition indicator plasmid pDJ33/440N1neoTNF has been previously described (28), as has the pHIV-Luc-ΔVif HIV-1 proviral indicator plasmid (2,21).
Retrotransposition and toxicity assays
HeLa cells (3 × 105) were co-transfected with 2 µg of an APOBEC3 expression plasmid, or the parental pK vector as a control, together with 2 µg of pDJ33/440N1neoTNF, pcDNA3 or a derivative of pDJ333/440N1neoTNF lacking a functional IAP pol gene (28). Forty-eight hours post-transfection, the transfected cells were harvested and 106 plated onto 10 cm dishes. A further 24 h later, the cells were subjected to selection using G418 (700 µg/ml) and maintained in G418 for a further 14 days. At this point, the cultures were either fixed and stained with crystal violet, and G418 resistant colonies counted, or the cells were used for DNA isolation using a DNAeasy kit (Qiagen). The entire IAP Gag open reading frame was then amplified by PCR and cloned into pcDNA3 prior to DNA sequencing.
Inhibition of HIV-1ΔVif infectivity by different APOBEC3 proteins was assayed as previously described by co-transfection into 293T cells (2,6,21). Progeny virions were harvested 48 h after co-transfection and used to infect CD4+ CCR5+ cells. Virus encoded firefly luciferase activity was then quantitated 24 h post-infection.
Sucrose gradient centrifugation and protein interaction assays
293T cells (2 × 106) were co-transfected with 20 µg of pDJ33/440N1neoTNF, or a control plasmid, together with 500 ng of pK, pK/hA3G or pK/hA3A. At 48 h post-transfection, cells were harvested into 1 ml of lysis buffer [50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.5% NP-40 and protease inhibitors (Roche)] and clarified by brief centrifugation. Lysates obtained from three plates were then pooled and layered onto a 9 ml 10–80% sucrose gradient. The gradients were subjected to centrifugation at 40000 r.p.m. for 2 h in an SW41 rotor. One ml fractions were collected and analyzed by western blot using either a 1:5000 dilution of a mouse anti-HA monoclonal (Covance) or a 1:2000 dilution of a rabbit anti-IAP Gag polyclonal antiserum (29). Blots were then treated with horseradish peroxidase-conjugated secondary antibodies (Amersham) and reactive proteins detected using chemiluminescence (Lumi-light, Roche). In parallel, cleared cell lysates obtained from transfected 293T cells were also used for analysis of in vivo protein binding by co-immunoprecipitation. ‘Input’ samples were derived directly from the cell lysates while ‘bound’ samples were obtained by incubating lysates in the presence of the rabbit polyclonal anti-IAP Gag antiserum and protein A Agarose (Invitrogen) for 2 h at 4°C. Western blots were performed as described above.
The ~73 kDa IAP Gag protein was expressed using an in vitro transcription/translation system and full-length protein isolated by elution after gel electrophoresis (29). The HA-tagged hA3A, hA3B and hA3G proteins were expressed by transfection of the relevant expression plasmids into 293T cells. At 48 h post-transfection, the cells were lysed in lysis buffer, clarified and recombinant IAP Gag protein (2 µg total) added. The lysate was then incubated at 4°C for 4 h, at which point the rabbit polyclonal anti-IAP Gag antiserum and protein A Agarose were added. After a further 30 min incubation at 4°C, the bound complexes were collected by centrifugation, washed and analyzed by western blot.
RESULTS AND DISCUSSION
To examine the ability of human APOBEC3 proteins to inhibit retrotransposition of IAP in human cells, we co-transfected HeLa cells with expression plasmids encoding various APOBEC3 proteins and the previously described IAP retrotransposition indicator construct pDJ33/440N1neoTNF. This construct contains a full-length IAP genome with the neo gene inserted in the antisense orientation (28). The neo gene is expressed under the control of an SV40 promoter and is disrupted by an intron in the sense orientation. Therefore, neo expression requires transcription and splicing of the IAP transcript, followed by reverse transcription and retrotransposition into the host cell genome.
