Abstract
Free full text

Adenovirus Virus-Associated RNA Is Processed to Functional Interfering RNAs Involved in Virus Production
Abstract
Posttranscriptional gene silencing allows sequence-specific control of gene expression. Specificity is guaranteed by small antisense RNAs such as microRNAs (miRNAs) or small interfering RNAs (siRNAs). Functional miRNAs derive from longer double-stranded RNA (dsRNA) molecules that are cleaved to pre-miRNAs in the nucleus and are transported by exportin 5 (Exp 5) to the cytoplasm. Adenovirus-infected cells express virus-associated (VA) RNAs, which are dsRNA molecules similar in structure to pre-miRNAs. VA RNAs are also transported by Exp 5 to the cytoplasm, where they accumulate. Here we show that small RNAs derived from VA RNAs (svaRNAs), similar to miRNAs, can be found in adenovirus-infected cells. VA RNA processing to svaRNAs requires neither viral replication nor viral protein expression, as evidenced by the fact that svaRNA accumulation can be detected in cells transfected with VA sequences. svaRNAs are efficiently bound by Argonaute 2, the endonuclease of the RNA-induced silencing complex, and behave as functional siRNAs, in that they inhibit the expression of reporter genes with complementary sequences. Blocking svaRNA-mediated inhibition affects efficient adenovirus production, indicating that svaRNAs are required for virus viability. Thus, svaRNA-mediated silencing could represent a novel mechanism used by adenoviruses to control cellular or viral gene expression.
Virus-associated (VA) RNAs are noncoding polymerase III-transcribed ~160-nucleotide RNAs that accumulate in the cytoplasm of adenovirus-infected cells (32). Adenovirus serotypes 2 and 5, like most human adenovirus serotypes, express two VA RNAs, VAI and VAII, whereas others carry a single VA RNA more related to VAI (32). While VAI is more abundant and has been better studied, little is known about VAII, which might carry out nonessential functions (27, 48) and compensate in part for the lack of VAI in VAI-mutated adenovirus (3, 4, 48). The main function of VAI is the blockade of RNA-dependent protein kinase (PKR), an interferon-inducible serine-threonine protein kinase activated in infected cells as part of the antiviral response. Adenovirus infection, as is the case with other viruses, could lead to the production of double-stranded RNA (dsRNA) through symmetrical transcription of the viral genome. dsRNA activates PKR, which phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF-2α) (21). Phosphorylation converts eIF-2α-GDP from a substrate to an inhibitor of its exchange factor eIF2B, so the complex eIF2 · GTP-Met · tRNA cannot be formed and translation initiation fails, leading to the shutoff of protein synthesis (9). However, adenovirus-produced VA RNAs bind to and block PKR activation and therefore protein synthesis inhibition (40). Also, VAI stabilizes ribosome-associated viral mRNAs, which could lead to enhanced levels of protein synthesis (39).
VA RNAs are single-stranded RNA molecules that fold into dsRNA (32). Although the nucleotide sequences of VA RNAs are poorly conserved between adenovirus serotypes, their secondary structure has been well maintained and can generally be divided into a terminal stem, a panhandle apical stem, and a more structured central domain (Fig. (Fig.1)1) (15, 32, 36). Mutagenic studies indicate that the apical stem-loop is required for binding to PKR and that the central domain is involved in the inhibition of PKR activation (35-37). The terminal stem is essential for binding to Exp 5, which mediates VA transport to the cytoplasm (16, 17). Although Exp5 binds dsRNA in a sequence-independent manner (5), the terminal stem is partially conserved across adenovirus serotypes, as it includes sequences essential for polymerase III transcription (32).
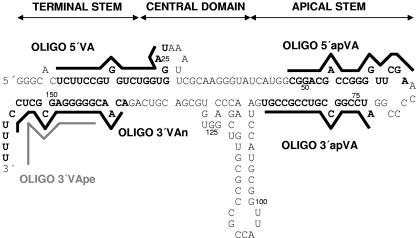
Predicted structure of VAI RNA from adenovirus type 2. The sequences that hybridize to oligonucleotides 5′VA, 3′VAn, 3′VApe, 5′apVA, and 3′apVA are indicated. The structure can be divided into a terminal stem, a central domain, and an apical stem. The sequence of VAI RNA from adenovirus type 5 is identical except for two nucleotides in the apical stem (A66G and G72U).
Minihelix RNAs and pre-microRNAs (pre-miRNAs) are also dsRNA molecules transported by Exp 5 to the cytoplasm before being further processed by Dicer to small interfering RNAs (siRNAs) or miRNAs, respectively (16, 18, 30, 38, 49). siRNAs and miRNAs are 21- to 25-nucleotide sequences that bind the RNA-induced silencing complex (RISC) to guide RNA silencing. RNA silencing allows the control of cellular gene expression and works as an alternative immune mechanism against viral infections in plant and insect cells (24). Recently, increasing evidence has supported the notion that RNA silencing plays an antiviral role in vertebrates also. Thus, expression of the retrovirus primate foamy virus type 1 (PFV-1) is downregulated by the cellular miRNA miR32 (23). Also, human immunodeficiency virus type 1 (HIV-1) and simian virus 40 (SV40) encode viral siRNA or miRNA precursors in their genomes (2, 46). Concomitantly, PFV-1 and HIV-1 evade cell control, expressing Tas and Tat proteins, respectively, that inhibit RNA silencing (2, 23). Similarly, other virus-encoded proteins, such as influenza virus NS1 protein or vaccinia virus E3L, affect the functionality of the miRNA pathway in heterologous systems (26). Notably, VAI RNAs can bind Dicer in vitro and have been described as competitors in the processing of both pre-miRNAs and short hairpin RNAs (29). Even if all these results indicate that some vertebrate viruses may need to block cellular silencing to guarantee their viability, viruses could also use the cellular silencing machinery to control gene expression. Thus, viral miRNAs that could regulate expression of both host and viral genes have been described in several viruses (6, 41, 42, 46). In the case of SV40, it has been shown that a viral miRNA could be involved in reducing susceptibility to cytotoxic T cells (46).
