Abstract
Free full text

Recruitment of the NCoA/SRC-1/p160 Family of Transcriptional Coactivators by the Aryl Hydrocarbon Receptor/Aryl Hydrocarbon Receptor Nuclear Translocator Complex
Abstract
The aryl hydrocarbon receptor complex heterodimeric transcription factor, comprising the basic helix-loop-helix-Per-ARNT-Sim (bHLH-PAS) domain aryl hydrocarbon receptor (AHR) and aryl hydrocarbon receptor nuclear translocator (ARNT) proteins, mediates the toxic effects of TCDD (2,3,7,8 tetrachlorodibenzo-p-dioxin). The molecular events underlying TCDD-inducible gene activation, beyond the activation of the AHRC, are poorly understood. The SRC-1/NCoA-1, NCoA-2/GRIP-1/TIF-2, and p/CIP/AIB/ACTR proteins have been shown to act as mediators of transcriptional activation. In this report, we demonstrate that SRC-1, NCoA-2, and p/CIP are capable of independently enhancing TCDD-dependent induction of a luciferase reporter gene by the AHR/ARNT dimer. Furthermore, injection of anti-SRC-1 or anti-p/CIP immunoglobulin G into mammalian cells abolishes the transcriptional activity of a TCDD-dependent reporter gene. We demonstrate by coimmunoprecipitation and by a reporter gene assay that SRC-1 and NCoA-2 but not p/CIP are capable of interacting with ARNT in vivo after transient transfection into mammalian cells, while AHR is capable of interacting with all three coactivators. We confirm the interactions of ARNT and AHR with SRC-1 with immunocytochemical techniques. Furthermore, SRC-1, NCoA-2, and p/CIP all associate with the CYP1A1 enhancer region in a TCDD-dependent fashion, as demonstrated by chromatin immunoprecipitation assays. We demonstrate by yeast two-hybrid, glutathione S-transferase pulldown, and mammalian reporter gene assays that ARNT requires its helix 2 domain but not its transactivation domain to interact with SRC-1. This indicates a novel mechanism of action for SRC-1. SRC-1 does not require its bHLH-PAS domain to interact with ARNT or AHR, but utilizes distinct domains proximal to its p300/CBP interaction domain. Taken together, these data support a role for the SRC family of transcriptional coactivators in TCDD-dependent gene regulation.
Most if not all of the toxic effects of the polycyclic aromatic hydrocarbon and halogenated aromatic hydrocarbon groups of chemical carcinogens, including 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD), are mediated through their activation of the heterodimeric transcription factor, consisting of the ligand-binding subunit aryl hydrocarbon receptor (AHR) and AHR nuclear translocator (ARNT) proteins (21). ARNT acts as a common dimerization partner for many of the basic helix-loop-helix (bHLH)-Per-ARNT-Sim (PAS) domain family of transcription factors (for a review, see reference 18), being capable of forming dimers with Sim1, Sim2 (15, 43), Hif-1α (53), MOP2 (also called HLF and Hif-2α) (16, 22), and Hif-3α (19). These heterodimeric complexes regulate an organism's adaptive responses to a variety of environmental and developmental signals, including environmental toxicants and oxygen tension. However, the molecular mechanisms underlying the ability of these proteins, in particular AHR and ARNT, to activate gene transcription are not well understood.
The ARNT-AHR complex (AHRC) activates the transcription of several genes upon AHR's association with TCDD, including those encoding several xenobiotic compound-metabolizing enzymes (21). Unliganded AHR exists in the cytoplasm as part of a multimeric complex containing two molecules of heat shock protein 90 (5, 20), the cochaperone p23 (27), and a 40-kDa protein termed hepatitis B virus X-associated protein 2 (XAP2) (7, 9, 36). Upon ligand binding, AHR translocates to the nucleus, where it associates with ARNT to form a functional transcription factor complex, the AHRC (42). This activated complex is capable of binding DNA at conserved motifs termed xenobiotic compound-responsive elements, thereby enhancing the transcription of specific target genes.
The p300 protein and its homologue, the CREB-binding protein (CBP), as well as the estrogen receptor-associated protein 140 (ERAP140) have been implicated in AHRC transcriptional activation via a direct interaction with ARNT (28, 40). These proteins were among the first described to act as transcriptional adaptors or coactivators (10, 13). Furthermore, p300 and CBP have been shown to support this role for a widely diverse group of transcription factors, including multiple nuclear/steroid hormone receptors, CREB, p53, NF-κB, Ets, and Hif-1α (3, 10, 25, 26, 35, 48, 55).
The nuclear receptor coactivator/steroid receptor coactivator (NCoA/SRC/p160) family of proteins represent a distinct class of coactivator proteins. The cloning of SRC-1/NCoA-1 (26, 40, 51), NCoA-2/GRIP-1/TIF-2 (23, 26, 41, 51, 52), and p/CIP/AIB/ACTR (2, 8, 51), hereafter referred to as SRC-1, NCoA-2 and p/CIP, respectively, revealed that these proteins are members of a single gene family encoding proteins of approximately 160 kDa. The NCoA/SRC/p160 family of proteins exhibit a number of important properties of bona fide transcriptional coactivators. Biochemical and crystallographic evidence suggests that each of these related proteins can interact directly with liganded steroid hormone receptors in an activation function-2 (AF-2)-dependent manner (11, 23, 38, 51). Furthermore, overexpression of individual NCoA/SRC/p160 proteins can enhance the transcriptional activities of several nuclear hormone receptors in response to their respective ligands (23, 26, 41, 51, 52).
These nuclear proteins have been shown to play a dual role by forming a “bridge” between several classes of transcription factors and the core transcription initiation complex as well as facilitating a more open chromatin configuration by virtue of their intrinsic histone acetyltransferase activities (8, 50). A large body of correlative evidence has supported the proposal that reversible acetylation of lysine residues within the amino-terminal domains of the core histones changes the nucleosomal conformation by loosening DNA-histone contacts (54). Structurally, these proteins contain a highly conserved domain that mediates the interactions with other coactivators such as CBP and pCAF (8, 26, 51). Of particular interest to us, these proteins contain regions homologous to the bHLH-PAS regions of ARNT and AHR. The SRC-1 protein has been implicated in AHRC-mediated transcription via a direct interaction with the AHR transcription activation domain (TAD) (28).
To further elucidate the molecular mechanisms underlying TCDD-dependent gene activation, we investigated the possibility that SRC-1, NCoA-2, and p/CIP are mediators of AHRC transactivation and explored their interaction with ARNT and AHR. We report here that ARNT is capable of interacting with SRC-1 and NCoA-2, while AHR interacts with all three coactivator proteins. These proteins enhance AHRC-mediated transactivation and associate with an AHRC-regulated enhancer region in a dioxin-dependent fashion. Furthermore, the ARNT/SRC-1 interaction is independent of the carboxy-terminal ARNT TAD but rather is mediated through the helix 2 region in the amino-terminal half of the mouse ARNT (mARNT) protein.
MATERIALS AND METHODS
Plasmid constructs and vectors.
ARNT, AHR, SRC-1, NCoA-2, and p/CIP mammalian expression constructs and SRC-1 yeast two-hybrid prey constructs were synthesized as described previously (26, 44, 51) or generated by PCR and cloned in frame into the yeast expression vector pJG0.4.2. The hemagglutinin (HA)-tagged mARNT cDNA expression construct was the kind gift of Jerry Pelletier (39). The bait plasmids pAHR-MP17c and pARNTΔQ-MP17c, encoding amino acids 1 to 687 of the ARNT protein, were generated by PCR with synthetic oligonucleotides and cloned into the same plasmid backbone as the other bait plasmids.
For glutathione S-transferase (GST) pulldown assays, SRC-1 fragments were generated with synthetic oligonucleotides or by restriction digestion and inserted in frame into the bacterial expression vector pGEX-5X-1 or pGEX-5X-2 (Pharmacia, Piscataway, N.J.). For the luciferase assays, a 4.2-kb fragment of the rat CYP1A1 gene containing 2.6 kb of sequence information 5′ of the transcriptional initiation site was cloned into the vector pGL3-Basic (Promega, Madison, Wis.). The GAL4 DNA-binding domain (DBD)-mARNT fusion expression construct was the kind gift of Yoshiaki Fujii-Kuriyama (49). GAL4-DBD-ARNT mutants were generated from existing ARNT mutants by PCR and cloned into the expression vector pBXG-1. The GAL4 upstream activation sequence (UAS)-driven luciferase reporter vector pG5E4T was the generous gift of Michael Carey. The enhanced green fluorescent protein (EGFP)-ARNT cDNA expression plasmid was generated by excising full-length mouse ARNT from pcDNAI/Neo-mARNT (44) with HindIII and XhoI and ligating this fragment in frame into pEGFP-C2 (Clontech, Palo Alto, Calif.). The vectors pEGFP-AHR, pEGFP-ARNTΔQ, and pEGFP-ARNTΔQDH2 were generated by PCR and cloned in frame into pEGFP-C2.
Transient transfections and reporter gene assays in mammalian cells.
