Abstract
Free full text

Age-Dependent Resistance to Lethal Alphavirus Encephalitis in Mice: Analysis of Gene Expression in the Central Nervous System and Identification of a Novel Interferon-Inducible Protective Gene, Mouse ISG12
Abstract
Several different mammalian neurotropic viruses produce an age-dependent encephalitis characterized by more severe disease in younger hosts. To elucidate potential factors that contribute to age-dependent resistance to lethal viral encephalitis, we compared central nervous system (CNS) gene expression in neonatal and weanling mice that were either mock infected or infected intracerebrally with a recombinant strain, dsTE12Q, of the prototype alphavirus Sindbis virus. In 1-day-old mice, infection with dsTE12Q resulted in rapidly fatal disease associated with high CNS viral titers and extensive CNS apoptosis, whereas in 4-week-old mice, dsTE12Q infection resulted in asymptomatic infection with lower CNS virus titers and undetectable CNS apoptosis. GeneChip expression comparisons of mock-infected neonatal and weanling mouse brains revealed developmental regulation of the mRNA expression of numerous genes, including some apoptosis regulatory genes, such as the proapoptotic molecules caspase-3 and TRAF4, which are downregulated during development, and the neuroprotective chemokine, fractalkine, which is upregulated during postnatal development. In parallel with increased neurovirulence and increased viral replication, Sindbis virus infection in 1-day-old mice resulted in both a greater number of host inflammatory genes with altered expression and greater changes in levels of host inflammatory gene expression than infection in 4-week-old mice. Only one inflammatory response gene, an expressed sequence tag similar to human ISG12, increased by a greater magnitude in infected 4-week-old mouse brains than in infected 1-day-old mouse brains. Furthermore, we found that enforced neuronal ISG12 expression results in a significant delay in Sindbis virus-induced death in neonatal mice. Together, our data identify genes that are developmentally regulated in the CNS and genes that are differentially regulated in the brains of different aged mice in response to Sindbis virus infection.
Age-dependent resistance to neurotropic viral infections was first described more than half a century ago (33, 48, 64). Younger mice are more susceptible than adult mice to several different families of neurotropic viruses, including rhabdoviruses, reoviruses, bunyaviruses, alphaviruses, and flaviviruses (reviewed in references 9, 14, 34, 50, and 64). This phenomenon is not restricted to mice, since several human neurotropic viruses also commonly cause more severe infections in children than in adults, including the most prevalent etiology of encephalitis worldwide, Japanese B encephalitis, and important causes of arthropod-borne encephalitides in the Western hemisphere, such as Western equine encephalitis virus and La Cross virus (reviewed in references 20 and 59). Children infected with these viruses have both a higher acute mortality rate, as well as a higher rate of neurologic sequelae, including mental retardation, paralysis, and seizure disorders.
The molecular determinants of increased susceptibility of younger hosts to severe viral encephalitis remain poorly defined. In murine models, several biologic variables have been postulated to contribute to the acquisition of resistance, including maturation of the reticuloendothelial system (15), development of anatomic barriers (60), changes in receptor availability (31), potentiation of interferon (IFN) production (19), acceleration of immune responses (53, 73), and decreases in systemic stress responses (69). In addition, studies with Sindbis virus (reviewed in reference 14), Semliki Forest virus (49, 50), and reovirus (65) have also suggested that neuronal maturation is a critical factor in the development of resistance to viral infection. Neuronal maturation correlates not only with resistance to viral replication but also is accompanied by increased cellular resistance to virus-induced apoptosis (reviewed in references 8, 34, and 35).
Studies to identify the molecular mechanisms underlying the biologic variables postulated to contribute to age-dependent resistance to fatal viral encephalitis have been largely inconclusive. In general, innate antiviral immune responses such as IFN production and induction of IFN-regulated genes are more robust in younger mice that have higher levels of replication and, therefore, the characterization of such responses has not provided insight into the molecular basis for increased disease severity in younger mice (7, 62, 69). Although acquired immune responses are delayed in younger mice, mechanisms independent of acquired immunity must be important since mice devoid of both B and T cells also exhibit age-dependent resistance to fatal Sindbis virus encephalitis (74). Furthermore, despite the postulated importance of age-dependent susceptibility to apoptosis in CNS pathogenesis (reviewed in references 8, 34, and 35) and the observation that expression levels of the antiapoptotic gene, bcl-2, correlates with susceptibility to Sindbis virus-induced apoptosis in cultured neurons in vitro (36), postnatal developmental changes in the expression of antiapoptotic Bcl-2 family members have not been described in vivo (reviewed in reference 12).
To investigate the molecular basis of age-dependent resistance to fatal viral encephalitis, we used a Sindbis virus mouse model system. Sindbis virus, the prototype member of the Alphavirus genus, is a single-strand positive-sense RNA virus. In mice, Sindbis virus produces encephalitis that serves as an animal model for the human arthropod-borne encephalitides caused by Eastern, Western, and Venezuelan equine encephalitis viruses. Age-dependent susceptibility to lethal Sindbis virus infection was originally described by Johnson et al. in 1972 (24) and has been characterized extensively (reviewed in reference 14). In neonatal mice, intracerebral inoculation of wild-type (strain AR339) and most laboratory strains of Sindbis virus result in rapidly fatal disease, characterized by high levels of intracerebral and systemic production of IFN and cytokines (29, 69, 70), and extensive neuronal apoptosis (36). In weanling and adult mice, most strains are avirulent, despite a similar tropism within the central nervous system (22; reviewed in reference 13). Single amino acid changes within the viral E2 envelope glycoprotein can overcome the age-dependent resistance to fatal disease (71), and infection with neurovirulent mutant strains is associated with increased viral replication, increased neuronal apoptosis, and increased induction of host inflammatory response genes in older mice (25). Although mutant strains of Sindbis virus that are neurovirulent in older mice may cause neuronal death by mechanisms independent of apoptosis (18), strong evidence exists that neuronal apoptosis plays a critical role in the susceptibility of neonatal mice to lethal disease. Enforced neuronal expression of several different cellular and virally encoded apoptotic regulatory gene products, including Bcl-2, Bax, CrmA, survival motor neuron protein, and peripheral benzodiazepine receptor, reduces mortality in neonatal mice (25, 28, 36, 37, 46).