In this report we show that VAI RNAs are processed to small RNAs (svaRNAs) in adenovirus-infected cells and in VA RNA-expressing cells. These svaRNAs interact with Argonaute 2, the functional endonuclease from RISC, and inhibit the expression of reporter genes with complementary sequences. This suggests that they could behave as functional siRNAs or miRNAs. Finally, blockade of VAI-derived small RNAs decreases adenovirus production, indicating that svaRNAs could control the expression of cellular or viral genes required for adenovirus viability.
MATERIALS AND METHODS
Cell lines and viruses.
Human 293 and HeLa cells and mouse fibroblast 3T3 cells were obtained from the ATCC. All cell lines were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 2 mM l-glutamine. Cell lines were cultured at 37°C under a 5% CO2 atmosphere. All cell culture reagents were obtained from Gibco-BRL/Life Technologies. Plasmids were transfected using calcium phosphate as described elsewhere (14). For stable transfections, clones were selected in 1 mg/ml of G418 (GIBCO) 3 days after transfection. Transfection with 2′-O-methyl (2ome) oligonucleotides (Sigma) was performed using Lipofectamine 2000 (Invitrogen) following the recommendations of the supplier. In brief, 100 pmol of oligonucleotide was mixed with 0.4 μg of pGL3-Promoter, which expresses luciferase and served as a transfection control, 0.2 μg of the Renilla constructs (see below), and 1.2 μg of plasmid pAdvantage (pAdv) or a control plasmid. The DNA mix was incubated with 4 μl of Lipofectamine 2000 in 1 ml of Opti-MEM (Gibco) and added to 3.5 × 105 cells. Cells were analyzed 2 days posttransfection. 2′-O-Methyl oligonucleotides used were 2omeMut (CGACGUCAGACAACGGGGGAGCGCUCCUUUU), 2omeAS3′ (AAAAGGAGCGCUCCCCCGUUGUCUGACGUCG), 2omeAS3′Mut (AAAAAGAGCGAUCCCACGUUGUCUGACGUCG), 2omeAS5′ (AAUUUAUCCACCAGACCACGGAAGAGUGCCC), and 2omeAS5′Mut (AAUUUAUCCACAAGACCACAGAAGAGUACCC).
Adenoviruses were amplified in 293 cells, purified by double cesium chloride gradient ultracentrifugation, and desalted in a G50 Sephadex column. The adenovirus titer was calculated with an AdenoX rapid titer kit (BD Biosciences) or by limiting dilution. 293 cells were infected in DMEM-2% fetal bovine serum with wild-type adenovirus type 5 (AdWT) or a mutant virus that does not express VA RNAs (AdΔVA) (7, 48) at a multiplicity of infection (MOI) of 3. Infection of 3T3 cells was carried out under the same conditions but at an MOI of 100. Similarly, stable HeLa cell lines were infected at an MOI of 30. Transfected HeLa cells were infected, at an MOI of 30, 1 day after transfection and were analyzed for luciferase expression 2 days postinfection. 293 cells transfected with 2′-O-methyl oligonucleotides were infected 1 day posttransfection, and the virus titer was evaluated from the cell supernatant or from both the cell supernatant and the pellet at 3 days postinfection.
Plasmids.
pAdvantage (Promega) contains VAI and VAII genes from adenovirus type 2 under the control of their own promoters. pFAgo2 expresses Argonaute 2 fused to Flag and hemagglutinin (HA) tags and was kindly donated by T. Tusch (34). pGL3-Promoter (Promega) expressing luciferase was used as a transfection control. pRL-SV40 (Promega) was used to construct the plasmids that express VAI RNA sequences in the 3′ untranslated region (3′ UTR) of Renilla mRNA. Appropriate oligonucleotide sequences were cloned into the XbaI site (in lowercase letters) of pRL-SV40 to obtain pRLVAint (tctagaTCGAACCCCGGTCGTCCGCCATGATctaga), pRL5VA (tctagACCCTTGCGAATTTATCCACCAGACCACGGAAGAGTGCCCGCTctaga), and pRL3′VA (tctagAGGAGCGCTCCCCCGTTGTCTGACGTCGCtctaga), which bind their complementary sequences at the central part, 5′ end, and 3′ end of VAI RNA, respectively. Sequences were cloned in the sense or antisense orientation as indicated. In pRL5VAx2 and pRL3VAx2, two identical sequences were cloned in tandem at the XbaI site. Positive clones were verified by DNA sequencing (ABI Prism 310 genetic analyzer; Perkin-Elmer).
Protein analysis.
Renilla and firefly luciferase activity was quantified using the Dual Luciferase System (Promega) as previously described (14) in a Berthold Luminometer (Lumat LB 9507). Western blots were hybridized to the anti-Flag (αFlag) antibody M2 (Sigma), diluted 1:1,000, and were visualized by using an anti-mouse antibody coupled to peroxidase (Sigma), diluted 1:1,000, and enhanced chemiluminescence (Perkin-Elmer). Immunoprecipitations were done by mixing extracts obtained from 106 cells with 10 μl of agarose-coupled αFlag antibody (Sigma) or a control antibody. After binding for 2 h at 4°C, beads were washed three times in 1 ml of immunoprecipitation buffer (13), and depleted extracts and beads were evaluated by Western blotting or were used to isolate RNA. Adequate amounts were loaded on the gel so comparisons could be made directly.