Hepa1c1c7 cells, hereafter referred to as Hepa-1 cells, were maintained in alpha minimal essential medium supplemented with 10% fetal calf serum and 100 U of streptomycin and penicillin per ml. Typically, 500 ng each of the CYP1A1 promoter-driven luciferase and pCMV-β-Gal reporter plasmids was cotransfected with either 2 or 5 μg of either pCMX-SRC-1, pCMX-NCoA-2, or pCMX-p/CIP into six-well plates. The final DNA concentration was adjusted to 6 μg per well with empty expression vector, and transfection was achieved with 15 μl of Superfect transfection reagent (Qiagen, Valencia, Calif.) according to the manufacturer's protocol. Three hours after transfection, cells were treated with either 5 × 10−9 M TCDD or 0.1% dimethyl sulfoxide (DMSO) and incubated for a further 18 h before harvesting. Cells were lysed in 500 μl of 1× harvest buffer (50 mM Tris/MES [morpholineethanesulfonic acid, pH 7.8], 1 mM dithiothreitol, and 0.1% Triton X-100) on ice.
Cell lysates were vortexed briefly, and cellular debris was pelleted by centrifugation. Then 15 μl of lysate was added to 185 μl of harvest buffer, 15 μl of luciferase assay cocktail [750 mM Tris/MES (pH 7.8), 150 mM magnesium acetate, and 40 mM ATP] and mixed with 100 μl of 1 mM luciferin. Luciferase activity was measured in a Monolight 2010 luminometer (Analytical Luminescence Laboratory, Sparks, Md.). β-Galactosidase activity was determined with standard protocols (45), and luciferase activity was normalized to these values.
For reporter gene studies with GAL4-DBD-ARNT fusions, 250 ng of pG5E4T and 250 ng of pCMV-β-galactosidase were cotransfected either alone or with a GAL4-DBD-ARNT construct and increasing amounts of coactivator expression plasmid into HEK 293T cells (hereafter referred to as 293T cells). Cells were harvested 20 h after transfection and analyzed as described above.
In vivo coimmunoprecipitation.
293T cells were transfected with 5 μg of HA-tagged mARNT, 5 μg of mAHR and 5 μg of SRC-1, NCoA-2, or p/CIP. Approximately 20 h after transfection, cells were exposed to 20 nM TCDD or vehicle (0.1% DMSO). Whole-cell extracts were prepared as follows. Confluent plates were lysed in 1× LysS buffer (50 mM Tris [pH 7.8], 300 mM NaC1, 0.5% NP-40, 1 mM EDTA, 10% glycerol, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine, and 10 μg each of leupeptin, antipain, and aprotinin per ml) for 30 min on ice. Cellular debris was pelleted by ultracentrifugation at 60,000 × g in a Beckman L-65 centrifuge with an SW55 Ti rotor for 30 min at 4°C. Lysates were assayed for protein concentration with Bio-Rad protein assay solution, following the manufacturer's protocol.
Affinity-purified antibodies directed against the influenza virus hemagglutinin (HA) tag epitope were purchased from Santa Cruz Biotechnologies Inc. (Santa Cruz, Calif.). Affinity-purified rabbit polyclonal antibodies generated against SRC-1, NcoA-2, and p/CIP have been described previously (51). For each reaction, 1 mg of cell lysate was incubated with 5 μg of affinity-purified anti-HA or the corresponding preimmune immunoglobulin G (IgG) overnight at 4°C. Complexes were then mixed with 50 μl of protein A-Sepharose beads (50% slurry) on a rotating wheel at 4°C for 1 h. The beads were then washed six times with LysS buffer, and proteins were eluted in sodium dodecyl sulfate sample buffer and boiled for 3 min. Complexes were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blotting was performed with either affinity-purified anti-SRC-1, anti-NCoA-2, or anti-p/CIP antibodies (51). Precipitated complexes were detected with horseradish peroxidase-linked goat anti-rabbit IgG and an enhanced chemiluminesence (ECL) kit (Pierce, Rockford, Ill.).
Green fluorescent protein assays.
In order to study the interaction between ARNT and SRC-1 in the intact cell, mouse ARNT-EGFP or deletion mutant forms of ARNT-EGFP (0.5 μg) were cotransfected into Hepa1 c4 mutant cells (33) (hereafter referred to as c4 cells) with 4 μg of either pcDNA3-LUC, HA-tag-SRC-1, SRC-1, or pcDNA3 (Invitrogen, Carlsbad, Calif.) expression plasmid and GenePorter2 (Gene Therapy Systems, San Diego, Calif.) according to the manufacturer's protocol. Similarly, in order to study the interaction between AHR and SRC-1, transfections were performed in an identical fashion but in wild-type Hepa1 cells. Twenty hours after transfection, cells were fixed in ice-cold methanol for 15 min. After fixing, cells were washed three times with phosphate-buffered saline (PBS) and incubated with anti-HA IgG for 1 h at room temperature, washed again three times with PBS, and incubated with rhodamine-conjugated anti-rabbit IgG (Molecular Probes, Eugene, Oreg.) for a further 45 min, washed three more times in PBS and once in water, and mounted on glass slides with standard protocols. Cells were visualized by fluorescent microscopy with an AX-70 (Olympus, Melville, N.Y.) upright microscope and photographed with Ektachrome ASA 400 film (Eastman Kodak, Rochester, N.Y.).
Single-cell microinjection assay.
Hepa1 cells were seeded on poly-l-lysine-treated glass coverslips at subconfluent density, and microinjections were performed as previously described (51) with minor modifications. Briefly, to allow for ligand solubility, injected cells were maintained in serumless Dulbecco's modified Eagle's medium supplemented with 3.7% bovine serum albumin. Immediately after injection, cells were treated with either 5 × 10−9 M TCDD or 0.1% DMSO and incubated for a further 18 h.
Chromatin immunoprecipitation assay.
Hepa-1 cells were treated with 10−8 M TCDD for 35 to 45 min. Cross-linking was achieved by adding formaldehyde to a final concentration of 1% at room temperature for 10 min. Cells were washed twice with ice-cold PBS and collected in 1 ml of ice-cold PBS. Cells were pelleted at 700 × g at 4°C and resuspended in 0.3 ml of cell lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% SDS, Roche complete protease inhibitor cocktail) and incubated on ice for 10 min. Cell lysates were sonicated to yield DNA fragments ranging in size from 200 to 900 bp. Samples were centrifuged for 10 min at 4°C. Supernatants were diluted 10-fold to a final solution containing 15 mM Tris-HCl (pH 8.1), 1% Triton X-100, 1 mM EDTA, 150 mM NaCl, 0.1% SDS, and complete protease inhibitor cocktail. The solutions were precleared with 50 μl of 50% slurry protein A-Sepharose containing 2.5 μg of sheared salmon sperm DNA for 2 h at 4°C.
Immunoprecipitation was performed overnight at 4°C with antibodies against the various coactivators; 50 μl of protein A-Sepharose containing 2.5 μg of salmon sperm DNA and 2 mg of bovine serum albumin per ml was then added to the solution and incubated for an additional 1 h. The beads were pelleted and washed in the following buffers: buffer A (20 mM Tris-HCl [pH 8.1], 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), buffer B (20 mM Tris-HCl [pH 8.1], 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), and LiCl-detergent buffer (10 mM Tris-HCl [pH 8.1], 0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA). Finally, they were washed twice with TE (Tris-EDTA) buffer. Immunocomplexes were extracted with 1% SDS-0.1 M NaHCO3 from the beads.
Cross-linking was reversed by heating the eluted material at 65°C overnight. The eluates were then digested with proteinase K at 45°C for 1 h. The solutions were extracted with phenol-chloroform-isoamyl alcohol. DNA was purified by ethanol precipitation and amplified by PCR over the murine CYP1A1 enhancer region (−1141 to −784 from the transcription initiation start site). Primer sequences were 5′-CTATCTCTTAAACCCCACCCCAA-3′ and 5′-CTAAGTATGGTGGAGGAAAGGGTG-3′.
In vitro GST pulldown assays.
[35S]methionine-labeled full-length AHR, ARNT, or mutant ARNT proteins were synthesized with a T7 TNT-Quick in vitro transcription-translation system (Promega) according to the manufacturer's protocol. GST-SRC-1 fusion proteins were expressed in the bacterial cell line BL21. Cells were lysed in 1× LysS buffer (50 mM Tris [pH 7.8], 1.0 M NaCl, 1.0 mM EDTA, 10% glycerol, β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine, and 10 μg each of leupeptin, antipain, and aprotinin per ml) for 30 min on ice. Cellular debris was pelleted by ultracentrifugation at 60,000 × g in an SW41 Ti rotor for 1 h at 2°C. Recombinant proteins were bound to glutathione-Sepharose beads by incubation at 4°C for 3 h on a rotating wheel. Beads were subsequently washed with 0.5× LysS buffer (except that the NaCl concentration was lowered to 300 mM).