The age-dependent resistance of mice to lethal infection with Sindbis virus and other neurotropic viruses led us to hypothesize that gene expression changes in the host CNS during development may play a role in the acquisition of resistance to neurotropic virus infection. To investigate this hypothesis, we used DNA microarray technology to compare CNS gene expression in mock-infected and Sindbis virus-infected neonatal and weanling mice. Our results identify specific host genes that are developmentally regulated in the CNS and candidate regulators of age-dependent CNS viral pathogenesis. Furthermore, we show that increased expression of one of these genes, mouse ISG12, exerts protective effects in neonatal Sindbis virus encephalitis.
MATERIALS AND METHODS
Recombinant virus strains.
The previously described recombinant strain of double-subgenomic Sindbis virus, dsTE12Q (23), was produced from a viral cDNA clone by in vitro transcription and RNA transfection of BHK-21 cells as previously described (36) and used for most experiments. To construct a recombinant chimeric Sindbis virus expressing mouse 1SG12, a 273-bp fragment containing the open reading frame of the ISG12 gene was amplified by PCR from cDNA prepared from dsTE12Q-infected mouse brains, adding BstEII restriction sites to the upstream and downstream primer and a flag epitope sequence to the upstream primer. The mouse ISG12 gene was cloned into the BstEII sites of dsTE12Q to generate the plasmid dsTE12Q/ISG12, and the correct sequence of the insert was confirmed by sequencing. A control chimeric viral cDNA, dsTE12Q/ISG12control, was also constructed in which the start codon of the ISG12 gene was deleted. Stock viruses were produced from viral cDNA clones as previously described (36) and viral titers were measured by plaque assay titer determination on BHK-21 cells.
Animal studies.
The right cerebral hemispheres of 1-day-old and 4-week-old CD1 mice (Charles River) were inoculated with 1,000 PFU of virus diluted in 0.03 ml of Hanks balanced salt solution (HBSS). A 0.03-ml volume of HBSS balanced was used for mock infections. For mortality studies, five to eight separate litters were inoculated with virus and the mice where observed daily for 21 days to monitor survival. For RNA isolation, brains were dissected at 1, 2, and 3 days (24, 48, and 72 h) after inoculation for reverse transcription-PCR (RT-PCR) analysis and at 3 days (66 h) postinoculation for GeneChip expression analysis. The left hemisphere of the brain was rapidly frozen and stored at −80°C. The right cerebral hemisphere was used to make a freeze-thaw 10% homogenate in HBSS for subsequent plaque assay on BHK-21 cells.
Histopathology.
The right cerebral hemispheres of paraformaldehyde-fixed mouse brains were embedded in paraffin, and a series of 4-μm-thick sagittal sections were cut from medial to lateral. For each dsTE12Q-infected brain, sequential sections were examined by hematoxylin and eosin (H&E) staining to detect histopathology, TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) staining to detect apoptosis, and immunoperoxidase staining to detect Sindbis virus antigen. TUNEL staining was performed by using the ApopTag peroxidase in situ apoptosis detection kit (Intergen). Immnunostaining to detect Sindbis virus antigen was performed by using a polyclonal anti-Sindbis virus antibody (1:400 dilution) (22) and the avidin-biotin peroxidase method (Vecta-Stain ABC kit; Vector Laboratories). A similar method was used to detect mouse fractalkine with a polyclonal goat antimurine fractalkine antibody (1:125 dilution; R&D Systems, Minneapolis, Minn).
GeneChip expression analysis.
RNA was prepared for GeneChip expression analysis as specified by the manufacturer (Affymetrix) and as described previously by our laboratory (25) and others (6, 32, 77). Total RNA was isolated from the left hemispheres of brains dissected 3 days after virus infection, and RNA obtained from six mice per treatment group was pooled. Biotin-labeled cRNA was generated from pooled RNA samples as described and hybridized to Murine 11K GeneChip subarrays A and B (Affymetrix) (25).
Data analysis.
The data collected after hybridization of biotin-labeled cRNA from each treatment group was analyzed by using GeneChip Suite Software. Internal controls for hybridization intensity and cRNA synthesis were included on each chip and have been discussed elsewere (6). Samples were normalized to glyceraldehyde phosphate dehydrogenase (GAPDH), actin, and 18S RNA levels. The Absolute Analysis algorithm was used to calculate whether genes were “absent” or “present” based on previously described parameters (77). To calculate the fold changes in genes in two different samples, the Comparison Analysis algorithm (Affymetrix) was used (77). Four different comparison analyses were performed, including one with cRNA from mock-infected brains of 1-day-old mice as the baseline sample and cRNA from dsTE12Q-infected brains of 1-day-old mice as the experimental sample (NSvNM; N, neonatal; S, Sindbis virus infected; M, mock infected), one with cRNA from mock-infected brains of 4-week-old mice as the baseline sample and cRNA from dsTE12Q-infected brains of 4-week-old mice as the experimental sample (WSvWM; W, weanling; S, Sindbis virus infected; M, mock infected), one with cRNA from dsTE12Q-infected brains of 1-day-old mice as the baseline sample and cRNA from dsTE12Q-infected brains of 4-week-old mice as the experimental sample (WSvNS), and one with cRNA from mock-infected 1-day-old mice as the baseline sample and cRNA from mock-infected brains of 4-week-old mice as the experimental sample (WMvNM). Genes that were both “absent” in the absolute analysis of the baseline sample and “decreasing” in the experimental sample on comparison analysis were excluded from analysis. Genes that were both “absent” in the absolute analysis of the experimental sample and “increasing” in the experimental sample upon comparison analysis were also excluded. In addition, genes that were altered in the comparison of NSvNM or WSvWM but not altered in the comparison of WSvWM or WMvNM were excluded from analysis.