Preparation and analysis of RNA.
Total RNA was purified using guanidium thiocyanate as described elsewhere (8). Nuclear and cytoplasmic RNA was purified as previously described (12). To isolate small RNA, 30 μg of total RNA was separated in a 15% polyacrylamide gel, and the RNA that ran between 20 and 30 bp was isolated. RNA from polyacrylamide, input extracts, or fractions bound or not bound to antibody beads, was isolated as previously described (13).
For Northern blotting, 20 μg of total RNA was mixed with 1 volume of loading sample buffer (95% formamide, 1 mM EDTA, 0.025% bromophenol blue and xylene cyanol dyes) and boiled at 95°C for 3 min. Samples were loaded onto a 15% polyacrylamide gel, and RNAs were separated by electrophoresis. Labeled DNA molecular weight markers that had been compared previously to the sizes of small RNAs such as let-7, miR16, and miR21 were also loaded. The gel was transferred to a Hybond + membrane (Amersham Pharmacia Biotech) in 25 mM NaxHPO4 (pH 6.5) and hybridized overnight to labeled oligonucleotides in 5% sodium dodecyl sulfate (SDS)-200 mM NaxHPO4 (pH 6.5). The membrane was then washed three times for 30 min with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS and exposed to Biomax film (Kodak) or to a Cyclone screen (Perkin-Elmer). Oligonucleotides were labeled with T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP (13). Oligonucleotide U6 (5′ TGCTAATCTTCTCTGTATCGT 3′) was used as a loading control. Oligonucleotides 5′VA (5′ ATCCACCAGACCACGGAAGA 3′) and 3′VAn (5′ AGGAGCGCTCCCCCGTTGT 3′) bind to the 5′ and 3′ ends of VAI RNAs, respectively. Oligonucleotides 5′apVA (5′ TCGAACCCCGGTCGTCCG 3′) and 3′apVA (5′ ACGGCGGACGGCCGGAT 3′) bind to sequences in the apical stem of VAI RNA, as shown in Fig. Fig.11.
Primer extensions were done as described previously (13). Oligonucleotides used were 5′VA, 3′VApe (AAAAGGAGCGCTCCCCC), let-7 (AAAACTATACAACCTACTAC), and U6. Six picomoles of γ-ATP-labeled oligonucleotides was incubated with 3 μg of total RNA or with the small or immunoprecipitated RNAs described above. Only 0.25 μg of RNA was used in the primer extensions with oligonucleotide U6. Samples were loaded onto a 14% polyacrylamide gel and separated by electrophoresis. Gels were dried and exposed to a screen that was developed in a Cyclone phosphorimager (Perkin Elmer).
RESULTS
The VAI RNA terminal stem is processed into svaRNAs.
To test whether VA RNAs could be processed to small RNAs, we used Northern blotting to analyze RNAs isolated from 293 cells that were either mock infected or infected with AdWT or AdΔVA (7, 48). The Northern blot was hybridized to oligonucleotides 5′VA, 3′VAn, 5′apVA, and 3′apVA, which recognize the longer dsRNA regions of VAI (Fig. (Fig.1)1) or to oligonucleotides that bind to similar sequences of VAII RNAs. We detected two small RNAs from the terminal stem of VAI RNA that were named svaRNAs (Fig. (Fig.2A).2A). As expected, svaRNAs were detected only in AdWT-infected cells (Fig. (Fig.2A,2A, lane 3), not in uninfected cells or in cells infected with the VA-deleted adenovirus (Fig. (Fig.2A,2A, lanes 1 and 2). The size of the signal obtained with oligonucleotide 3′VA was 23 nucleotides (Fig. (Fig.2A,2A, lane 3, upper panel). Oligonucleotide 5′VA detected a weaker signal of approximately 29 nucleotides, which is higher than the regular 21 to 25 nucleotides observed for most miRNAs (Fig. (Fig.2A,2A, lane 3, middle panel). However, we could not detect small RNAs that correspond to the apical stem of VAI RNAs or to any VAII sequences (data not shown). Since VAII RNAs are expressed 10- to 100-fold less than VAI RNAs (44), a putative signal could exist that is below the detection limit of the technique.
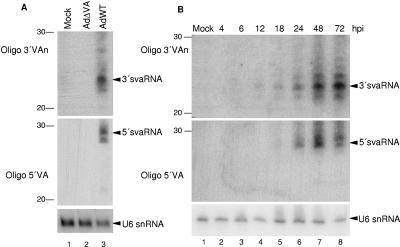
svaRNAs are found in adenovirus-infected 293 cells.(A) Northern blotting was carried out with RNA isolated from 293 cells that were either mock infected or infected with AdWT or AdΔVA. (B) 293 cells were mock infected or infected with wild-type adenovirus, and Northern blotting was done with RNAs isolated at different hours postinfection (hpi) as indicated at the top. The Northern blots were hybridized to oligonucleotide 3′VAn (upper panels), oligonucleotide 5′VA (middle panels), or a U6 snRNA oligonucleotide as a loading control (lower panels). The number of nucleotides is indicated on the left. The positions of 3′svaRNA, 5′svaRNA, and U6 snRNA are indicated.
VA RNAs are first detected in the cytoplasm of infected cells 18 h after infection (20). A Northern blot of RNA isolated at different times postinfection showed that svaRNAs were also clearly detected at 18 h postinfection (Fig. (Fig.2B,2B, lane 5). svaRNA accumulation then increased as infection proceeded, following a pattern similar to that of cytoplasmic accumulation of VA RNAs (Fig. (Fig.2B,2B, lanes 6 to 8). Thus, the highest accumulation is observed at 72 h postinfection, which corresponds to the end of the adenovirus cycle in 293 cells.