Small samples of each recombinant GST-SRC-1 fusion protein were eluted off the beads in 5× SDS sample buffer (62.5 mM Tris [pH 6.8], 10% glycerol, 2% SDS, 700 mM β-mercaptoethanol, and 0.00125% bromophenol blue) and subjected to SDS-PAGE. Protein concentrations were determined by comparison to bovine serum albumin standards as assessed by Coomassie staining. For GST pulldowns, equimolar amounts of each GST-SRC-1 fusion protein (approximately 10 μg) were incubated with 4 μl of in vitro-translated ARNT for 2 h at 37°C. Bead volumes for each sample were standardized with blank glutathione-Sepharose beads, and reaction volumes were brought up to 200 μl with 1× PPI buffer (20 mM HEPES [pH 7.9], 300 mM NaCl, 1 mM EDTA, 4 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, 0.1% NP-40, 0.5 mM phenylmethylsulfonyl fluoride). Beads were washed six times with PPI buffer, and protein complexes were eluted with SDS sample buffer and boiled for 3 min. Proteins were subjected to SDS-PAGE, and gels were dried and subjected to autoradiography.
Yeast two-hybrid interaction assays.
The yeast strain L40C has been described previously (43) and was a generous gift of John Colicelli. SRC-1 mutant constructs or the empty pJG.4.2 plasmid was transformed into the yeast strain L40C containing either plasmid pARNTΔQ-MP17c or pAHR-MP17c. For interaction studies involving pAHR-MP17c, cultures were grown in the presence or absence of 1 mM β-napthaflavone (β-NF). Single colonies were picked and grown individually in synthetic complete medium lacking leucine and tryptophan.
To assay for dimerization ability, equal amounts of plasmid-containing L40C were resuspended in 500 μl of modified Z-buffer (40 mM NaH2PO4, 60 mM Na2HPO4, 10 mM KCl, pH 7.0). Then 50 μl of 0.1% SDS and three drops of CHCl3 were added, and samples were vortexed for 1 min. Then 200 μl of 4-mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG) in 0.1 M NaPO4 (pH 7.0) was added to each sample, and reaction mixtures were incubated at 37°C. Cellular debris was pelleted by centrifugation, and the absorbance of supernatants was measured at 420 nm (4). All values were derived from two or more transformations; each determination was performed in triplicate, and final values are the result of four separate experiments.
RESULTS
Transcriptional coactivation of the AHRC.
To investigate whether the bHLH-PAS family of nuclear coactivators are involved in AHRC-dependent transactivational processes, we chose to study the effect of overexpression of SRC-1, NCoA-2, and p/CIP on CYP1A1 induction by the AHRC. Toward this end we cotransfected nuclear coactivator cDNAs in mammalian expression vectors into Hepa-1 cells with a CYP1A1 promoter-driven luciferase construct. TCDD treatment alone enhanced CYP1A1 promoter-driven luciferase activity nearly 25-fold in cells not transfected with coactivator expression plasmids (Fig. (Fig.1A).1A). Cotransfection of each coactivator expression plasmid potentiated the TCDD-dependent transactivation of the CYP1A1 promoter-luciferase construct.
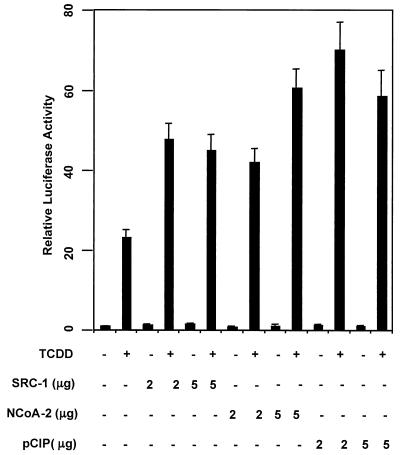
SRC-1, NCoA-2, and p/CIP function as transcriptional coactivators of TCDD-dependent gene activation. Hepa-1 cells were transfected with a CYP1A1 promoter-driven luciferase construct, and 2 to 5 μg of either empty vector or SRC-1, NCoA-2, or p/CIP expression construct. Cells were treated with 5 nM TCDD or vehicle for 24 h. Lysates were then assayed for luciferase activity.
SRC-1, NCoA-2, and p/CIP were equipotent with regard to coactivation potential (two- to threefold greater levels seen in their presence than in their absence). Cotransfection of the coactivator constructs did not significantly alter transactivation in the absence of TCDD. These results strongly suggest a transcriptional coactivator function for SRC-1, NCoA-2, and p/CIP for induction of the CYP1A1 gene. Western blot analysis of the Hepa-1 cell extract revealed an abundance of each coactivator in untransfected cells (data not shown). Therefore, endogenous coactivators may partially satisfy the AHRC's need for coactivator in this assay, resulting in the high activity observed in cells not transfected with any coactivator protein in the presence of ligand.
In vivo coimmunoprecipitation.
The above results indicate that dioxin-dependent activation of the AHRC and the concomitant increase in transcription of CYP1A1 and potentially other dioxin-responsive genes is modulated by the activity of the NCoA/SRC/p160 family of transcriptional coactivators. In order to address whether a physical interaction occurs between ARNT or AHR and these coactivators, whole-cell extracts from 293T cells transiently cotransfected with both HA-tagged mARNT and mAHR, together with either SRC-1, NCoA-2, or p/CIP, and treated with either TCDD or vehicle were used for in vivo coimmunoprecipitation studies.
Affinity-purified polyclonal rabbit antibodies generated against the influenza virus hemagglutinin tag (HA), and therefore capable of immunoprecipitating HA-tagged mARNT complexes, coimmunoprecipitated exogenous SRC-1 and NCoA-2 but not p/CIP (Fig. (Fig.2).2). Cotransfection with the mAHR expression plasmid and treatment with 20 nM TCDD did little to alter the ability of the anti-HA antibody to precipitate either SRC-1 or NCoA-2 (lane 4 in Fig. Fig.2),2), suggesting that ARNT can dimerize with AHR and bind SRC-1 at the same time. Anti-HA IgG failed to precipitate coactivator in cells transfected with mAHR and coactivator expression plasmid but not HA-tagged mARNT, demonstrating an absolute requirement for the latter protein (data not shown). Affinity-purified antibodies to AHR coimmunoprecipitated all three coactivators from cells grown in either the presence or absence of TCDD.
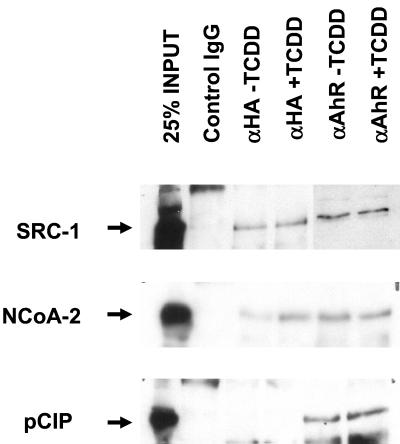
Coimmunoprecipitation of the SRC family of coactivators with AHR and ARNT. 293T cells were transfected with 5 μg each of AHR and HA-tagged ARNT expression vector and 5 μg of either the SRC-1, NCoA-2, or p/CIP expression vector. Twenty hours after transfection, cells were treated with either 20 nM TCDD or vehicle. Two hours later, cells were harvested and total cellular protein was extracted. Extracts were incubated with either affinity-purified anti-HA IgG or preimmune control IgG bound to protein A-Sepharose beads, and complexes were fractionated by SDS-PAGE. Complexes were then transferred to nitrocellulose, and membranes were probed with affinity-purified rabbit anti-SRC-1, anti-NCoA-2, or anti-p/CIP. Detection was performed with an ECL kit. The left lane of each panel contained 200 μg of HEK 293T whole-cell lysate (25% of input).
Interaction of ARNT with coactivators in mammalian cells analyzed by stimulation of an ARNT-dependent reporter gene.
In order to further investigate a potential interaction between ARNT and the coactivators, we employed a GAL4 UAS-thymidine kinase (TK)-luciferase reporter gene (pG5E4T) driven by a fusion of the GAL4 DBD and mARNT and tested whether the coactivators could stimulate the ARNT-dependent reporter gene activity. Reporter gene activity was enhanced by cotransfection of GAL4-DBD-mARNT and increasing amounts of SRC-1 expression plasmid in 293T cells (Fig. (Fig.3A).3A). SRC-1 maximally increased ARNT-dependent transcription approximately 6.5-fold, while empty expression vector had no effect on reporter activity. These observations support the notion that SRC-1 can interact with ARNT. The enhancement of luciferase activity is most likely due to the endogenous activation function of SRC-1 (26, 41). Furthermore, NCoA-2 but not p/CIP was capable of enhancing transcription of pG5E4T by GAL4-DBD-ARNT in 293T cells (Fig. (Fig.3B),3B), consistent with the coimmunoprecipitation data. The ability of p/CIP to enhance dioxin-dependent transcription (Fig. (Fig.1A)1A) is most likely due to p/CIP's ability to interact with mAHR.