Cloning of mouse ISG12.
5′ and 3′ RACE (rapid amplification of cDNA ends) was performed by using the Marathon protocol and Marathon-Ready mouse brain cDNA according to the manufacturer's instructions (Clontech) with primers derived from the expressed sequence tag (EST) of GenBank accession no. AA145371 (forward primer for 3′ RACE, AACATGTTGGGAACACTGTT; reverse primer for 5′ RACE, CTGCTGATTGGAGTGTGGCT). The 5′ and 3′ RACE products were subcloned into the TA cloning vector, and multiple subclones were sequenced. The 5′ and 3′ RACE sequences were aligned with the EST sequence (GenBank accession no. AA145371) to obtain a predicted complete cDNA sequence of mouse ISG12.
Analysis of CNS gene expression by RT-PCR.
Total RNA was extracted by using TRIzol Reagent, and poly(A) RNA was isolated by using an Oligotex Direct mRNA kit (Qiagen) from triplicate mouse brains on days 1, 2, and 3 after mock infection or infection with 1,000 PFU of dsTE12Q intracerebrally. A 1-μg portion of poly(A)+ RNA was used to synthesize cDNA with avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim) and an oligo(dT) primer (Boehringer Mannheim), and PCR was performed as described previously (74) to detect the Sindbis virus E2 glycoprotein gene (plus-strand primer [positions 8607 to 8626], 5′-GGATCGTCTGGCAGAAGCAA-3′; minus-strand primer [positions 8893 to 8912], 5′-AAGCCTTCTACACGGTCCTG-3′), murine fractalkine (plus-strand primer, 5′-GACGAAATGCGAAATCATGTGC-3′; minus-strand primer 5′-TCCAATGCTCTGAGGCTTAGC-3′), murine ISG12 (plus-strand primer 5′-GTTGGGAACACTGTTTGGCTC-3′; minus-strand primer 5′-TAGGATGGCATTTG TTGATGTG-3′), murine TRAF4 (plus-strand primer, 5′-GCAGGAGTTTCTCAGTGAAGG-3′; minus-strand primer, 5′-CACTCAGACGTGGCATGCTG-3′), and the constitutively expressed cellular gene, GAPDH (plus-strand primer, 5′-ACCACCATGGAGAAGGCTGG-3′; minus-strand primer, 5′-CTCAGTGTAGCCC AGGATGC-3′). Serial dilutions of positive controls for each gene of interest were amplified at 20, 25, 30, and 35 cycles to determine the optimal number of amplification cycles that resulted in a linear relationship between the initial RNA input and PCR product. Twenty-five cycles of amplification was used in each PCR in the data shown in Fig. Fig.33 and and44.
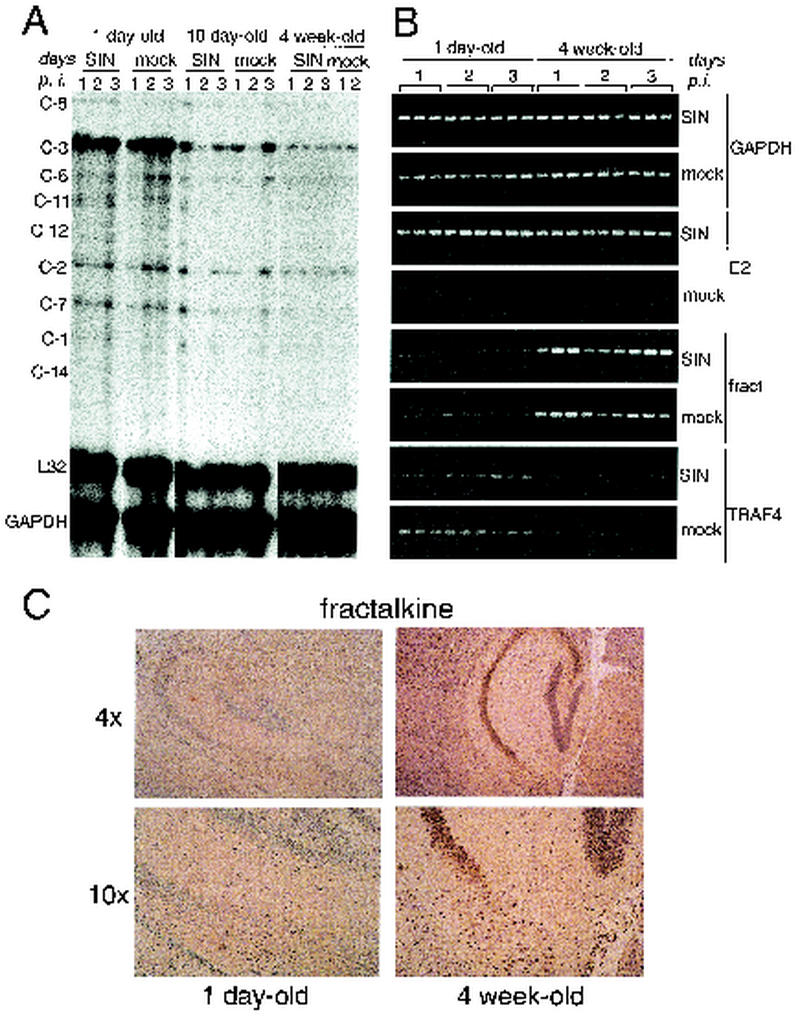
Caspase-3, TRAF4 and fractalkine expression differs in the brains of 1-day-old versus 4-week-old CD1 mice. (A) RiboQuant Apo-1 Multi-Probe RNase protection assay measuring caspase mRNA expression in the brains of dsTE12Q- or mock-infected mice of different ages at serial time points after infection. RNA was pooled from three mice per time point per treatment group. (B) RT-PCR analysis of RNA samples from triplicate mouse brains harvested at serial time points after infection with dsTE12Q or mock infection to detect GAPDH, Sindbis virus E2 envelope glycoprotein, fractalkine, and TRAF4. (C) Immunoperoxidase staining to detect fractalkine expression in mock-infected 1-day-old and 4-week-old brain tissue. Representative sections from the hippocampus by using a ×4 (upper panels) or ×10 (lower panels) objective lens. C-, caspase; p.i., postinfection; fract, fractalkine.