As summarized in Figure 1, and demonstrated in Figure 2A, hA3G expression indeed reduced IAP retrotransposition by ~4-fold, as previously reported by Esnault et al. (26). Similarly, both hA3C and hA3F reduced IAP retrotransposition by 2- to 3-fold. In contrast, hA3B reduced IAP retrotransposition by ~20-fold while hA3A reduced IAP retrotransposition by 50- to 100-fold (Figures 1 and and2A).2A). These differences were not due to major discrepancies in the level of expression of each APOBEC3 protein in transfected cells (Figure 1). Moreover, this inhibition did not result from non-specific toxicity, as none of the APOBEC3 proteins had any effect on the number of G418 resistant colonies obtained upon simple co-transfection of HeLa cells with a neo expression plasmid (Figure 2B).
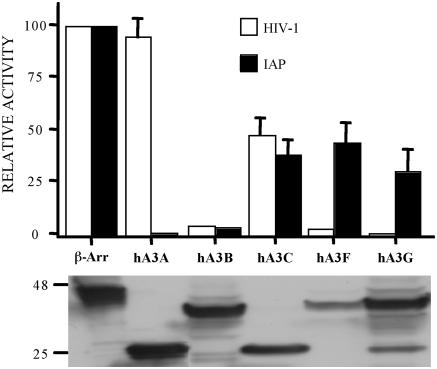
Effect of human APOBEC3 proteins on IAP retrotransposition and HIV-1 infectivity. Upper panel: IAP retrotransposition was quantified by co-transfection of HeLa cells with the IAP retrotransposition indicator construct pDJ33/440N1neoTNF and the indicated APOBEC3 expression plasmids. After selection in G418, resistant colonies were counted. The effect of the same APOBEC3 proteins on HIV-1ΔVif infectivity was determined by co-transfection into 293T cells together with the pHIV-Luc-ΔVif indicator construct. Released HIV-1 virions were collected at 48 h and used to infect CD4+, CCR5+ cells. Induced, virus encoded luciferase activity was quantified 24 h later. In both cases, data are presented as a percentage of the activity seen in cultures co-transfected with a plasmid expressing the irrelevant β-arrestin gene. Average of three independent experiments with standard deviation indicated. Lower panel: western analysis of APOBEC3 protein expression in transfected 293T cells using an anti-HA tag specific mouse monoclonal. hA3A and hA3C are predicted to be ~22 kDa in size while hA3B, hA3F and hA3G are ~40 kDa.
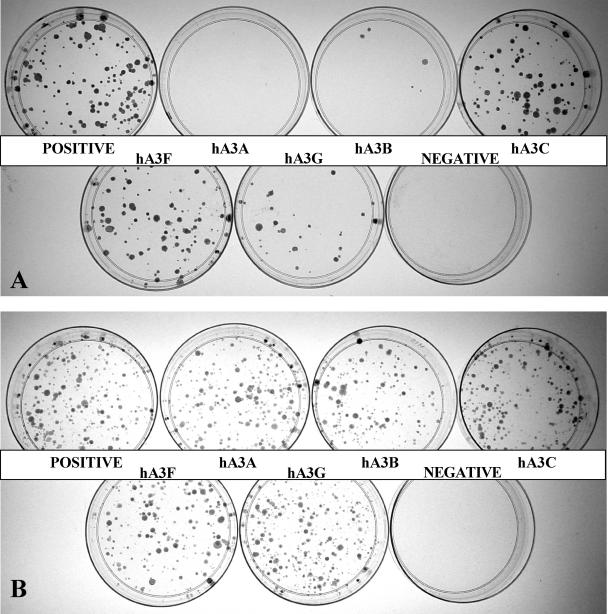
Effect of APOBEC3 proteins on IAP retrotransposition and cell viability. (A) HeLa cells were transfected with the IAP retrotransposition indicator plasmid pDJ33/440N1neoTNF, together with an APOBEC3 expression plasmid or a control plasmid expressing β-arrestin. A derivative of pDJ33/440N1neoTNF lacking a functional pol gene served as the negative control. Cells were subjected to selection in G418 and resistant colonies counted 17 days after transfection. This experiment is representative of the data compiled in Figure 1. (B) Same as (A) except that HeLa cells were co-transfected with pcDNA3, which encodes a neo cDNA, instead of with pDJ33/440N1neoTNF. The negative in this case is a mock transfection.