Neither viral replication nor viral protein expression is required for VA processing.
Even if pre-miRNAs are similar in structure to VA RNAs, the latter are longer and include a stem-loop-rich central domain. Thus, we hypothesized that a productive adenovirus infectious cycle or viral proteins could be required for VA RNA processing into svaRNAs. Because mouse cells are nonpermissive for adenovirus replication, we isolated RNA from 3T3 mouse fibroblasts infected with adenovirus at different times. Northern blot analysis of the RNAs indicated that even in the absence of viral replication, svaRNAs accumulated after 18 h of infection (Fig. (Fig.3A,3A, lanes 5 to 8). In agreement with the results obtained with 293 cells, we detected small RNAs only with sequences corresponding to the terminal stem of VAI RNA.
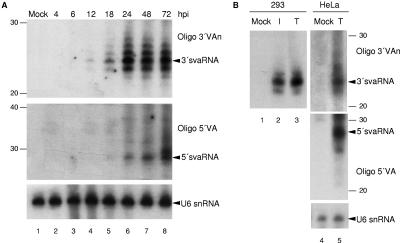
svaRNAs are produced in the absence of viral proteins or adenovirus replication. (A) Nonpermissive 3T3 cells were infected as for Fig. Fig.2B,2B, and Northern blotting was carried out accordingly. (B) 293 or HeLa cells were either mock transfected, transfected with pAdvantage (T), or infected (only 293 cells) with wild-type adenovirus (I). RNA was isolated and analyzed by Northern blotting by hybridization to oligonucleotide 3′VAn, 5′VA, or U6 snRNA as indicated. The number of nucleotides is indicated on the sides. The positions of 3′svaRNA, 5′svaRNA, and U6 snRNA are indicated.
Several adenovirus proteins have been implicated in the transcription of VA RNAs. E1B is a transcription activator, while E1A acts as an activator in infected cells and as a repressor in transfected cells (45). However, svaRNAs were detected by Northern blotting in E1-expressing 293 cells transfected with pAdv, which contains VA genes from adenovirus type 2 (Fig. (Fig.3B,3B, lane 3). Similarly, HeLa or 3T3 cells transfected with pAdv accumulated svaRNAs (Fig. (Fig.3B,3B, lane 5) (data not shown), indicating that adenovirus proteins are not required for VAI small RNA processing.
Characterization of svaRNAs.
Intact VAI RNAs can be detected in transfected or infected cells even at late times of infection. Thus, not all VAI RNA is cleaved into svaRNA. To calculate the efficiency of VAI RNA processing, RNA isolated from 293 cells infected with adenovirus was analyzed by Northern blotting with oligonucleotide 3′VAn. The result is shown in Fig. Fig.4A,4A, where the svaRNA part (lower panel) was exposed 10 times longer than the VAI RNA part (upper panel). The amounts of VAI RNA and 3′svaRNA were quantified, indicating that only 2 to 5% of the total VAI RNA was processed into svaRNA. Similar results were observed for HeLa cells transfected with pAdv or for infected 3T3 cells (data not shown).
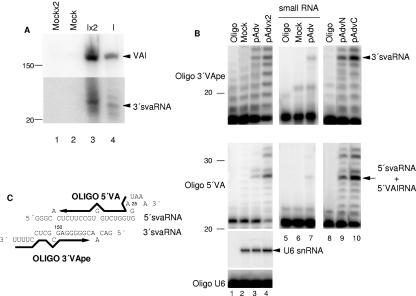
Characterization of svaRNAs. (A) Efficiency of svaRNA processing. RNA isolated from mock-infected or infected (I) 293 cells was analyzed by Northern blotting by hybridization to oligonucleotide 3′VAn. Single and double (×2) amounts of RNA were used to ensure proper quantitation. Positions of intact 160-nucleotide VAI RNA and 3′svaRNA are indicated. Note that the lower panel shows a 10-times-longer exposure. (B) Sequence and localization of svaRNAs. RNA was isolated from total, nuclear, or cytoplasmic fractions of HeLa cells that were either mock transfected or transfected with pAdv. Total RNA was separated in a polyacrylamide gel, and 20- to 30-nucleotide small RNAs were purified. Buffer (Oligo) (lanes 1, 5, and 8), total RNA (lanes 2 to 4), small RNAs (lanes 6 and 7), nuclear RNA (lane 9), or cytoplasmic RNA (lane 10) was analyzed by primer extension with oligonucleotide 3′VApe (upper panel) or 5′VA (middle panel) or with a U6 snRNA oligonucleotide as a loading control (lower panel). A double amount of RNA was used where indicated (×2). The primer extension corresponding to 3′svaRNA, 5′svaRNA, and U6 snRNA is indicated. The number of nucleotides is given on the left. (C) Schematic representation of svaRNAs. Sequences of 5′ and 3′ svaRNAs are shown together with the oligonucleotides used for primer extension.