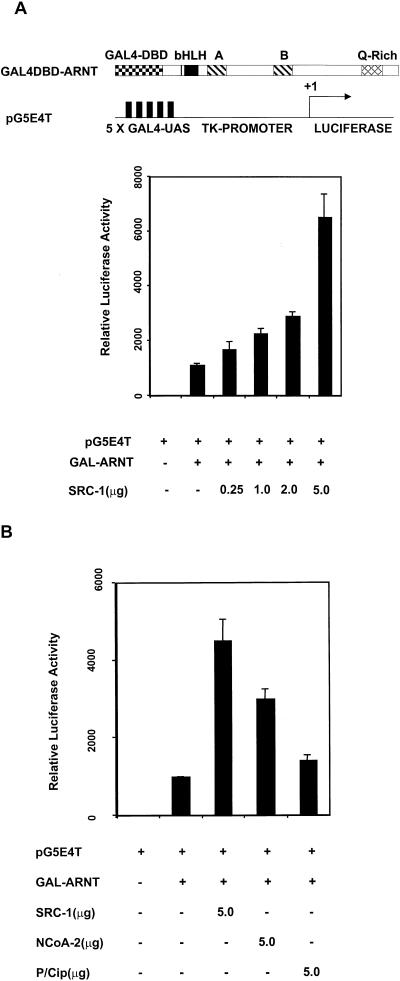
Interaction between ARNT and SRC-1 in mammalian cells as appraised by reporter gene analysis. (A) SRC-1 modulation of ARNT-dependent activation of a GAL4 UAS-TK-driven luciferase reporter. 293T cells were cotransfected with GAL4-DBD-mARNT with either empty vector or increasing amounts of pCMX-SRC-1. (B) A comparison of the coactivator properties of SRC-1, NCoA-2, and p/CIP in the GAL4-DBD-mARNT-responsive luciferase assay.
Fluorescent visualization and colocalization of ARNT/SRC-1 and AHR/SRC-1 complexes.
Coimmunoprecipitation assays are subject to a potential artifactual association of proteins occurring after cellular disruption. Colocalization experiments in mammalian cells in vivo do not suffer from this potential artifact. Such experiments were therefore undertaken in order to further substantiate the interaction of ARNT and AHR with SRC-1. We cotransfected a cDNA plasmid encoding a mARNT-enhanced green fluorescent fusion protein together with either pcDNA3, luciferase expression vector (pCMV-Luc), or pCMX-SRC-1 into the ARNT-deficient Hepa-1 mutant c4 cell line to study how overexpression of SRC-1 might affect the subcellular distribution of mARNT. The c4 cell line was chosen to avoid competition for expressed SRC-1 protein by endogenous ARNT that might potentially confound the interpretation of results. Experiments were performed with equivalent amounts of vector DNA, with pcDNA3 plasmid as a supplement. Representative results of these experiments are presented in Fig. Fig.4A.4A.
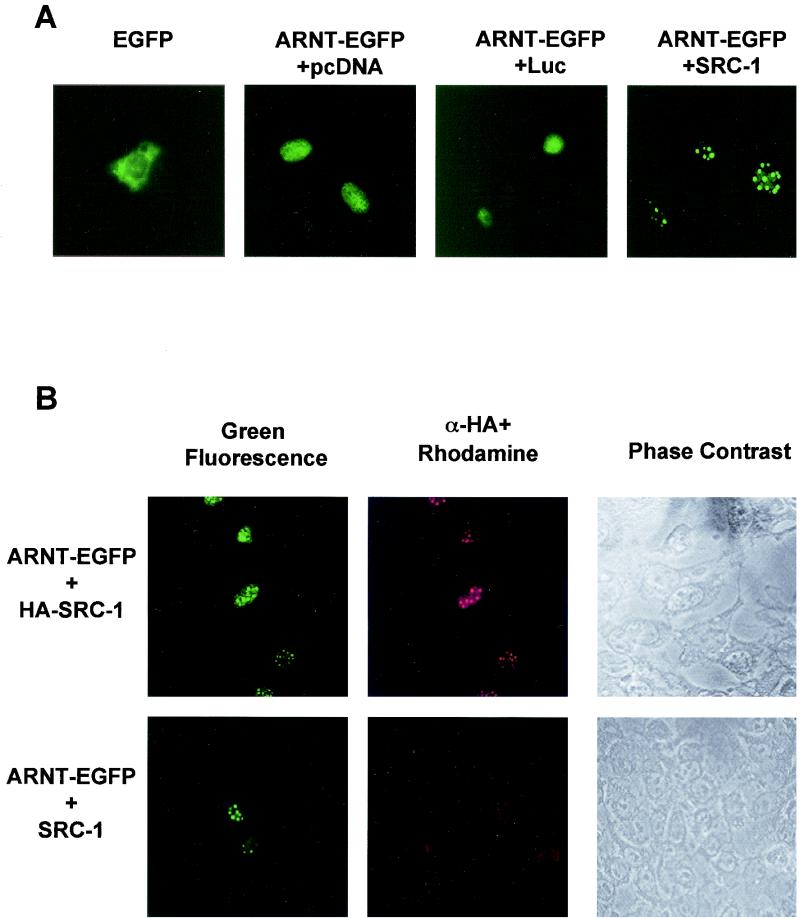
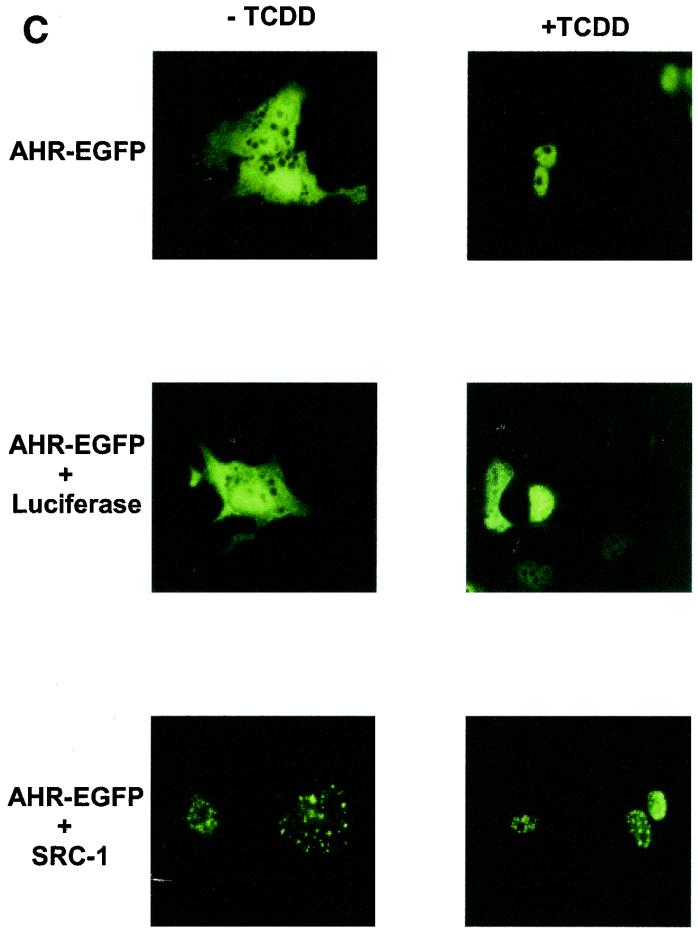
Intranuclear distribution of ARNT is altered by overexpression of SRC-1. (A) Hepa-1 mutant c4 cells were transiently transfected with pEGFP (0.5 μg) or mARNT-EGFP (0.5 μg) together with 4 μg of either pcDNA3, pLUC-DNA3, or pCMX-SRC-1 and observed for EGFP fluorescence. (B) Hepa-1 mutant c4 cells were transfected with mARNT-EGFP (0.5 μg) and either HA-tagged SRC-1 (4 μg) or untagged SRC-1 (4 μg) expression vector. Fixed cells were incubated with rabbit anti-HA IgG and rhodamine-conjugated goat anti-rabbit IgG. (C) Hepa-1 cells were transiently transfected with the indicated expression vectors, untreated or treated with TCDD for 18 h, as indicated, and observed for EGFP fluorescence.
When pEGFP-C2, encoding green fluorescent protein, was transfected into asynchronous c4 cells, green fluorescent protein distributed throughout the cell, although predominantly in the cytosol. When mARNT fused to enhanced green fluorescent protein was transfected into the c4 cell line, the mARNT-EGFP fusion moiety distributed to the nuclear compartment and appeared to distribute in a homogenous fashion throughout the nucleus. Overexpression of firefly luciferase as a negative control did not alter the expression pattern of mARNT-EGFP. However, expression of SRC-1 caused the redistribution of mARNT-EGFP into discrete foci within the nucleus. This was seen in a large proportion but not all of the transfected cells. The nature of these foci is not clear, although they do not appear to be nucleoli.
In order to verify that the redistribution of mARNT-EGFP was due to the presence of mSRC-1 protein, colocalization experiments were performed. Hepa-1 mutant c4 cells were cotransfected with mARNT-EGFP and either a HA-tagged SRC-1 or untagged SRC-1 expression vector (pCMX-HA-SRC-1 or pCMX-SRC-1). HA was detected with rabbit anti-HA IgG and rhodamine-conjugated goat anti-rabbit IgG. It was found that cells possessing green fluorescent intranuclear foci (indicating the presence of mARNT-EGFP) were also positive for rhodamine fluorescence when they had been cotransfected with the HA-tagged SRC-1 expression vector. Green fluorescence and rhodamine fluorescence (and thus mARNT-EGFP and HA-tagged SRC-1) colocalized to the same foci in these nuclei (Fig. (Fig.4B).4B). In cells cotransfected with the untagged SRC-1 cDNA, there was a complete absence of rhodamine fluorescence. These results support the notion that SRC-1 is directly responsible for causing redistribution of mARNT-EGFP into the nuclear bodies.