It is interesting to compare the effect of human APOBEC3 proteins on IAP retrotransposition with their effect on HIV-1 infectivity. As shown in Figure 1, and previously reported (1–6,21), hA3G, hA3F and hA3B can potently inhibit the infectivity of HIV-1ΔVif virions produced in their presence. In contrast, hA3C has at most a modest inhibitory effect on HIV-1ΔVif infectivity while hA3A has no detectable phenotype. These data demonstrate that there is no correlation between the effect of human APOBEC3 proteins on IAP retrotransposition and on HIV-1ΔVif infectivity and again argue against any non-specific toxicity induced by hA3A.
We next used PCR to clone IAP genomic sequences from the G418 resistant colonies that arose in the HeLa cultures co-transfected with the IAP indicator plasmid and expression vectors encoding hA3A, hA3B, hA3G or β-arrestin, which was used as a control (Figure 2A). No C to T mutations were noted upon sequencing 4320 bp of the IAP genome sequences derived from control cultures (data not shown). In contrast, we detected 79 C to T mutations after sequencing 4550 bp of IAP sequence obtained from the hA3G co-expressing cultures (Table 1) and 186 C to T changes after sequencing 5800 bp of IAP sequence obtained from the hA3B co-expressing cultures (Table 1). The ~2-fold higher level of C to T mutations detected in the hA3B expressing cultures, when compared with the hA3G expressing cultures, may correlate with the ~5-fold more severe inhibition of IAP retrotransposition induced by hA3B when compared with hA3G (Figures 1 and and2A2A).
Table 1
Relative levels of C to T editing induced in retrotransposed IAP genome sequences by co-expressed APOBEC3 proteins
hA3A (22 200) 200) | A | T | C | G | hA3B (5800) | A | T | C | G | hA3G (4550) | A | T | C | G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | – | 1 | 0 | 0 | A | – | 0 | 0 | 2 | A | – | 0 | 0 | 0 |
| T | 0 | – | 4 | 0 | T | 0 | – | 2 | 0 | T | 0 | – | 0 | 0 |
| C | 0 | 3 | – | 0 | C | 0 | 186 | – | 0 | C | 1 | 79 | – | 0 |
| G | 0 | 0 | 0 | – | G | 1 | 0 | 0 | – | G | 0 | 0 | 0 | – |
G418 resistant colonies obtained upon co-transfection of HeLa cells with the pDJ33/440N1neoTNF IAP indicator construct, and plasmids expressing hA3A, hA3B or hA3G, were harvested and used to prepare DNA. IAP genomic sequences were then PCR amplified, cloned and sequenced. The total number of bases sequenced is shown in parentheses, with mutations indicated in the grid. Predicted bases are shown on the left and the observed bases across the top.
Analysis of the sequence context of these C to T mutations showed that 73 (92%) of the 79 mutations induced by hA3G were in the sequence context CC* (where the asterisk indicates the edited residue) while the other 6 (8%) were in the sequence context TC*. In the case of hA3B, 166 (89%) of the 186 C to T mutations were in the sequence context TC*, 16 (9%) in the sequence context CC* and 2 each (1%) in the context AC* or GC*. This sequence preference for editing of IAP reverse transcripts closely mirrors the hA3G and hA3B sequence preference previously observed using HIV-1 (2,3,5,6,17,19).
Surprisingly, sequencing of 22200 bp of IAP sequence derived from G418 resistant colonies obtained from the cultures co-transfected with the IAP retrotransposition indicator construct and the hA3A expression plasmid revealed only background levels of mutagenesis, i.e. we did not observe significant levels of C to T editing (Table 1). This result therefore strongly suggests that the potent inhibition of IAP retrotransposition induced by hA3A (Figures 1 and and2A)2A) is not due to editing of nascent IAP reverse transcripts.