Given the low processing efficiency, we decided to analyze if the svaRNAs were functional, so we first characterized them further. Thus, their 5′ ends were analyzed by primer extension. Oligonucleotide 3′VApe, a short oligonucleotide that interacts with the last 17 nucleotides of VAI RNA (Fig. (Fig.4C),4C), could be preferentially extended 6 nucleotides when hybridized to RNA isolated from HeLa cells transfected with pAdv (Fig. (Fig.4B,4B, lanes 3 and 4, upper panel). Since the total length of the 3′svaRNA is 23 nucleotides (Fig. (Fig.22 and and3),3), we concluded that the 3′svaRNA sequence comprises the last 23 nucleotides of VAI RNA (Fig. (Fig.4C).4C). Similarly, oligonucleotide 5′VA (Fig. (Fig.4C)4C) could be extended 6 nucleotides to the 5′ end of VAI RNA (Fig. (Fig.4B,4B, lanes 3 and 4, middle panel). A fainter extension of 9 nucleotides was also detected that could correspond to VAI RNAs initiated in a less efficient transcription start site (31). However, these 5′VA bands could represent mostly intact VAI RNAs. Therefore, small RNAs were isolated from the 20- to 30-nucleotide band excised from a polyacrylamide gel where control RNA or RNA from pAdv-transfected cells had been separated. When the small RNAs were used in the primer extension reactions, both oligonucleotides 3′VA and 5′VA were extended 6 nucleotides (Fig. (Fig.4B,4B, lanes 6 and 7, upper and lower panels). This result confirms that 5′svaRNAs comprise the first 29 nucleotides of VAI RNA (Fig. (Fig.4C),4C), as indicated by Northern blotting (Fig. (Fig.22 and and3).3). Finally the subcellular localization of VA RNAs was addressed. RNA was isolated from nuclear and cytoplasmic fractions of HeLa cells transfected with pAdv, and primer extension was carried out. Proper fractionation was verified by hybridization to U6 snRNAs and let-7 probes (data not shown). svaRNAs accumulate preferentially in the cytoplasmic fraction, even if 20% of these RNAs can also be found in the nucleus (Fig. (Fig.4B,4B, lanes 9 and 10, upper and lower panels).
Functional miRNAs bind the RISC endonuclease Ago2 (28, 34, 43). To analyze if svaRNAs also bind Ago2, we made use of a plasmid that encodes Ago2 sequences fused to a Flag tag (pFAgo2 [34]). Expression of Flag-tagged Ago2 (FAgo2) could easily be detected by Western blotting with an anti-Flag antibody in pFAgo2-transfected HeLa cells (Fig. (Fig.5A,5A, lane 2). Also, using immunoprecipitation, tagged Ago2 protein could be found bound to the anti-Flag antibody but not bound to a control antibody (Fig. (Fig.5A,5A, lanes 3 to 6). Thus, extracts from HeLa cells cotransfected with pAdv and either a control plasmid or pFAgo2 were immunoprecipitated with an anti-Flag or a control antibody. The input, supernatant, and bound fractions of each immunoprecipitation were used to isolate RNA that was then analyzed by primer extension using oligonucleotide let-7, 3′VApe, or 5′VA. As expected, let-7 and miR16, abundant endogenous miRNAs in HeLa cells, were detected bound to the anti-Flag antibody in cells transfected with pFAgo2 (Fig. (Fig.5B,5B, lane 10, upper panel) (data not shown) but were not detected bound to a control antibody or to any antibody in cells lacking FAgo2 (Fig. (Fig.5B,5B, lanes 3, 5, and 8, upper panel) (data not shown). 3′svaRNA and 5′svaRNA were detected in the same fractions as let-7, indicating that svaRNAs bind to Ago2 (Fig. (Fig.5B,5B, lane 10, middle and lower panels). The amount of RNA extended with oligonucleotide 5′VA in the fraction bound to Ago2 was very small, although clearly detectable (Fig. (Fig.5B,5B, compare lanes 9 and 10). This result supports the possibility that only 5′svaRNA is bound to Ago2, since oligonucleotide 5′VA detects both intact VAI RNA and 5′svaRNA. The amount of 3′svaRNA bound to FAgo2 was greater than the amount of let-7 (Fig. (Fig.5B,5B, compare lanes 9 and 10 in upper and middle panels). This could be due to a relatively low efficiency of transfection. Thus, we isolated stable HeLa cell lines expressing FAgo2 as detected by Western blotting or by immunoprecipitation with an anti-Flag antibody (Fig. (Fig.5C,5C, clones 1 and 4 in lanes 3 and 6). These stable transformants showed that Ago2 binding efficiencies to let-7 and 3′svaRNA were similar (data not shown) (Fig. (Fig.5D).5D). The stable clones were also used to analyze the binding to Ago2 of svaRNAs produced in adenovirus-infected cells. The result, shown in Fig. Fig.5D,5D, is similar to that shown in Fig. Fig.5B.5B. Both 3′svaRNA and 5′svaRNA were detected in the same fractions as let-7, indicating that svaRNAs produced in adenovirus-infected cells also bind to Ago2 (Fig. (Fig.5D,5D, lanes 11 and 16). This binding is specific, as the terminal ends of other abundant and structured RNAs, such as U1 snRNA or 5S rRNA, were not found in the Ago2 fractions (data not shown).

svaRNAs coimmunoprecipitate with Ago2. (A) Flag-tagged Ago2 protein can be efficiently immunoprecipitated with an anti-Flag antibody. Extracts from cells transfected with pAdv or cotransfected with pAdv and a plasmid that encodes a Flag-tagged Ago2 protein (pAdvFAgo2) were immunoprecipitated with a control antibody (αMock) or with an anti-Flag antibody (αFlag). Input, bound, and unbound fractions were separated by SDS-polyacrylamide gel electrophoresis, and Western blotting was carried out with anti-Flag antibody. The position of Flag-tagged Ago2 (FAgo2) is indicated. (B) Ago2 binds svaRNAs. RNA was isolated from the fractions described for panel A, and a primer extension reaction was carried out. The oligonucleotides used revealed let-7 miRNA (upper panel), 3′svaRNA (middle panel), or 5′svaRNA (lower panel). Ub, unbound fractions; B, bound fractions; I, input; O, oligonucleotide-only reaction. The primer extension corresponding to let-7, 3′svaRNA, and 5′svaRNA is indicated. (C) Characterization of HeLa clones expressing Flag-tagged Ago2. Extracts from HeLa cells or cells transfected with a plasmid expressing FAgo2 transiently (FAgo2) or stably (clones C1 to C4) were immunoprecipitated with αFlag antibody. Total extracts (upper panel) or extracts bound to αFlag (lower panel) were analyzed by Western blotting with an αFlag antibody. The position of FAgo2 is indicated. (D) svaRNAs from adenovirus-infected cells bind Ago2. Extracts from HeLa cells transfected with pAdv and pFAgo2 (pAdvFAgo2) or from adenovirus-infected HeLa clones 1 (C1 + AdWT) and 4 (C4 + AdWT) that express Ago2 were immunoprecipitated with an αFlag or a control antibody. RNA was isolated from the input, bound, and unbound fractions of the immunoprecipitates, and a primer extension reaction was carried out.