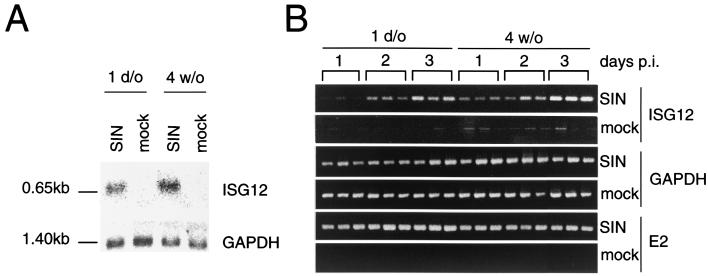
Age-related difference in upregulation of mouse ISG12 expression after dsTE12Q infection. (A) Northern blot analysis of ISG12 and GAPDH mRNA expression in mouse brains harvested at 60 h after intracerebral mock or virus infection of 1-day-old or 4-week-old mice. (B) RT-PCR analysis of ISG12, GAPDH, and Sindbis virus E2 envelope glycoprotein gene in RNA harvested from triplicate mouse brains at serial time points after mock or virus infection. SIN, Sindbis virus, strain dsTE12Q; d/o, day-old; p.i., postinfection.
Northern blot analysis.
A total of 20 μg of total RNA per sample (pooled from three mouse brains) was electrophoresed on a 1.2% agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized with a [32P]dCTP randomly labeled mouse ISG12 or mouse GAPDH probe by using Express Hyb hybridization solution (Clontech) according to the manufacturer's instructions.
RNase protection assay.
RNase protection assays to measure RNA expression of caspases in mouse brain samples were performed by using the RiboQuant Apo-1 Multi-Probe RNase Protection Assay kit (PharMingen) according to the manufacturer's instructions.
RESULTS
The dsTE12Q strain of Sindbis virus strain produces age-dependent lethal disease in mice.
Wild-type Sindbis virus (and recombinant strains of Sindbis virus derived from molecular clones that are similar to wild-type virus) are known to produce lethal disease in neonatal mice and asymptomatic infection in weanling and adult mice. Because of the utility of using double subgenomic strains of Sindbis virus as vectors to express host genes in vivo (reviewed in reference 16), we evaluated whether the double subgenomic strain of Sindbis virus, dsTE12Q, also produces age-dependent fatal disease. We found that intracerebral inoculation of 1,000 PFU of dsTE12Q resulted in 100% mortality of 1-day-old infected mice within 8 days of infection, whereas no mortality or clinically apparent disease was observed in 4-week-old infected mice (Fig. (Fig.1A1A and data not shown). The absence of clinical symptoms of infection in 4-week-old mice cannot be explained by the inability of Sindbis virus to replicate in weanling mice, since mean peak viral titers were ca. 108 PFU per g of brain in the 4-week-old infected mice (Fig. (Fig.1B).1B). In 1-day-old-infected mice, mean viral titers were somewhat higher and peaked ca. 109 PFU per g of brain by day 2 postinfection, when all of the mice were moribund. These data demonstrate that Sindbis virus replicates both in 1-day-old mice and in 4-week-old infected mice (albeit less efficiently) but only causes fatal disease in 1-day-old mice.
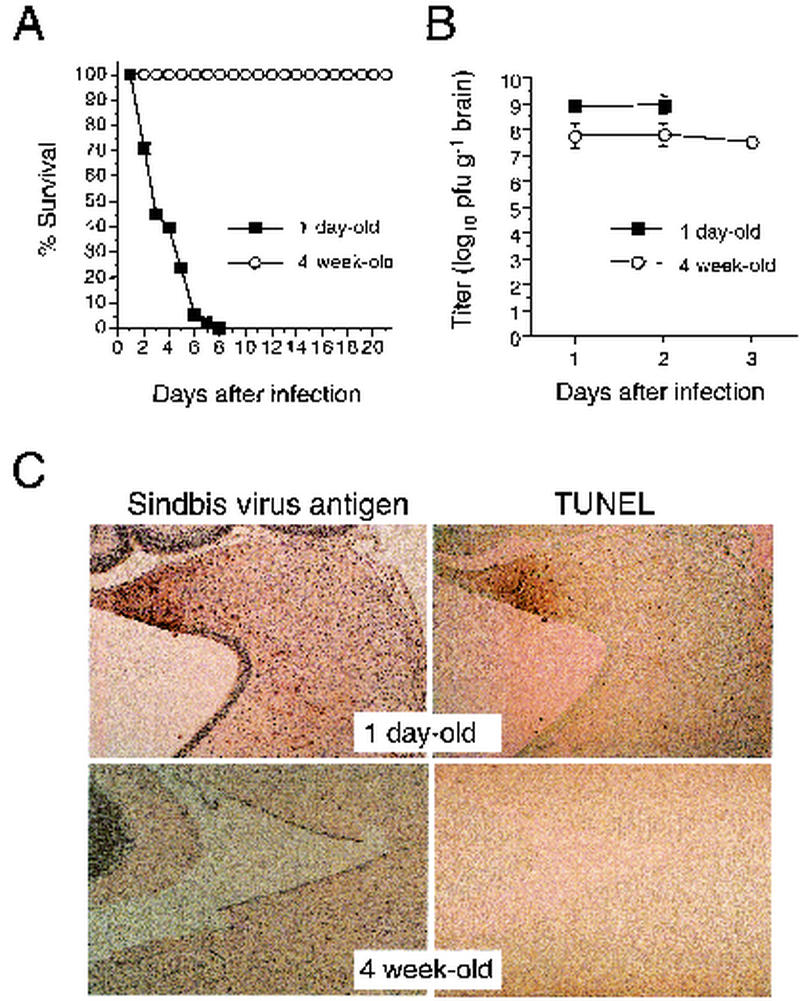
dsTE12Q infection produces more severe CNS disease in 1-day-old CD1 mice than in 4-week-old CD1 mice. (A) Survival curves of 1-day-old and 4-week-old CD1 mice infected intracerebrally with 1,000 PFU Sindbis virus, strain dsTE12Q. The data represent combined survival probabilities for five independent litters with 10 to 12 mice per litter. (B) Viral growth of dsTE12Q in brains of CD1 mice infected at 1 day and 4 weeks of age. Each datum point represents the geometric mean viral titer and standard error of the mean for three mouse brains. (C) Immunoperoxidase labeling to detect Sindbis virus antigen and TUNEL labeling to detect apoptotic nuclei in the brains of mice infected at 1 day or four weeks of age. Representative regions are shown from the colliculus 60 h after infection. Magnification, ×194.