The APOBEC3 protein family, as well as its more distant relatives APOBEC1, APOBEC2 and AID, contain either one or two copies of a cytidine deaminase active site, with the consensus sequence His-X-Glu-X23-28-Pro-Cys-X2-4-Cys, that also functions as a zinc coordinating region (30). Mutational disruption of either of the two active sites in hA3G can inhibit the ability of hA3G to block HIV-1 replication (16). Moreover, we have previously shown that mutation of active site residues 95-SPC-97 or 286-SPC-288 of hA3G to alanine disrupts the ability of hA3G to inhibit retrotransposition by Ty1 in yeast cells (24). We therefore introduced this same inactivating mutant into the same three active site residues in hA3A (99-SPC-101 to AAA) and asked whether this would affect the ability of hA3A to inhibit IAP retrotransposition. As shown in Figure 3, the hA3A (SPC-AAA) mutant was expressed at a level that was comparable with wild-type hA3A in transfected cells and, more importantly, fully retained its ability to block IAP retrotransposition. These data are therefore fully consistent with the hypothesis that disruption of IAP retrotransposition by hA3A occurs independently of editing.
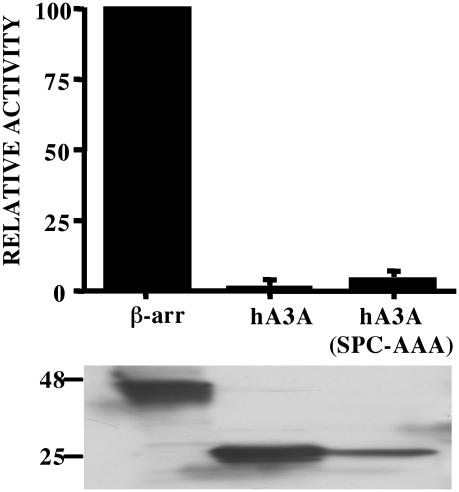
An hA3A mutant bearing a defective cytidine deaminase active site remains able to inhibit IAP retrotransposition. These data were derived and are presented as described in Figure 1 and again used a primary antibody specific for the HA epitope tag, present on both the β-arrestin and hA3A proteins, in the lower panel. The hA3A (SPC-AAA) mutant bears alanine residues in place of the active site residues 99-SPC-101.
Inhibition of retrovirus replication, and of Ty1 retrotransposition, by specific APOBEC3 proteins is mediated by their selective incorporation into retroviral or retrotransposon particles due to a specific interaction with the Gag protein (6,10–14,22,24,31). We therefore used co-immunoprecipitation to ask whether hA3A, hA3B and hA3G also specifically interact with the IAP Gag protein in vivo. In this experiment, 293T cells were co-transfected with an expression vector expressing an HA-tagged APOBEC3 or control protein (β-arrestin), together with an IAP Gag expression plasmid (28). Proteins bound to IAP Gag in vivo were collected by immunoprecipitation at 48 h after transfection using an IAP Gag-specific antiserum (29). As shown in Figure 4A, we observed a readily detectable interaction between either hA3B or hA3G and IAP Gag. In contrast, we did not observe co-immunoprecipitation of IAP Gag with hA3A or β-arrestin, a human cytoplasmic protein used as a negative control, in several independent experiments.