When AHR-EGFP was cotransfected with the SRC-1 expression plasmid into Hepa-1 cells in the absence of ligand, SRC-1 caused AHR-EGFP to redistribute into discrete foci within the cytosol. (Presumably SRC-1 can be cytosolic, at least when overexpressed [Fig. [Fig.4C].)4C].) In the presence of TCDD, AHR-EGFP was almost completely nuclear and was distributed in a homogenous fashion throughout the nuclear compartment. However, as seen with ARNT, AHR redistributed into discrete foci within the nucleus in the presence of TCDD when SRC-1 was overexpressed, but not when luciferase was overexpressed.
These experiments confirm that both ARNT and AHR can interact with SRC-1 in vivo. It should be noted, however, that these and the previously described experiments were all obtained with transfected cDNAs. It is possible that proteins overexpressed in this manner may not behave identically to the corresponding endogenous proteins. The following experiment studied endogenous proteins and does not suffer from this potential artifact.
Requirement of SRC-1/p160 proteins for TCDD-dependent transactivation.
In order to determine the function of SRC-1, NCoA-2, and p/CIP in TCDD-dependent gene activation, we injected Hepa-1 cells with affinity-purified IgGs specific for each coactivator. The reporter gene lacZ was placed under the control of the p36 minimal promoter and four copies of a classic xenobiotic compound-responsive element. Injection of either anti-SRC-1 or anti-p/CIP IgG prevented TCDD from activating the xenobiotic compound-responsive element-dependent transcription unit (Fig. (Fig.5A).5A). Since the cocktail injected in each case contained IgG, injected cells could be scored by immunofluorescence after staining with a rhodamine-conjugated secondary antibody (Fig. (Fig.5B).5B). Anti-NCoA-2 IgG failed to affect TCDD-dependent transactivation at this reporter (data not shown). This is not surprising, however. The ability of NCoA-2 to interact with several members of the nuclear receptor family of transcription factors is well documented (23, 26, 41, 51, 52), yet this antibody failed to block retinoic acid receptor-mediated activation in a similar assay (51), indicating that this antibody may not be suitable as an inhibitory factor in this assay.
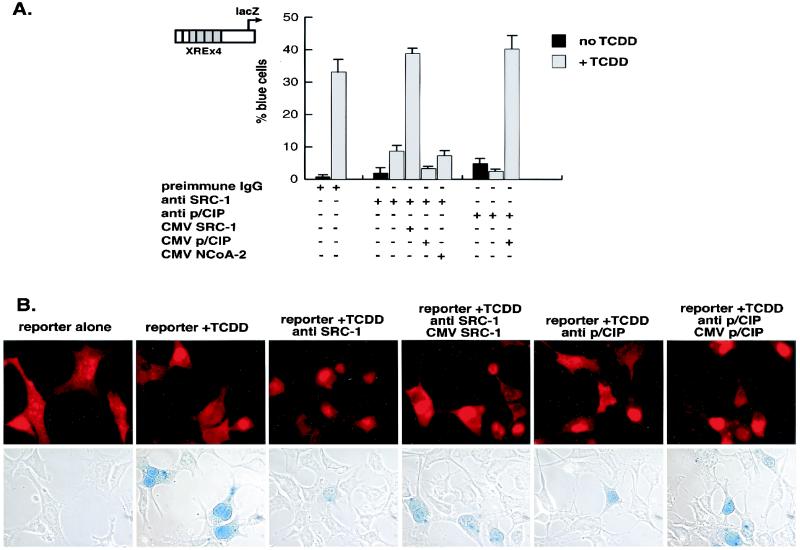
Role of SRC-1/p160 proteins in AHR/ARNT function as assessed by microinjection of antibodies into the nuclei of single cells. (A) Nuclei of Hepa-1 cells incubated with or without TCDD were microinjected with control rabbit IgG, with or without either anti-SRC-1 or anti-p/CIP affinity-purified IgG and, where indicated, SRC-1, NCoA-2, or p/CIP expression vector. (B) The cells were probed with rhodamine-conjugated goat anti-rabbit IgG to identify injected cells and stained for β-galactosidase activity. Photomicrographs of rhodamine-stained cells and the corresponding light micrographs after 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining are shown.
The inhibitory effect of anti-SRC-1 and anti-p/CIP IgG could be reversed by coinjection of the SRC-1 and p/CIP expression vector, respectively. Unlike retinoic acid receptor-responsive reporter gene activity, rescue from the effects of anti-p/CIP IgG did not require coinjection of CBP expression vector (51), suggesting that the AHRC recruits a distinct p/CIP regulatory complex rather than that of the retinoic acid receptor. Coinjection of either the NCoA-2 or p/CIP expression vectors could not restore the transcriptional activity inhibited by anti-SRC-1. This possibly reflects the inability of NCoA-2 and p/CIP to compete for large macromolecular complexes sequestered by the anti-SRC-1 IgG because of lack of specificity for anti-SRC-1 IgG or may indicate a distinct role for each coactivator in ARHC-dependent transcription, but this remains to be determined.
CYP1A1 enhancer occupation by the SRC-1/p160 proteins.
We performed chromatin immunoprecipitation assays to test if SRC-1, NCoA-2, and p/CIP could be recruited to the CYP1A1 enhancer in a TCDD-dependent fashion (Fig. (Fig.6).6). Formaldehyde-fixed protein-DNA immunocomplexes precipitated with affinity-purified antibodies raised against SRC-1, NCoA-2, or p/CIP were denatured and subjected to PCR with oligonucleotides flanking the CYP1A1 enhancer. In the absence of TCDD, the CYP1A1 enhancer DNA could not be amplified. However, in the presence of TCDD, the CYP1A1 enhancer region could be amplified after immunoprecipitation with all three coactivator antibodies, demonstrating the presence of the coactivators at the enhancer. Regions flanking the CYP1A1 enhancer could not be amplified by PCR, indicating that these proteins were associated with the CYP1A1 enhancer region and not the proximal promoter (data not shown). These data unambiguously demonstrate TCDD-dependent recruitment of the endogenous SRC-1, NCoA-2, and p/CIP coactivator proteins to the CYP1A1 enhancer in vivo. Differences in the intensities of the SRC-1-, NCoA-2-, and p/CIP-specific bands may reflect the avidity of the antibodies for their respective proteins and do not necessarily indicate a greater affinity of one coactivator for the AHRC complex over another.
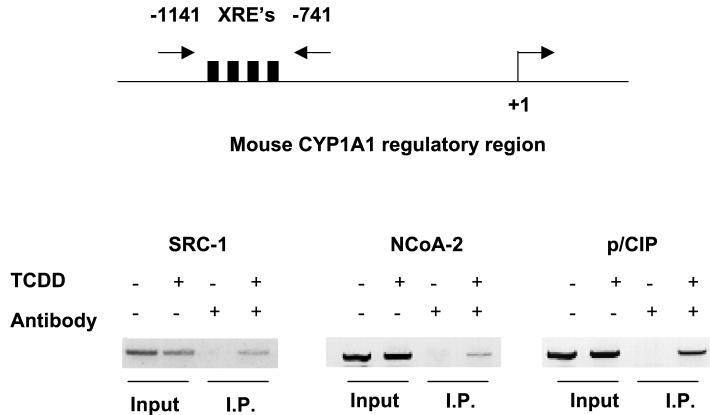
Chromatin immunoprecipitation of the CYP1A1 enhancer region. PCR amplification of the CYP1A1 enhancer by affinity-purified antibodies raised against SRC-1, NCoA-2, and p/CIP. Total input DNA for each PCR can be visualized in the first two lanes of each panel. Precipitation (I.P.) reactions performed in the absence and presence of TCDD are shown in lanes 3 and 4 of each panel, respectively. XRE's, xenobiotic compound-responsive elements.
Determination of ARNT and AHR interaction domains within SRC-1.
In order to determine the ARNT and AHR interaction domains within SRC-1, B42 activation domain-SRC-1 and LexA DNA-binding domain (LexA-DBD)-ARNT/AHR fusion constructs were used in the yeast two-hybrid system to test for protein-protein interactions. Attempts to clearly define the ARNT interaction domain within the SRC-1 protein with the yeast two-hybrid β-galactosidase method were hampered by the ability of ARNT's transcriptional activation domain to confer high-level β-galactosidase activity in cells cotransformed with only empty prey vector. Deletion of sequences encoding amino acids 688 to 791 of the mARNT protein (comprising its carboxy-terminal TAD) in the bait vector abolished the high constitutive activity observed with the full-length protein.