Analysis of svaRNA function in the control of gene expression.
svaRNAs could control the expression of cellular or viral genes in infected cells. To test if svaRNAs can interfere with gene expression, we constructed plasmids expressing Renilla luciferase (RL) mRNA that include different sequences from VAI RNAs. The sequences chosen hybridize to the apical stem of VAI RNAs (nucleotides 40 to 65; RLVAint) or to the 5′ (RL5VA) or 3′ (RL3VA) terminal stem. The target sequences were included once or twice (RLxVAx2) in the 3′ UTR of RL mRNA and in the sense (RLs) or antisense (RLas) orientation (Fig. (Fig.6A).6A). Note that the sense sequence of one end of VAI RNA contains the antisense sequence of the other end with two mutations that disrupt base pairing (RLasMut) (11) (Fig. (Fig.1).1). HeLa cells were cotransfected with each Renilla construct together with pAdv or a control plasmid. A firefly luciferase expression plasmid was also included in all cases as a transfection and expression control. Luciferase activities were measured 2 days posttransfection, and RL values were corrected by firefly luciferase activity, even if the latter showed similar levels in all samples, indicating that transfection efficiencies were homogeneous. The percentage of RL expression relative to the expression of RL without VA sequences was plotted (Fig. (Fig.6B).6B). Cotransfection of the RL constructs with the control plasmid yielded similar luciferase activities in all cases, indicating that the VA sequences introduced did not alter RL mRNA expression, stability, or translation (Fig. (Fig.6B).6B). pAdv is commercialized to increase expression from cotransfected plasmids, as VAI expression should block PKR activation and therefore increase gene expression (22, 47). pAdv transfections need to be optimized to obtain a VA-mediated increase of expression. Using our regular cotransfection conditions, expression of Renilla was not increased by the presence of VA RNAs except in the case of RLas5VAx2Mut, which was expressed at 1.8-fold-higher levels (Fig. (Fig.6B).6B). However, expression of Renilla with VA terminal antisense sequences decreased in the presence of pAdv, suggesting that small VA RNAs could be functional interfering RNAs. The highest decrease was observed in Renilla genes containing 1 or 2 copies of the VAI 3′-end sequence (Fig. (Fig.6B).6B). This inhibition is specific, as we did not observe decreased expression of RLas3VAx2Mut or RLas5VAx2Mut, whose binding to 3′svaRNA or 5′svaRNA, respectively, is abrogated by two disrupting mutations (Fig. (Fig.1).1). Similar results have been observed upon transfection of 293 cells or 3T3 cells (data not shown) or upon infection with wild-type adenovirus (Fig. (Fig.6C).6C). Again, RL expression from RL or RLVAint constructs was very similar in wild-type adenovirus-infected cells. However, expression from Renilla genes containing 1 or 2 copies of the VAI 3′-end sequence was reduced to approximately 20% (Fig. (Fig.6C).6C). No differences were observed when cells were infected with an adenovirus that does not express VA sequences (AdΔVA; Fig. Fig.6C6C).
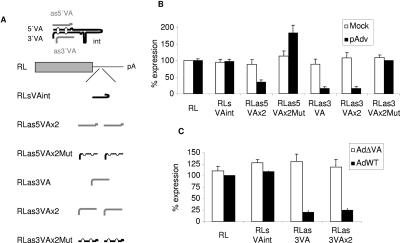
Expression of RL mRNAs that bind to VAI RNA terminal sequences is inhibited in cells expressing VAI RNA. (A) Schematic representation of the reporter genes used for this study. VAI RNA (diagramed at the top) sequences were expressed in the 3′ UTR of RL. Sequences chosen were VA internal sequences (VAint) and either the sense (s) or antisense (as) 5′ (5VA) or 3′ (3VA) end. Note that the sense sequence of one end of VAI RNA contains the antisense sequence of the other end with two mutations that disrupt base pairing (Mut). Sequences were inserted once or twice (x2). (B) HeLa cells were cotransfected with the RL constructs shown and a control plasmid (Mock) or pAdv, and luciferase activity was measured 2 days posttransfection. (C) HeLa cells transfected with the RL constructs shown in panel A were infected with AdWT or AdΔVA. Luciferase activity was measured 3 days posttransfection. Percentages of RL expression relative to the expression of the unmodified RL construct are indicated. Error bars, standard deviations. Results shown are averages from four different experiments.
Blocking of svaRNAs affects adenovirus production.
If svaRNAs are essential for adenovirus viability, mutations in the terminal stem of VAI should result in defects in viral amplification. It has been demonstrated that VAI RNA functions primarily to block PKR function, as eIF2α mutants that cannot be phosphorylated complement the growth defect of an adenovirus deleted in the VAI gene (10). However, substitutions of the sequences at the 3′ termini of VAI were introduced into adenoviruses that express VAI molecules that are able to be properly folded and transported to the cytoplasm of infected cells. Even though these VAI 3′-end mutants should be functional in blocking PKR, the mutant viruses showed a 10-fold decrease in viral replication (3). This indicates that the 3′ end of VAI could play a different role in adenovirus viability. Unfortunately, mutations in the 5′ end of VAI affect VAI RNA expression, since these sequences overlap with the polymerase III promoter sequences (32).