Histopathologic analyses of 1-day-old and 4-week-old dsTE12Q-infected brains revealed marked differences in CNS pathology. Consistent with the lower viral titers, few Sindbis virus antigen positive cells were detected in the brains of infected 4-week-old mice (Fig. (Fig.1C).1C). In contrast, numerous large foci of Sindbis virus antigen-positive cells were found in different regions of the brains of dsTE12Q-infected 1-day-old mice, most commonly in the supraventricular zone, olfactory bulb, hippocampus, colliculus, hippocampus, thalamus, striatum, and posterior neocortex. Although the number of Sindbis virus antigen-positive cells was markedly decreased in dsTE12Q-infected 4-week-old mice, the virus displayed a similar cellular tropism for neurons and similar regional tropism in 1-day-old and 4-week-old mice. In regions of viral antigen staining in the brains of 1-day-old mice, numerous pyknotic nuclei were observed by H&E staining (data not shown) and numerous apoptotic nuclei were observed by TUNEL straining (Fig. (Fig.1C).1C). In contrast, the brains of infected 4-week-old mice had no TUNEL-positive nuclei or histologic evidence of neuronal destruction, a finding consistent with previous reports of age-dependent resistance to alphavirus-induced neuronal apoptosis (1, 22, 38).
Despite extensive neuronal destruction, there was no detectable inflammatory cell infiltration observed in the brains of virus-infected 1-day-old mice. The lack of inflammatory changes characteristic of “encephalitis” in the brains of moribund neonatal mice is consistent with our previous observations in 10-day-old mice infected with the virulent dsTE12H strain of Sindbis virus (25) and findings by Trgovcich et al. in neonatal mice infected with the virulent TRSB strain of Sindbis virus (70). Similar to previous studies of Sindbis virus pathogenesis in weanling mice, classic histologic features of encephalitis such as perivascular cuffing and lymphocytic infiltration were, however, observed in the brains of dsTE12Q-infected 4-week-old mice (data not shown).
In summary, these data demonstrate that intracerebral infection of dsTE12Q in neonatal mice results in lethal disease accompanied by high levels of CNS replication and extensive neuronal death in the absence of histologically detectable inflammation. Morever, intracerebral dsTE12Q infection in weanling mice results in an asymptomatic “encephalitis” that is associated with lower levels of viral replication and no evidence of neuronal destruction. Thus, dsTE12Q infection provides a useful model for studying the molecular determinants of age-dependent resistance to lethal CNS viral infection.
Apoptosis-related genes are developmentally regulated in the mouse CNS.
To identify developmental changes in CNS gene expression that are candidate regulators of age-dependent susceptibility to fatal viral infection, we used DNA microarray technology to compare gene expression in the brains of mock-infected 1-day-old and mock-infected 4-week-old mice (Table (Table1).1). The mRNA expression levels of 77 genes (including 67 known genes and 10 ESTs) increased >5-fold in the brains of 4-week-old mock-infected mice compared to 1-day-old mock-infected mice, whereas the mRNA expression of only 18 genes (including 14 known genes and four ESTs) decreased more than 5-fold in the brains of the 4-week-old mice. Many of these genes encode proteins are either specifically expressed in the nervous system or that play specific roles in neuronal function, such as protein involved in myelination, neurotransmission, neuronal receptors, synaptic vesicle transport, and ion channels. Also, several genes encoding proteins involved in the cytoskeleton, calcium-binding, extracellular matrix and adhesion, protein degradation, metabolism, and lipid transport (e.g., apolipoprotein D) were also found to be developmentally regulated.
TABLE 1.