We therefore employed a bait construct (ARNTΔQ) encoding amino acids 1 to 687 of the mARNT protein. Constructs were transformed into the yeast strain L40C and tested for their ability to activate the β-galactosidase reporter gene. Among the SRC-1 deletion mutants tested, the one encoding amino acids 763 to 1100 conferred the highest activity on the lacZ gene, as assayed by β-galactosidase activity (250-fold over basal levels) when cotransformed with ARNTΔQ (Fig. (Fig.7A).7A). This region encompasses the CBP interaction domain on SRC-1. Deletion of amino acids 763 to 895 significantly decreased activity in this assay, as evidenced by the reduced interaction of SRC-1896-1200 with ARNTΔQ compared with SRC-1763-1100. Deletion of amino acids 1034 to 1100 (represented by SRC-1763-1033) from SRC-1763-1100 resulted in only a slight decrease in β-galactosidase activity.
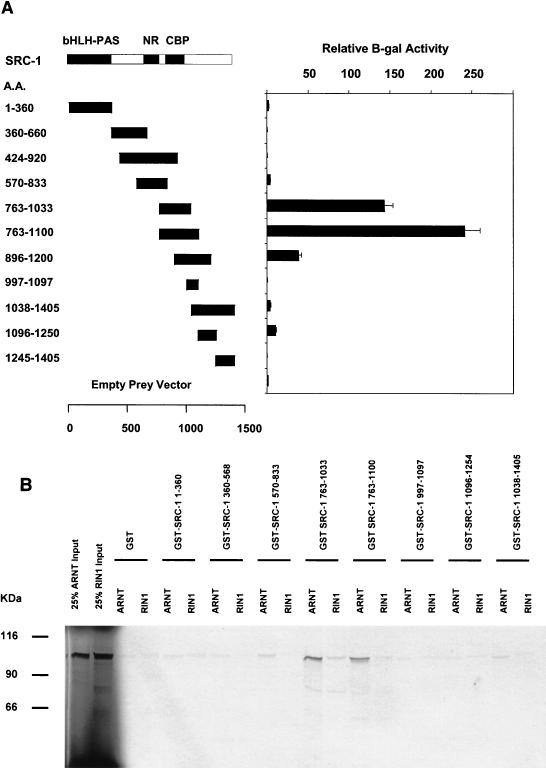
Identification of interaction domains on SRC-1 for ARNT with yeast two-hybrid and GST pulldown assays. (A) SRC-1 deletion mutants were fused to the B42 activation domain in pJG4.5 and transformed into the yeast strain L40C along with ARNTΔQ fused to the LexA DNA-binding domain and assayed for β-galactosidase activity. Results are expressed as increases (n-fold) over mean values obtained for the empty prey vector. The interaction domains previously identified within SRC-1 for the transcriptional coactivator CBP and for several nuclear hormone receptors (10) are indicated. (B) SRC-1 mutants were fused to glutathione S-transferase and expressed in Escherichia coli. Expressed fusion proteins from bacterial cell lysates were put on glutathione-agarose beads. Approximately 10 μg of protein coupled to beads was incubated with in vitro-translated [35S]methionine-labeled ARNT or RIN1, and absorbed proteins were subjected to SDS-PAGE. The two left lanes contained 1 μl of in vitro-translated RIN1 and 1 μl of in vitro-translated mARNT (25% input) for comparison.
We sought to confirm the location of the ARNT interaction domain with an in vitro GST pulldown assay using full-length ARNT together with eight mutant GST-SRC-1 fusion constructs, each encoding a discrete portion of the protein and together encompassing its entire length. The results of these experiments are presented in Fig. Fig.7B.7B. All GST-SRC-1 fusion moieties were quantified on SDS-PAGE with bovine serum albumin standards so that equimolar amounts of each fusion were used in this assay. RIN1, a protein of the same approximate size as mARNT and not likely to interact with SRC-1 in vivo could not be pulled down by any GST-SRC-1 fusion protein in this assay. Firefly luciferase was used as another negative control for this assay. None of the SRC-1 mutant proteins pulled down 35S-labeled luciferase (data not shown). GST-SRC-1763-1100 and GST-SRC-1763-1033 were equally effective in precipitating in vitro-translated full-length ARNT, confirming that GST-SRC-1763-1033 contains the minimal ARNT interaction domain.
The interaction between AHR and SRC-1 was also studied by yeast two-hybrid and GST pulldown analyses. Unlike ARNT, full-length AHR was not constitutively active in yeast cells in the absence of the AHR ligand β-NF. The B42-SRC-1 fusion protein, encoding amino acids 896 to 1200 of the SRC-1 protein, was capable of interacting with AHR in yeast cells in a ligand-dependent fashion, although the severalfold activation of the lacZ reporter in yeast cells by SRC-1 was only approximately 4-fold, compared to approximately 250-fold for the ARNT-SRC-1 interaction. This may be due to AHR's TAD, the known interaction site for SRC-1 (29), being at least partially masked in yeast cells. The same region of SRC-1 was capable of interacting with AHR in the GST interaction assay, but in this case the interaction was ligand independent (Fig. (Fig.8A8A and B). SRC-1763-1100, which interacted maximally with ARNT, was completely incapable of recruiting AHR in both assays.
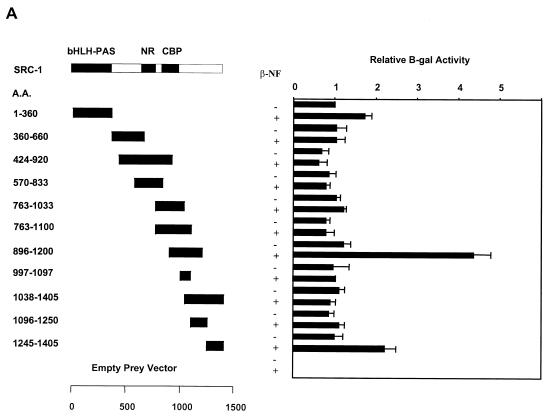
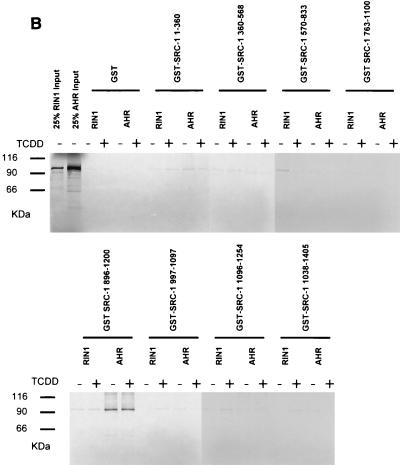
Identification of AHR interaction domains within SRC-1. (A) Yeast two-hybrid analysis of the interaction between AHR and various deletion mutants of SRC-1. The experiments were performed in the presence or absence of 1 mM β-NF, as indicated. (B) GST pulldown assay of [35S]methionine-labeled AHR and GST-SRC-1 deletion mutant fusion proteins.
The results from the yeast liquid culture and GST pulldown assays are congruent and indicate that the ARNT interaction domain is encompassed within amino acids 763 to 1033 of mSRC-1 and that the AHR interaction domain is encompassed within amino acids 896 to 1200. GST control protein and the fragments encoding regions flanking but not overlapping the above ARNT and AHR interaction domains on SRC-1 were ineffective in precipitating ARNT and AHR, respectively. Therefore, our data indicate that the ARNT and AHR interaction domains within SRC-1 may overlap but are distinct. SRC-11-360 contains regions of homology with ARNT and AHR's bHLH and PAS domains, which are known dimerization regions for other bHLH-PAS proteins that interact with ARNT, but surprisingly this fragment was incapable of interacting with ARNT or AHR, as determined by GST pulldown and yeast two-hybrid assays. Recently, it has been reported that the transcriptional coactivator CBP/p300 is recruited by ARNT for dioxin-dependent transactivation, and the respective interaction domains have been identified (28). Interestingly, GST-CBP1892-2441, encoding the region of CBP that interacts with SRC-1, was not capable of competing with in vitro-translated ARNT for binding to GST-SRC-1763-1100 (data not shown). These data, taken together, suggest that ARNT's interaction with these two coactivators is not mutually exclusive.
Determination of SRC-1 interaction domains within ARNT.
As shown above, the bait vector pARNTΔQ-MP17c was capable of interacting with the SRC-1763-1100 deletion mutant (Fig. (Fig.7A).7A). To define the interaction domain within ARNT that mediates its interaction with SRC-1, in vitro-translated mutants of the mARNT protein (Fig. (Fig.9A9A ) were tested with GST-SRC-1763-1100 to determine if they could be precipitated in the GST pulldown assay. None of the ARNT mutants that lacked the protein's helix 2 domain were capable of interacting with SRC-1 in the GST pulldown assay, including a mutant in which helix 2 alone (amino acids 128 to 142) was deleted (Fig. (Fig.9B).9B). Furthermore, the ARNT bHLH fragment alone (amino acids 70 to 142) was pulled down efficiently by SRC-1. The results are internally consistent and indicate that the SRC-1 interaction domain in ARNT encompasses the helix 2 domain.