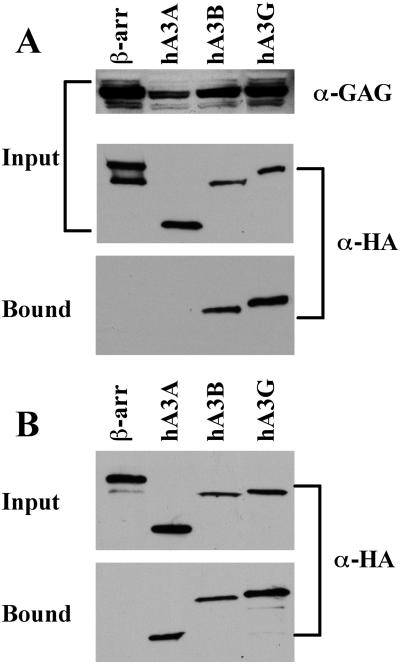
Binding of IAP Gag by APOBEC3 proteins in vivo and in vitro. (A) 293T cells were co-transfected with the IAP Gag expression plasmid pDJ33/440N1neoTNF and vectors expressing HA-tagged hA3A, hA3B, hA3G or β-arrestin, the latter as a negative control. After 48 h, the cells were lysed and an aliquot retained to analyze input protein levels. The remaining lysate was incubated with a rabbit anti-IAP Gag antiserum and protein A agarose. Bound, as well as input, proteins were separated by gel electrophoresis and visualized by western blot using a mouse monoclonal specific for the HA tag present on the APOBEC3 and β-arrestin proteins, or using the anti-IAP Gag antiserum. (B) 293T cells were transfected with plasmids expressing HA-tagged hA3A, hA3B, hA3G or β-arrestin and lysates prepared 2 days later. After incubation in the presence of purified full-length recombinant IAP Gag, Gag protein complexes were collected using a rabbit polyclonal anti-Gag antiserum and protein A agarose. Bound and input proteins were then visualized by western blot using an anti-HA monoclonal. For both panels, input lanes contain 2% of the starting material while bound lanes contain 25% of the immunoprecipitated protein fraction.
While the experiment shown in Figure 4A looked at the ability of hA3A, hA3B and hA3G to interact with the IAP Gag protein in vivo, we also wished to test whether these proteins could interact in vitro. For this purpose, we prepared full-length, recombinant IAP Gag (29) and then added this purified protein to cell lysates prepared from 293T cells transfected with plasmids expressing hA3A, hA3B, hA3G or β-arrestin. Protein complexes containing IAP Gag were then collected and analyzed by western blot. As shown in Figure 4B, we again observed a readily detectable interaction between hA3B or hA3G and IAP Gag. Remarkably, however, we also consistently observed specific binding of hA3A, but not of the β-arrestin control protein, by IAP Gag in this in vitro assay. No binding to the protein A Sepharose matrix was observed if the recombinant IAP Gag protein was omitted from the reaction (data not shown). Therefore, it appears that hA3A, hA3B and hA3G can all interact with IAP Gag in vitro (Figure 4B) but that hA3A differs from hA3B and hA3G in being unable to form a stable complex with IAP Gag in vivo (Figure 4A).
In yeast cells expressing hA3G, the hA3G protein migrates at its predicted, <100 kDa size on sucrose gradients in the absence of Ty1 Gag but moves to high molecular mass (HMM) fractions when Ty1 Gag is co-expressed (24). Presumably, this reflects incorporation of hA3G into Ty1 retrovirus-like particles. We therefore asked whether the migration of hA3A or hA3G on a similar sucrose gradient would be affected by co-expression of IAP Gag, and vice versa. As shown in Figure 5, all of the IAP Gag protein migrated to near the bottom of the sucrose gradient, consistent with the efficient formation of IAP retrovirus-like particles (29), and this migration was unaffected by hA3G or hA3A co-expression. Analysis of the hA3A protein showed that hA3A resided almost exclusively near the top of the sucrose gradient, well away from the HMM fractions containing the IAP Gag protein, regardless of whether IAP Gag was co-expressed or not (Figure 5). This result is consistent with our inability to detect an in vivo IAP Gag:hA3A interaction by co-immunoprecipitation (Figure 4A).

Sucrose gradient fractionation of IAP Gag and APOBEC3 proteins. 293T cells were transfected with pDJ33/440N1neoTNF, which expresses IAP Gag, with vectors expressing hA3G or hA3A, or with appropriate control plasmids. After 48 h, the cells were lysed and the lysates fractionated across 10–80% sucrose gradients. Fractions were then collected, with the top of the gradient at left, and subjected to western analysis using anti-IAP Gag and anti-HA tag antibodies, as indicated. M, mock transfected cell lysate; I, input cell lysate (0.5% of total).
In contrast to hA3A, hA3G was found to migrate into the HMM segment of the sucrose gradient and was, in fact, found in almost exactly the same fractions as the IAP Gag protein (Figure 5). Remarkably, the mobility of hA3G on this sucrose gradient was not affected by IAP Gag co-expression, i.e. it migrated at an HMM regardless of whether IAP Gag was co-expressed (Figure 5).