Genes that change by ≥5-fold in 4-week-old compared to 1-day-old uninfected mouse brains
| Gene family and gene name | Accession no. | Fold change | Gene family and gene name | Accession no. | Fold change | |
|---|---|---|---|---|---|---|
| Annexins | ||||||
    Annexin V Annexin V | U29396 | 5.6 | ||||
| Actin/cytoskeleton | ||||||
    Tctex-1 Tctex-1 | W15873 | −8.2 | ||||
    Beta-tubulin Beta-tubulin | AA098307 | −5.2 | ||||
    Connexin-30 Connexin-30 | Z70023 | 6.3 | ||||
    Alpha-tubulin Alpha-tubulin | M13444 | 6.5 | ||||
    Gelsolin Gelsolin | J04953 | 7.4 | ||||
| Apoptosis related | ||||||
    Caspase-3 Caspase-3 | U54803 | −12.8 | ||||
| Calcium-binding proteins | ||||||
    Calbindin-D28K Calbindin-D28K | M21531 | 5 | ||||
    Calcyclin Calcyclin | M37761 | 6.4 | ||||
    S100 beta protein S100 beta protein | L22144 | 7.4 | ||||
| Cell cycle related | ||||||
    CDC25MM CDC25MM | L20899 | 7.5 | ||||
| Chemokines | ||||||
    Fractalkine Fractalkine | U92565 | 9.8 | ||||
| Extracellular matrix and cell adhesion | ||||||
    Nidogen Nidogen | X14194 | −7.7 | ||||
    Tenascin Tenascin | X56304 | −5.1 | ||||
    Similar to human cell adhesion kinase beta Similar to human cell adhesion kinase beta | AA175767 | 8.8 | ||||
| Nervous system related | ||||||
    1. Neurotransmitters 1. Neurotransmitters | ||||||
        Proenkephalin A Proenkephalin A | M13227 | 6.3 | ||||
        Telencephalin Telencephalin | U06483 | 10.5 | ||||
        GABA GABA | X51986 | 36.0 | ||||
    2. Synaptic function 2. Synaptic function | ||||||
        SNAP25 SNAP25 | M22012 | 5.2 | ||||
        Syntaxin 4 Syntaxin 4 | U76832 | 5.6 | ||||
    3. Other 3. Other | ||||||
        Unc-33-like phosphoprotein (Ulip) Unc-33-like phosphoprotein (Ulip) | X87817 | −16.1 | ||||
        Similar to human Doc-2 Similar to human Doc-2 | W37024 | −5.7 | ||||
        N-copine N-copine | R74842 | 5.2 | ||||
        P24 P24 | D83206 | 5.7 | ||||
        Neurofilament component H Neurofilament component H | M35131 | 5.9 | ||||
        GABA-a/BZ receptor GABA-a/BZ receptor | M63436 | 12.6 | ||||
        Neuronal pentraxin 1 (NP-1) Neuronal pentraxin 1 (NP-1) | U62021 | 14.3 | ||||
        Myelin proteolipid protein (PLP) Myelin proteolipid protein (PLP) | X07215 | 14.7 | ||||
        Myelin-associated glycoprotein Myelin-associated glycoprotein | M31811 | 15.3 | ||||
        Purkinje cell protein 2 (L7) Purkinje cell protein 2 (L7) | M21532 | 19.4 | ||||
        ZnT-3 ZnT-3 | U76007 | 22.2 | ||||
        Myelin-oligodendrocyte glycoprotein Myelin-oligodendrocyte glycoprotein | U64572 | 23.1 | ||||
        P400 protein P400 protein | X15373 | 27.4 | ||||
        Similar to human MOBP Similar to human MOBP | U81317 | 29.6 | ||||
        Oligodendrocyte-specific protein (OSP) Oligodendrocyte-specific protein (OSP) | U19582 | 30.7 | ||||
| Nucleotide exchange factors | ||||||
    R-ras3 R-ras3 | AB004879 | 5.1 | ||||
    Tiam-1 Tiam-1 | U05245 | 7 | ||||
    SKD-2 SKD-2 | U10120 | 8.7 | ||||
| Growth factors | ||||||
    CD-MPR CD-MPR | X64068 | 5.7 | ||||
    Colony-stimulating factor 1 receptor Colony-stimulating factor 1 receptor | X06368 | 5.9 | ||||
| IFN-pathway related | ||||||
    Similar to IFN-inducible protein 1-8U Similar to IFN-inducible protein 1-8U     (human) (human) | W20873 | −5.2 | ||||
    Ran Ran | L32751 | 13.9 | ||||
| Ion channels | ||||||
    Shaker K+ channel beta Shaker K+ channel beta | X97281 | 5.4 | ||||
    mbGIRK3 K+ channel mbGIRK3 K+ channel | U11860 | 6.3 | ||||
    Voltage-gated Na+ channel (beta 1 subunit) Voltage-gated Na+ channel (beta 1 subunit) | L48687 | 8.2 | ||||
    TYRO3 TYRO3 | U05683 | 10.1 | ||||
    Voltage-dependent K+ channel beta subunit Voltage-dependent K+ channel beta subunit | U31908 | 12.2 | ||||
    Voltage-gated K+ channel protein KV1.1 Voltage-gated K+ channel protein KV1.1 | Y00305 | 17.3 | ||||
| Lipocalins | ||||||
    Apolipoprotein D Apolipoprotein D | L39123 | 26.1 | ||||
| Metabolism | ||||||
    Polysialic acid synthase Polysialic acid synthase | X83562 | −16.2 | ||||
    5-Aminolevulinic acid synthase 5-Aminolevulinic acid synthase | M63244 | −11.1 | ||||
    Protein phosphatase 2A Protein phosphatase 2A | AA065915 | −6.8 | ||||
    Glutathione S-transferase Glutathione S-transferase | AA638086 | 5.1 | ||||
    Aldehyde dehydrogenase Aldehyde dehydrogenase | M74570 | 5.7 | ||||
    Aspartate aminotransferase Aspartate aminotransferase | J02623 | 5.8 | ||||
    N1-acetyltransferase SSAT N1-acetyltransferase SSAT | L10244 | 6.1 | ||||