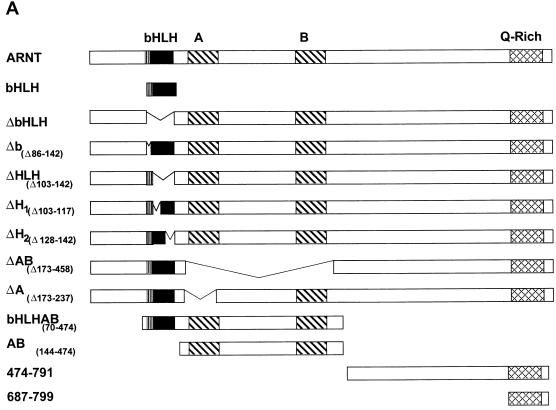
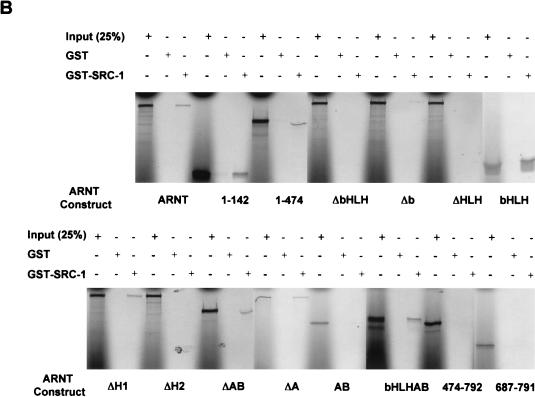
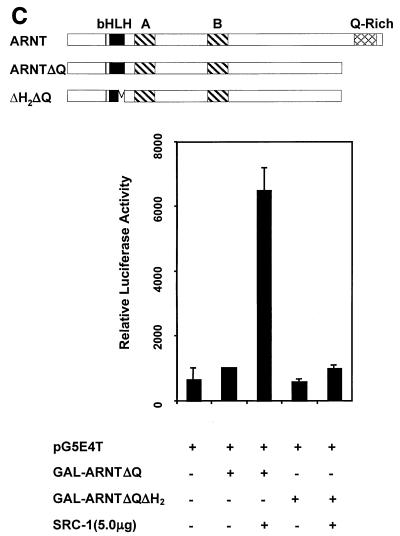
Identification of SRC-1 interaction domains within ARNT with the yeast two-hybrid and GST pulldown assays. (A) Schematic of the mARNT mutants used in the GST pulldown assay. Mutants were cloned into pcDNAI/Neo or pcDNA3.1/His C. Critical amino acids deleted or included are indicated in parentheses. (B) GST pulldown of ARNT minimal mutants by GST-SRC-1763-1100. Complexes were fractionated by SDS-PAGE on gels ranging in acrylamide concentration from 7.5 to 10%. (C) Effect of the loss of the ARNT TAD and helix 2 on recruitment of SRC-1 in the ARNT-dependent luciferase assay. Above appears a schematic of mARNT deletion mutants fused to the GAL4 DBD.
It is interesting that carboxy-terminal sequences of the mARNT protein, residues 474 to 687 and 687 to 791, were incapable of interacting with SRC-1. ARNT interacts with its transcription factor dimerization partners, AHR and SIM1, through their respective bHLH-PAS domains (43, 44). Our data indicate that, while SRC-1 contains a bHLH-PAS domain, it does not utilize this domain to interact with ARNT, whereas ARNT requires it own bHLH domain for interaction with SRC-1. Most importantly, these studies indicate that the mARNT TAD is not required for interaction with SRC-1, unlike many of the nuclear hormone receptors (26, 41, 51), HIF-1α (6), and AHR (29).
These surprising and novel observations led us to pursue another avenue of investigation to confirm the dependence of mARNT helix 2 for interaction with SRC-1. Figure Figure9C9C demonstrates that GAL4-DBD-ARNTΔQ could not activate transcription at a GAL4 UAS-responsive promoter in mouse Hepa-1 cells, consistent with studies indicating the requirement for the mARNT carboxy-terminal TAD for activated transcription (34, 49). Cotransfection with the SRC-1 expression plasmid increased luciferase activity by approximately sevenfold. However, addition of SRC-1 did not increase luciferase activity when an ARNT construct was used that had a deletion of helix 2 as well as the TAD (GAL-ARNTΔH2ΔQ). This result supports the notion that helix 2 of ARNT is required for interaction with SRC-1.
DISCUSSION
The aryl hydrocarbon receptor nuclear translocator (ARNT) is the dimerization partner of a growing family of structurally related transcription regulators. Responses to a diverse array of signals, including hypoxia and environmental pollutants, are mediated by the ARNT protein dimerized with other bHLH-PAS proteins (18). These heterodimers bind distinct DNA response elements in the upstream regulatory regions of specific target genes. One such partner, the aryl hydrocarbon receptor, only binds ARNT after it has been activated by a wide range of chemical carcinogens, most notably halogenated and polycyclic aromatic hydrocarbons. The induction of CYP1A1 provides a model system for the study of ARNT-mediated transcriptional processes. The characteristics of the ARNT-coactivator interactions that we have defined here may thus be applicable to many or all of the ARNT-dependent transcription systems besides those mediated by the AHR/ARNT dimer.
The principal mechanism by which polycyclic aromatic hydrocarbons cause cancer is understood in broad terms. Polycyclic aromatic hydrocarbons induce CYP1A1 and CYP1B1 via AHR. These cytochrome P450s then metabolize the polycyclic aromatic hydrocarbons to electrophilic derivatives that can mutate DNA, thereby activating proto-oncogenes or inactivating tumor suppressor genes (21). Unlike polycyclic aromatic hydrocarbons, TCDD is not genotoxic but acts as a tumor promoter. However, it is the most carcinogenic compound ever tested (24). Furthermore, TCDD has a wide range of effects besides carcinogenicity, including teratogenesis, suppression of the immune system, adverse effects on the reproductive system, and modulation of various hormonal effects. Most if not all of these effects depend upon ARNT as well as AHR (reviewed in references 17 and 47). This wide range of toxic responses suggests that the AHRC signaling pathway may interact with other signal transduction pathways. One way this may occur is via direct protein-protein interactions between AHR and ARNT and a common pool of modulatory proteins, such as SRC-1, NCoA-2, and p/CIP studied here.
In the present study, we investigated the role of the NCoA/SRC/p160 family of transcriptional coactivators in the modulation of AHRC-mediated gene regulation. We demonstrated the ability of SRC-1, NCoA-2, and p/CIP to coactivate transcription of TCDD-responsive genes. Furthermore, with a single-cell microinjection reporter assay, we demonstrated the requirement for SRC-1 and p/CIP for AHRC-dependent gene activation. We also demonstrate by coimmunoprecipitation experiments that ARNT can interact with SRC-1 and NCoA-2, but not p/CIP. This is in contrast to the situation with AHR, which can interact with all three NCoA/SRC/p160 coactivators. The interaction of ARNT with SRC-1 and NCoA2 was confirmed by reporter gene analysis in mammalian cells and colocalization experiments in mammalian cells. The latter type of experiment also confirmed the interaction between AHR and SRC-1.
We also showed that the endogenous coactivators associate with AHRC-regulated chromatin in a TCDD-dependent manner in vivo. Finally, we identified the regions within SRC-1 that are responsible for direct interactions with AHR and ARNT and the region within ARNT responsible for interaction with SRC-1. These represent important steps in our understanding of the role that coactivators play in gene regulation by the AHRC. Previously, it was proposed that SRC-1 does not interact with ARNT, based on the observations that deletion of the ARNT TAD had little effect on SRC-1-mediated coactivation of AHRC-dependent transcription and that anti-ARNT antibodies poorly immunoprecipitated 35S-labeled SRC-1 in vitro (29). We cannot fully reconcile the discrepancies between these data and ours except to note that that our studies focused on the amino-terminal helix 2 domain of ARNT.
In most of our assays, AHR interacted with SRC-1 in a ligand-independent fashion. It is possible that the unliganded AHR does genuinely interact with SRC-1 in vivo. Alternatively, the absence of a ligand effect may be an artifact of our experimental systems, particularly when AHR was overexpressed, perhaps being due to the absence or insufficiency of XAP2, p23, or other chaperone proteins that are associated with AHR in its unliganded state and which modulate its folding and functionality.
Activator-coactivator interactions appear to be extremely weak compared to interactions within activator homo- and heterodimers, so much so that studies demonstrating a direct interaction between the endogenous components are rare. Previous studies concerning interactions of CBP and the NCoA/SRC/p160 family of coactivators with nuclear hormone receptors have relied on coexpression experiments and in vitro pulldown assays to demonstrate interaction (8, 26, 28, 40, 51). The interactions we observed between ARNT and SRC-1 and NCoA-2 were not negated by ARNT's dimerization with ligand-bound AHR (Fig. (Fig.2),2), consistent with the notion that SRC-1 and NCoA-2 play a role in gene regulation by the AHRC. Furthermore, negation of activity of a xenobiotic compound-responsive element-driven reporter by anti-SRC-1 and anti-p/CIP IgG suggests that these coactivators are part of complexes that are absolutely required for AHRC-dependent transcription (Fig. (Fig.55).