It is interesting to contrast this result, obtained in 293T cells, with the situation in yeast, where hA3G migrated with an HMM in the presence of Ty1 Gag but at a low molecular mass in its absence (24). These yeast data argue that hA3G, which is clearly active in yeast cells (24,25), is only recruited into an HMM complex, at least in yeast cells, when a target Gag protein is co-expressed. Yet, in transfected 293T cells, hA3G is in an HMM complex even in the absence of IAP Gag co-expression (Figure 5). This result confirms the recent observation by Chiu et al. (32) that hA3G is found in a large, >700 kDa protein complex in actively growing human cell lines, although hA3G did migrate closer to its predicted molecular mass in resting primary human T-cells and monocytes. Chiu et al. (32) proposed, based on these data, that hA3G was incorporated into an HMM complex in dividing cells that perhaps functions to block inappropriate editing of cellular DNA by hA3G during host cell DNA synthesis.
Based on the data presented in Figure 5, and previously obtained in yeast cells (24), we propose that the HMM complex containing hA3G actually results from incorporation of hA3G into retrovirus-like particles derived from endogenous retroviruses and retrotransposons. Human cells contain ~230 000 endogenous retroviruses (26) and many of these are transcriptionally active, especially in dividing cells (33–37). We know that hA3G is able to interact with a wide range of diverse retroviral and retrotransposon Gag proteins, including HIV-1, MLV, PFV, Ty1 and IAP Gag (10–14,22,24,31) (Figure 2); so it seems likely that hA3G would also interact with many endogenous retroviral Gag proteins. Of note, Chiu et al. (32) showed that the HMM complex containing hA3G is disrupted by RNase treatment. This is striking, as RNase also disrupts the ability of hA3G to interact with HIV-1 Gag, MLV Gag and Ty1 Gag (10,12,14,24,31,38). We therefore hypothesize that the HMM complex formed by hA3G in actively replicating human cells results from the formation of a heterogeneous mixture of endogenous retrovirus Gag:hA3G complexes that, in cells co-expressing hA3G and IAP Gag, also include IAP Gag:hA3G complexes (Figure 3).
000 endogenous retroviruses (26) and many of these are transcriptionally active, especially in dividing cells (33–37). We know that hA3G is able to interact with a wide range of diverse retroviral and retrotransposon Gag proteins, including HIV-1, MLV, PFV, Ty1 and IAP Gag (10–14,22,24,31) (Figure 2); so it seems likely that hA3G would also interact with many endogenous retroviral Gag proteins. Of note, Chiu et al. (32) showed that the HMM complex containing hA3G is disrupted by RNase treatment. This is striking, as RNase also disrupts the ability of hA3G to interact with HIV-1 Gag, MLV Gag and Ty1 Gag (10,12,14,24,31,38). We therefore hypothesize that the HMM complex formed by hA3G in actively replicating human cells results from the formation of a heterogeneous mixture of endogenous retrovirus Gag:hA3G complexes that, in cells co-expressing hA3G and IAP Gag, also include IAP Gag:hA3G complexes (Figure 3).
The most surprising result reported in this manuscript is that hA3A is an extremely potent inhibitor of IAP retrotransposition (Figures 1 and and2A).2A). Remarkably, hA3A has not been previously observed to significantly inhibit the infectivity of any retrovirus tested (2,5,7,22,31); so this result represents the first report of a biological activity for hA3A. While hA3A is actually a more potent inhibitor of IAP retrotransposition than either hA3B or hA3G, hA3A differs from both these proteins in that it neither induced C to T hypermutation of the IAP genome (Table 1) nor detectably interacted with Gag in co-expressing cells (Figures 4A and and5).5). We therefore do not understand the molecular basis for this inhibition, although we do know that hA3A is not a non-specific inhibitor of HeLa transformation to G418 resistance (Figure 2B) and does not significantly affect IAP Gag expression in co-transfected cells (Figure 5). Of note, a previously described (24) inactivating mutation of the cytidine deaminase active sites in hA3G did not block the ability of hA3A to inhibit IAP retrotransposition when introduced into hA3A (99-SPC-101→AAA) (Figure 3), a result which is fully consistent with the lack of any detectable editing of the IAP genome by wild-type hA3A.