    Similar to human platelet 6-phosphofructokinase Similar to human platelet 6-phosphofructokinase | AA072252 | 6.4 | ||||
    Similar to phosphatidylinositol 4,5-bisphosphate Similar to phosphatidylinositol 4,5-bisphosphate | AA638530 | 6.6 | ||||
    Hypoxanthine-guanine Hypoxanthine-guanine     phosphoribosyltransferase phosphoribosyltransferase | J00423 | 8.1 | ||||
    Carbonic anhydrase II Carbonic anhydrase II | K00811 | 10.4 | ||||
    Glycerol-3-phosphate dehydrogenase Glycerol-3-phosphate dehydrogenase | M13366 | 10.4 | ||||
    Similar to human glyoxalase II Similar to human glyoxalase II | W08455 | 13.3 | ||||
| Protease inhibitors | ||||||
    Cystatin B Cystatin B | U59807 | 5.1 | ||||
    Similar to human flastase inhibitor Similar to human flastase inhibitor | AA145127 | 5.6 | ||||
    Similar to human HtrA Similar to human HtrA | AA059662 | 7.3 | ||||
    Inter-alpha trypsin inhibitor, heavy chain 3 Inter-alpha trypsin inhibitor, heavy chain 3 | X70393 | 7.5 | ||||
| Protein degradation | ||||||
    Cathepsin C Cathepsin C | U89269 | 5.3 | ||||
    Cathepsin S Cathepsin S | AA255186 | 7.5 | ||||
| MISCELLANEOUS | ||||||
    Parotid secretory protein (PSP) Parotid secretory protein (PSP) | X01697 | −19.6 | ||||
    Salivary protein 1 Salivary protein 1 | X00349 | −18.9 | ||||
    CD24 antigen CD24 antigen | X53825 | −14.4 | ||||
    Retinol-binding protein Retinol-binding protein | AA049662 | 5 | ||||
    Human mitochondrial 75-kDa FeS protein Human mitochondrial 75-kDa FeS protein | V00835 | 5.1 | ||||
    Prion protein Prion protein | M13685 | 5.3 | ||||
    P-selectin glycoprotein ligand-1 (PSGL-1) P-selectin glycoprotein ligand-1 (PSGL-1) | X91144 | 6.1 | ||||
    FK506-binding protein 13 (FKBP13) FK506-binding protein 13 (FKBP13) | M77831 | 6.9 | ||||
    Hrp12 Hrp12 | U50631 | 8.2 | ||||
    Histone H2A Histone H2A | Z30940 | 8.8 | ||||
    Alpha B crystallin Alpha B crystallin | M63170 | 9.1 | ||||
    Prealbumin Prealbumin | D89076 | 9.9 | ||||
    Parvalbumin Parvalbumin | X59382 | 37.3 | ||||
| Unknown | ||||||
    EST EST | W53443 | −10.9 | ||||
    EST EST | AA152671 | −8.4 | ||||
    EST EST | AA516966 | −5.4 | ||||
    EST EST | AA408420 | −5 | ||||
    EST EST | AA166452 | 5.4 | ||||
    EST EST | AA521958 | 5.5 | ||||
    EST EST | W41070 | 5.5 | ||||
    EST EST | R74735 | 6.1 | ||||
    EST EST | AA139510 | 8.3 | ||||
    EST EST | AA266791 | 8.4 | ||||
    EST EST | R74641 | 11.1 | ||||
    EST EST | R75354 | 19.8 | ||||
    EST EST | R75030 | 20.9 | ||||
    EST EST | AA673405 | 21.4 |
Although developmental regulation of genes involved in neuronal function may play a role in determining age-related susceptibility to infection with Sindbis virus, there is insufficient understanding at present of the molecular details of virus-neuron interactions to postulate which genes may be candidate regulators of viral pathogenesis. In contrast, apoptosis-related genes that are developmentally regulated are identifiable as candidate determinants of age-dependent differences in Sindbis virus pathogenesis since apoptosis plays a critical role for apoptosis in the pathogenesis of several different CNS viral infections. Therefore, we compared the expression of all known antiapoptotic bcl-2 family members, all cell death signaling molecules, and all cell death proteases (caspases) included in the Affymetrix 11K GeneChip (Fig. (Fig.2).2). Of the apoptosis-related genes included in the analysis, CNS mRNA expression of Bcl-2 family members did not differ between mock-infected 1-day-old and 4-week-old mice. We confirmed this by RNase protection assay using the Apo-2 Multi-Probe RNase protection assay kit (PharMingen) (data not shown). Among the cell death signaling molecules, developmental regulation was observed only for TRAF4, a TRAF family member that promotes apoptosis through interactions with the p75NTR neurotrophin receptor (76). TRAF4 mRNA expression was decreased by nearly 12-fold in the brains of 4-week-old compared to 1-day-old mice. Only three caspases were included in the GeneChip analysis, two of which are involved primarily in inflammation, caspase-1 and -11, as well as caspase-3, a common death effector molecule that is downstream in multiple different apoptotic signaling pathways (reviewed in reference 72). Interestingly, caspase-3 mRNA expression was decreased by almost 12-fold in the brains of the weanling mice compared to the brains of the neonatal mice.
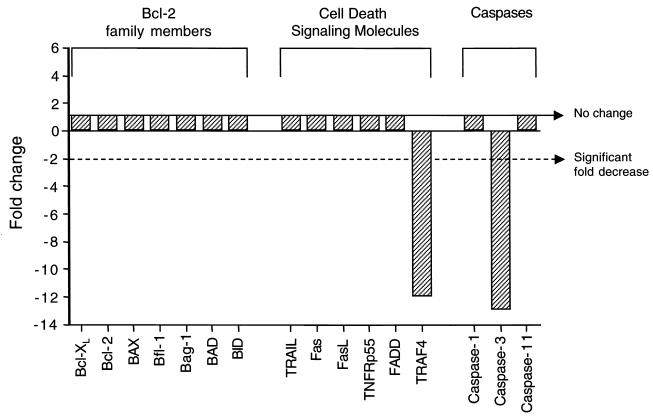
mRNA expression patterns of apoptosis-related genes in the brains of mock-infected 4-week-old mice compared with mock-infected 1-day-old mice.