An alternative to evaluating the role of svaRNAs in adenovirus infection would be to block svaRNA interactions with hybridizing 2ome modified oligonucleotides (19, 33). 2ome oligonucleotides were designed with antisense sequences to 3′svaRNA (2omeAS3′) or 5′svaRNA (2omeAS5′). Similar sequences with three point mutations that abrogate binding to 3′svaRNA (2omeAS3′Mut) or 5′svaRNA (2omeAS5′Mut) were used as negative controls. A control oligonucleotide was also designed with sense sequences of 3′svaRNA (2omeMut). To evaluate the functionality of 2ome oligonucleotides, 2omeAS3′ or 2omeMut was cotransfected with pAdv and an RL containing 3′svaRNA antisense sequences (RLas3VA). Cells transfected with the 2omeMut oligonucleotide showed RL expression similar to that of mock-transfected cells. However, cells transfected with the 2omeAS3′ oligonucleotide showed increased RL activity, suggesting that 3′svaRNA-mediated inhibition had been partially blocked (Fig. (Fig.7A7A).

Blocking of svaRNAs affects adenovirus viability. (A) svaRNA-mediated inhibition can be blocked with antisense 2′-O-methyl oligonucleotides. Cells were transfected with pAdv and RLas3VA either alone (Mock) or in combination with 2′-O-methyl oligonucleotides with sense (2omeMut) or antisense (2omeAS3′) sequences of 3′svaRNA. Luciferase expression, measured 3 days posttransfection, is plotted as the percentage of activity relative to an unmodified RL construct. (B) Cells transfected with 2′-O-methyl oligonucleotides with 3′svaRNA sense sequences (2omeMut), with 5′svaRNA (2omeAS5′) or 3′svaRNA (2omeAS3′) antisense sequences, or with mutated 5′svaRNA (2omeAS5′Mut) or 3′svaRNA (2omeAS3′Mut) antisense sequences were infected with wild-type adenovirus. Cell supernatants were collected at 3 days postinfection, and virus titers were evaluated and plotted. Error bars, standard deviations. Results shown are averages from three different infections.
Thus, cells transfected with 2ome oligonucleotides were infected with AdWT or AdΔVA, and 3 days postinfection the supernatant was collected to titer adenovirus with the AdenoX rapid titer kit (Fig. (Fig.7B).7B). The result shows that AdWT production was higher in cells with functional svaRNAs. Cells transfected with any of the 2omeMut control oligonucleotides produce more AdWT than cells transfected with the 2ome oligonucleotides with antisense sequences to 3′svaRNA or 5′svaRNA. Also, AdWT production was more affected in cells with nonfunctional 3′svaRNA than in cells with blocked 5′svaRNA (Fig. (Fig.7B).7B). However, no differences were observed in virus titer when the cells transfected with the 2ome oligonucleotides were infected with AdΔVA, which lacks VA sequences (data not shown). This shows that 2omeAS3′ and 2omeAS5′ do not affect adenovirus production in the absence of VA sequences. Finally, a similar result was obtained when virus was titered from both supernatant and cellular fractions, indicating that the difference in AdWT production is not at the level of virus cell release (data not shown).
DISCUSSION
We have shown that the terminal stem of VAI RNA is processed to 5′ and 3′ svaRNAs. Even though the processing is not very efficient, these svaRNAs interact with the silencing machinery and are capable of inhibiting gene expression. During the process of review of our work, a complementary study was published concluding that not only VAI RNA but also VAII RNA is processed by Dicer to small RNAs (1). In agreement with our results, the authors also show that these small RNAs produced in vitro can inhibit gene expression when cotransfected with a sensor reporter and that extracts from infected cells can cleave reporters with 5′ or 3′ VA sequences. These data lead to the suggestion that VA-derived small RNAs comprise the terminal stem of VA RNAs (1).
Our experiments extend further in the analysis of the svaRNAs. The combination of Northern blotting and primer extension with purified small RNAs suggests that the 3′svaRNA sequence comprises the last 23 nucleotides of VAI RNA while 5′svaRNAs consist of the first 29 nucleotides (Fig. (Fig.2,2, ,3,3, and and4B).4B). Thus, the length of 3′svaRNA is similar to that of cellular miRNAs. However, 5′svaRNA is 29 nucleotides long, too long compared with regular miRNAs. This greater length could be explained if the stem formed by nucleotides 24 to 30 of VAI RNA forces an aberrant cleavage of Dicer (Fig. (Fig.1).1). Also, we generally observe higher accumulation of 3′svaRNA than of 5′svaRNA (Fig. (Fig.2,2, ,3,3, and and4B).4B). This higher accumulation correlates with higher binding to Ago2 (Fig. 5B and D), better inhibition of target reporter gene expression (Fig. (Fig.6B),6B), and stronger involvement in adenovirus production (Fig. (Fig.7B7B).
The VAI RNA terminal stem is processed to svaRNAs that bind Ago2 (Fig. (Fig.5)5) and inhibit the expression of reporter genes with complementary sequences (Fig. 6B and C). This inhibition is partly suppressed with antisense 2ome oligonucleotides (Fig. (Fig.7A).7A). Also, blocking of svaRNAs drastically affects AdWT production (Fig. (Fig.7B),7B), and adenoviruses that express properly folded VAI RNA with 3′-end mutations show a decrease in viral replication (3). These data strongly suggest that svaRNAs use the silencing machinery to inhibit gene expression and therefore improve adenovirus viability. svaRNA-mediated inhibition of gene expression should occur early during infection, as adenovirus infection suppresses RNA interference (RNAi) at late times of infection, probably to avoid silencing of viral genes (1, 29). This blockade of RNAi could also be caused by saturation of the cellular silencing pathway with svaRNAs. Alternatively, svaRNAs could control gene expression by other means. svaRNAs could act as miRNAs that bind to the terminal stem of VAI RNA and aid localization to ribosomes, where VAI RNA is found at late times of infection (39).