While hA3A failed to detectably interact with IAP Gag in vivo (Figures 4A and and5),5), we did observe a specific interaction between hA3A and recombinant IAP Gag protein in vitro (Figure 4B). This result raises the intriguing possibility that hA3A may transiently interact with IAP retrovirus-like particles either during their assembly or during the disassembly that is predicted to occur prior to the chromosomal integration of full-length IAP reverse transcripts. Unfortunately, we have not been able to quantify the level of IAP reverse transcripts in transfected cells in the absence of selection (data not shown) so we do not know whether hA3A acts before or after IAP reverse transcription. In any event, we can only hypothesize that hA3A utilizes a novel mechanism for inhibition of IAP retrotransposition that does not involve DNA editing. In this context, we note that Newman et al. (20) have proposed that inhibition of HIV-1 replication by hA3G can still be observed with certain hA3G mutants that lack a functional cytidine deaminase active site. Moreover, inhibition of Hepatitis B virus replication by hA3G may occur in the absence of significant editing (39,40), although this issue remains controversial (41). Whether this currently undefined second inhibitory mechanism is also used by hA3A to inhibit IAP retrotransposition is not currently known.
Acknowledgments
The authors thank Thierry Heidmann for reagents used in this research. This research was funded by NIH grants AI065301 and AI057099 to B.R.C. Funding to pay the Open Access publication charges for this article was provided by NIH grant AI65301.
Conflict of interest statement. None declared.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkj416
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/34/1/89/7128382/gkj416.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Revealing the hidden RBP-RNA interactions with RNA modification enzyme-based strategies.
Wiley Interdiscip Rev RNA, 15(3):e1863, 01 May 2024
Cited by: 1 article | PMID: 39392204 | PMCID: PMC11469752
Review Free full text in Europe PMC
Distinguishing preferences of human APOBEC3A and APOBEC3B for cytosines in hairpin loops, and reflection of these preferences in APOBEC-signature cancer genome mutations.
Nat Commun, 15(1):2369, 18 Mar 2024
Cited by: 4 articles | PMID: 38499553 | PMCID: PMC10948833
Large overlap in neutrophil transcriptome between lupus and COVID-19 with limited lupus-specific gene expression.
Lupus Sci Med, 11(1):e001059, 31 Jan 2024
Cited by: 1 article | PMID: 38302132 | PMCID: PMC10831459
Evolutionary potential of the monkeypox genome arising from interactions with human APOBEC3 enzymes.
Virus Evol, 9(2):vead047, 02 Aug 2023
Cited by: 2 articles | PMID: 37577211 | PMCID: PMC10422979
Unraveling the Enzyme-Substrate Properties for APOBEC3A-Mediated RNA Editing.
J Mol Biol, 435(17):168198, 11 Jul 2023
Cited by: 3 articles | PMID: 37442413 | PMCID: PMC10528890
Go to all (194) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
APOBEC3 proteins inhibit LINE-1 retrotransposition in the absence of ORF1p binding.
Ann N Y Acad Sci, 1178:268-275, 01 Oct 2009
Cited by: 25 articles | PMID: 19845642 | PMCID: PMC4475406
APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses.
Nature, 433(7024):430-433, 01 Jan 2005
Cited by: 242 articles | PMID: 15674295
APOBEC3A and APOBEC3B Activities Render Cancer Cells Susceptible to ATR Inhibition.
Cancer Res, 77(17):4567-4578, 11 Jul 2017
Cited by: 73 articles | PMID: 28698210 | PMCID: PMC5609510
Roles of APOBEC3A and APOBEC3B in Human Papillomavirus Infection and Disease Progression.
Viruses, 9(8):E233, 21 Aug 2017
Cited by: 61 articles | PMID: 28825669 | PMCID: PMC5580490
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (5)
Grant ID: AI65301
Grant ID: AI057099
Grant ID: R01 AI057099
Grant ID: AI065301
Grant ID: R01 AI065301