The chromatin immunoprecipitation assays presented here demonstrate in an unambiguous fashion that endogenous SRC-1, NCoA-2, and p/CIP are recruited to transcriptionally active CYP1A1 chromatin in a TCDD-dependent fashion. Combined, these experiments are the first demonstration that endogenous P160 coactivator protein is recruited for a functional role during activated transcription by the AHRC. Further studies on the kinetics of these phenomena would potentially help elucidate the mechanisms by which these activators and coactivators direct tissue- and target-specific gene activation. Interestingly, coinjection of CBP expression vector with p/CIP expression vector was not required to rescue reporter gene activity after injection with anti-p/CIP IgG (Fig. (Fig.5).5). This is in direct contrast with the results obtained with a retinoic acid receptor-responsive reporter (51), implying that the ARHC recruits a different p/CIP-containing complex or that it recruits the p/CIP-CBP complex for a different function. Not surprisingly, this is consistent with the notion that ARNT is responsible for recruitment of CBP (28) and does not appear to interact with p/CIP, whereas AHR can.
Studies with enhanced green fluorescent protein and rhodamine immunofluorescence indicate that ARNT and SRC-1 colocalize in the nucleus. The punctate redistribution of mARNT-EGFP within the nucleus after overexpression of SRC-1 indicates a direct interaction between the two proteins in vivo. This evidence is bolstered by the colocalization of SRC-1 in the same bodies (Fig. (Fig.4B).4B). Furthermore, these foci do not appear to be nucleoli, but may be promyelocytic leukemia protein (PML) bodies (31, 37). This punctate distribution is very similar to the observations made with GFP-HIF-1α after overexpression of SRC-1 and TIF2 (6) and GFP-AHR after overexpression of RIP140 (30). Furthermore, Voegel and colleagues have reported that TIF2 (NCoA-2) is present in similar dot-like structures within the nucleus (52).
It has been suggested that these TIF2-containing dot-like structures are PML bodies (37). These bodies have also been shown to contain p300/CBP, which is known to interact with ARNT (32) and retinoblastoma protein, which is known to interact with AHR (1). Furthermore, these bodies have been associated with RNA polymerase II activity, the production of nascent RNA, and the modulation of multiple hormone signaling pathways (12, 32). Therefore, it appears that SRC-1 directs ARNT to multimeric transcriptional complexes, and its ability to modulate AHRC-dependent gene transcription may not therefore be dependent on its intrinsic histone acetyltransferase activity or its transactivation function. This may signify a novel mechanism of action for SRC-1 with regard to gene regulation.
We undertook studies to determine the domains within SRC-1 and within AHR and ARNT required for their mutual interactions. Studies in yeast cells and in in vitro GST pulldown assays indicate that amino acids 763 to 1033 define the ARNT interaction domain on SRC-1, a region encompassing the CBP interaction site (see Fig. Fig.7).7). This domain contains two signature LXXLL motifs (where L is leucine and X is any amino acid) that have been shown to be critical for interaction with CBP, and other similar motifs within SRC-1 are responsible for interaction with nuclear hormone receptors (11, 38). However, deletion of amino acids 763 to 895 of the mSRC-1 protein strongly diminished its ability to interact with the mARNTΔQ bait in the yeast two-hybrid system. The amino acid sequences deleted do not contain LXXLL motifs, suggesting that these LXXLL motifs may not be critical for mSRC-1/mARNT interaction.
Surprisingly, two-hybrid and GST pulldown assays demonstrated that amino acids 896 to 1200 of SRC-1 define the AHR interaction domain (Fig. (Fig.8).8). No other flanking fragment of SRC-1 that does not overlap this region, in particular SRC-1763-1100, was capable of interacting with AHR. Thus, the interaction domain on SRC-1 for AHR is distinct from that for ARNT. This raises the possibility that one molecule of SRC-1 is capable of interacting with each subunit of the AHRC heterodimer. However, we have no direct data that would give insight into the stoichiometry of this interaction. Interestingly, a previous study has demonstrated that SRC-1 interacts with AHR through AHR's TAD, in particular the Q-rich region (30). Determination of the exact amino acid residues in each protein responsible for this interaction will provide a powerful means with which to study cross talk between the ARNT- and AHR-dependent pathways and other signal transduction pathways.
Studies to determine the SRC-1 interaction domain within mARNT revealed an absolute requirement for helix 2. This would appear to be a rare case of a transcription factor that can recruit a coactivator with motifs outside of its TAD and may signify a novel mode of action for SRC-1. Our example is not without precedent. The recently described LXXLL coactivator CIA modulates endoplasmic reticulum-dependent signaling in an AF-2 independent fashion (46), and more recently, Elferink and colleagues have demonstrated that enhancement of AHRC-dependent gene transcription is modulated via a direct interaction between retinoblastoma protein and the PAS B domain of AHR (14, 19). Furthermore, like the interaction between HIF-1α and SRC-1, ours is another example of two bHLH-PAS proteins that do not require both bHLH-PAS domains to interact (6).
The interactions between ARNT and SRC-1 and NCoA-2 were not negated by ARNT's dimerization with ligand-bound AHR (Fig. (Fig.2),2), consistent with the notion that SRC-1 and NCoA-2 play a role in gene regulation by the AHRC. The fact that ARNT and SRC-1 interact in a nonclassical fashion for bHLH-PAS proteins may allow for AHR/ARNT dimerization when ARNT is associated with SRC-1, despite the fact that ARNT's helix 2 is required for its interaction with both AHR and SRC-1. Much of our work attempted to characterize the direct interaction between SRC-1 and ARNT. We do not rule out that SRC-1 may modulate ARNT's action in an indirect fashion. It has been established that SRC-1 can form complexes with CBP (26), and an interaction between CBP and ARNT's TAD has been reported (28). Therefore, it is possible that a myriad number of proteins may affect ARNT-dependent transcriptional processes through indirect mechanisms of interaction.
In conclusion, we have demonstrated that AHR and ARNT can interact with the NCoA/SRC-1/p160 transcriptional coactivator proteins. Furthermore, the coactivator proteins enhance TCDD-dependent transcription. We have identified domains within SRC-1 and ARNT and SRC-1 and AHR that are critical for their mutual interactions. ARNT is the dimerization partner of numerous bHLH-PAS proteins, including HIF-1α, EPAS1, SIM1, and SIM2. Our observations regarding the interactions between the bHLH-PAS coactivators and ARNT may apply when ARNT interacts with these other proteins as well as AHR. The continuation of these studies should culminate in a more realistic model of gene activation in response to chemical carcinogens and provide insight into several phenomena, such as cross talk between signal transduction pathways and the pleiotropic effects of AHRC activation.
Acknowledgments
This work was supported by Public Health Service grant CA-28868 from the National Cancer Institute. T.V.B. and S.W. were supported in part by training fellowships from the University of California Toxic Substances Research and Teaching Program's Lead Campus Program.
We gratefully acknowledge the help of Irine Prastio and Rose Estrada in the preparation of the manuscript and Hongdiem Nguyen for excellent technical assistance.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.22.12.4319-4333.2002
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc133867?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.22.12.4319-4333.2002
Article citations
Exploring aryl hydrocarbon receptor expression and distribution in the tumor microenvironment, with a focus on immune cells, in various solid cancer types.
Front Immunol, 15:1330228, 12 Apr 2024
Cited by: 0 articles | PMID: 38680496 | PMCID: PMC11045933
p160 nuclear receptor coactivator family members and their role in rare fusion‑driven neoplasms (Review).
Oncol Lett, 27(5):210, 14 Mar 2024
Cited by: 0 articles | PMID: 38572059 | PMCID: PMC10988192
Review Free full text in Europe PMC
Dimerization Rules of Mammalian PAS Proteins.
J Mol Biol, 436(3):168406, 16 Dec 2023
Cited by: 0 articles | PMID: 38109992 | PMCID: PMC10922841
Review Free full text in Europe PMC
Implications of xenobiotic-response element(s) and aryl hydrocarbon receptor in health and diseases.
Hum Cell, 36(5):1638-1655, 17 Jun 2023
Cited by: 3 articles | PMID: 37329424
Review
When AHR signaling pathways meet viral infections.
Cell Commun Signal, 21(1):42, 24 Feb 2023
Cited by: 3 articles | PMID: 36829212 | PMCID: PMC9951170
Review Free full text in Europe PMC
Go to all (141) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Nuclear receptor coactivator SRC-1 interacts with the Q-rich subdomain of the AhR and modulates its transactivation potential.
Gene Expr, 8(5-6):273-286, 01 Jan 1999
Cited by: 69 articles | PMID: 10947077 | PMCID: PMC6157383
Hypoxia perturbs aryl hydrocarbon receptor signaling and CYP1A1 expression induced by PCB 126 in human skin and liver-derived cell lines.
Toxicol Appl Pharmacol, 274(3):408-416, 16 Dec 2013
Cited by: 45 articles | PMID: 24355420 | PMCID: PMC3919493
Aryl hydrocarbon receptor expression and activity in cerebellar granule neuroblasts: implications for development and dioxin neurotoxicity.
Toxicol Sci, 83(2):340-348, 10 Nov 2004
Cited by: 70 articles | PMID: 15537747
Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor.
Arch Biochem Biophys, 433(2):379-386, 01 Jan 2005
Cited by: 173 articles | PMID: 15581594
Review
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: R01 CA028868
PHS HHS (1)
Grant ID: CS-28868