Future work is required to identify svaRNA target genes. Database searches have failed to find genes with perfect homology to VAI terminal sequences, suggesting that inhibition could affect the translation of target genes and not their mRNA stability. Recently, several highly valuable bioinformatic tools, such as TargetScan, have been described that scan for putative microRNA target mRNAs (25). A TargetScan search on the Ensembl release 31.35d 3′ UTR database (based on the NCBI 35 assembly of the human genome) using 3′svaRNA as the test sequence yielded a list of 198 putative targets. Also, we modified the TargetScan program to admit the 29-nucleotide 5′svaRNAs, and the result was a list of 863 putative targets. Nine putative targets were found in both comparisons. Several cellular processes are well represented in the lists of putative targets. However, some of them are more abundant, as indicated by the presence of 21 phosphatases, 42 kinases, 11 ubiquitin-related proteins, and 35 G binding proteins or proteins related to small GTPases (such as Rho, Ras, or Ran). There are 38 factors related to transcription and 7 related to translation initiation. Also, there are cellular factors involved in VA RNA processing or function. Thus, there are three genes related to silencing and four genes related to interferon activation, including 2′-5′oligoadenylate synthase and RNase L. Unfortunately, the list of targets cannot be cured by comparison to orthologue sequences in mice or rats, because adenovirus is nonreplicative in rodents. We need to find alternative methods for target identification. This is of great importance not only for the understanding of adenovirus biology but because it may have implications for human health care. First-generation adenovirus vectors are widely proposed for gene therapy approaches. Thus, several human gene therapy trials are ongoing with these vectors. However, first-generation adenovirus vectors express normal levels of VA RNAs (data not shown), so they may induce a change in cellular gene expression whose toxicity needs to be evaluated.
Viral miRNAs have been detected in several viruses such as HIV-1, SV40, and herpesviruses including Epstein-Barr virus (EBV) (2, 6, 41, 42, 46). Even if EBV miRNAs are encoded elsewhere in the genome, EBV and other animal viruses express dsRNA structures similar to VA RNAs, which could also be processed into miRNAs. Thus, processing of VA RNA-like structures may represent a new mechanism used by several viruses to produce viral miRNAs. Such miRNAs should not be detected with miRNA prediction protocols, as they normally scan sequences of 50 to 100 nucleotides, while VAI-like RNAs are longer (41). Regarding viral miRNA functionality, it has been demonstrated only for SV40 viral miRNAs, which are involved in reduced susceptibility to cytotoxic T cells (46). This work shows that adenovirus svaRNAs could have a functional role in viral production. Therefore, RNA silencing could be used by several viruses to control viral or cellular gene expression in order to improve virus viability.
Acknowledgments
We are grateful to T. Tuschl for plasmid pFAgo2, to R. Alemany for providing AdΔVA, to R. Hernández-Alcoceba for help in adenovirus amplification, to C. Olague and E. Casales for technical assistance, and to J. Prieto, C. Smerdou, and R. Hernández-Alcoceba for critical reading of the manuscript.
This work was supported by CICYT (SAF2003-01804), FIS (01/1310 and 01/0843), Instituto Carlos III C03/02, and the Education and Health Department of the Navarra Government and through the agreement between FIMA and the “UTE project CIMA.” O.A. is an FPI fellow.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.80.3.1376-1384.2006
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1346933?pdf=render
Citations & impact
Impact metrics
Article citations
Quantitative Virus-Associated RNA Detection to Monitor Oncolytic Adenovirus Replication.
Int J Mol Sci, 25(12):6551, 14 Jun 2024
Cited by: 0 articles | PMID: 38928259
Anti-Adenoviral Effect of Human Argonaute 2 Alone and in Combination with Artificial microRNAs.
Cells, 13(13):1117, 28 Jun 2024
Cited by: 0 articles | PMID: 38994969 | PMCID: PMC11240694
Human Adenovirus Gene Expression and Replication Is Regulated through Dynamic Changes in Nucleoprotein Structure throughout Infection.
Viruses, 15(1):161, 05 Jan 2023
Cited by: 6 articles | PMID: 36680201 | PMCID: PMC9863843
Review Free full text in Europe PMC
Viral miRNA regulation of host gene expression.
Semin Cell Dev Biol, 146:2-19, 30 Nov 2022
Cited by: 10 articles | PMID: 36463091 | PMCID: PMC10101914
Review Free full text in Europe PMC
Multi-Omic Factors Associated with Frequency of Upper Respiratory Infections in Developing Infants.
Int J Mol Sci, 24(2):934, 04 Jan 2023
Cited by: 1 article | PMID: 36674462 | PMCID: PMC9860840
Go to all (96) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sequence-specific interference by small RNAs derived from adenovirus VAI RNA.
FEBS Lett, 580(6):1553-1564, 03 Feb 2006
Cited by: 63 articles | PMID: 16472808
Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities.
Plant Cell Physiol, 48(7):1050-1060, 13 Jun 2007
Cited by: 124 articles | PMID: 17567638
Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes.
J Virol, 81(19):10540-10549, 25 Jul 2007
Cited by: 70 articles | PMID: 17652395 | PMCID: PMC2045446
Post-transcriptional gene silencing by siRNAs and miRNAs.
Curr Opin Struct Biol, 15(3):331-341, 01 Jun 2005
Cited by: 324 articles | PMID: 15925505
Review