In addition to Bcl-2 family members, cell death signaling molecules, and caspases, certain chemokines, such as fractalkine, MDC, MIP-1β, RANTES, and SDF-1α, have also been shown to play a role in vitro in regulating susceptibility to neuronal apoptosis (reviewed in reference 34). Therefore, it is also of potential interest that fractalkine, a chemokine expressed by neurons in both a membrane-bound and soluble form (reviewed in reference 43), was developmentally upregulated postnatally in mouse brain. In our GeneChip expression analysis, fractalkine mRNA expression was nearly 10-fold greater in the brains of 4-week-old than in the brains of neonatal mice. Consistent with the lack of inflammation-associated changes in neuronal fractalkine expression in rats with experimental autoimmune encephalomyelitis (61), we found that fractalkine mRNA expression, unlike that of several other chemokines, was unaltered by Sindbis virus infection in either neonatal or weanling mice (Tables (Tables11 and and22).
TABLE 2.
Genes that change ≥5-fold in 1-day-old or 4-week-old Sindbis virus-infected mouse brainsa
| Gene family and gene name | Accession no. | Fold changea
| |||
|---|---|---|---|---|---|
| NSvNM | WSvWM | WSvNS | WMvNM | ||
| Antigen presentation | |||||
    MHC class II, Fas-BP Daxx MHC class II, Fas-BP Daxx | AA727845 | 7.5 | NC | −2.7 | NC |
    MHC class I, beta-2 microglobulin MHC class I, beta-2 microglobulin | X01838 | 8.5 | 4.1 | NC | 2.1 |
    MHC class I, heavy chain precursor H-2K MHC class I, heavy chain precursor H-2K | U47329 | 8.5 | 5.3 | −1.3 | 1.9 |
    MHC class I, H2-D MHC class I, H2-D | X00246 | 9.2 | 6.8 | −1.3 | NC |
    MHC class I, H-2M3 MHC class I, H-2M3 | U18797 | 9.4 | 1.9 | 2.8 | 2.7 |
    MHC class I, Qa-1a MHC class I, Qa-1a | L00606 | 9.9 | 9.9 | NC | NC |
    Tap-1 Tap-1 | M55637 | 11.6 | 3.4 | −4.9 | NC |
    MHC class I, Q8/9d MHC class I, Q8/9d | M29881 | 13.5 | 10.7 | −2 | NC |
    MHC class I, H2-T23 MHC class I, H2-T23 | AA060835 | 17.7 | NC | −9.6 | NC |
    MHC class I, H2-TL-T10-129 MHC class I, H2-TL-T10-129 | M35244 | 22.4 | 7.6 | −5 | NC |
    MHC class I, H-2 Dr MHC class I, H-2 Dr | M69070 | 22.6 | 17.6 | −2.4 | NC |
    MHC class II-associated invariant chain MHC class II-associated invariant chain | X00496 | 25.0 | 5.7 | −6.3 | NC |
    MHC class I, Qa-Tla MHC class I, Qa-Tla | M11284 | 30.1 | 7.4 | −2.1 | NC |
    MHC class I, A.CA/J(H-2K-1) MHC class I, A.CA/J(H-2K-1) | M69069 | 66.3 | 10.6 | −2.1 | −1.1 |
| Calcium-binding proteins | |||||
    Calgranuin B Calgranuin B | M83219 | 5.3 | NC | −6 | NC |
    Calcyclin Calcyclin | M37761 | 6.2 | NC | NC | 6.4 |
    Calgranuin A Calgranuin A | M83218 | 9.2 | −10.9 | NC | |
| Chemokines | |||||
    Growth-regulated protein (GRO1)/KC Growth-regulated protein (GRO1)/KC | J04596 | 8.4 | −10.3 | ||
    MCP-5 MCP-5 | U50712 | 10.1 | NC | NC | |
    Crg-2/IP-10 Crg-2/IP-10 | M33266 | 19.8 | 2.4 | −6.1 | NC |
    JE/MCP-1 JE/MCP-1 | M19681 | 24.6 | 10.3 | −8.1 | −4.2 |
    RANTES RANTES | U02298 | 35.4 | 10.2 | −14.4 | NC |
| Complement | |||||
    Complement component C4 Complement component C4 | M17440 | 7.6 | 2.1 | NC | 2.6 |
    Complement C2 component factor B Complement C2 component factor B | K01496 | 8.4 | NC | 2.7 | NC |
    Complement component C2 gene Complement component C2 gene | M60579 | 8.4 | −2.2 | ||
| Extracellular matrix and cell adhesion | |||||
    Entactin Entactin | X14194 | NC | 7.7 | −4.1 | −7.7 |
    Vascular cell adhesion molecule 1 (VCAM-1) Vascular cell adhesion molecule 1 (VCAM-1) | X67783 | 5 | 2.3 | −2.3 | −2.3 |
    Intercellular adhesion molecule 1 (ICAM-1) Intercellular adhesion molecule 1 (ICAM-1) | X52264 | 5.1 | −7 | ||
    Macrophage colony-stimulating factor 1 (CSF-1) Macrophage colony-stimulating factor 1 (CSF-1) | M21952 | 5.8 | −7.1 | NC | |
    Galectin-9 Galectin-9 | U55060 | 12.4 | 2.6 | −3.6 | NC |
    Proteoglycan, secretory granule Proteoglycan, secretory granule | X16133 | 12.5 | 3.5 | NC | 3.8 |
| Hematopoietic cell surface molecules | |||||
    CD82 antigen CD82 antigen | D14883 | 5.9 | NC | NC | 4.8 |
    Lymphocyte antigen Ly-6C Lymphocyte antigen Ly-6C | D86232 | 7.9 | 5.3 | NC | NC |
    Lymphocyte antigen Ly-6.2 Lymphocyte antigen Ly-6.2 | M18184 | 64.9 | 7.9 | −1.2 | 6.6 |
    High-affinity Fc receptor for IgG High-affinity Fc receptor for IgG | M31314 | 6.1 | 5.3 | NC | |
| IFN-pathway related | |||||
    IFN-activated 203 protein IFN-activated 203 protein | AA170251 | 5 | 6 | NC | NC |
    GBP2 GBP2 | AA153021 | 5.9 | 5.5 | NC | NC |
    Intercrine Intercrine | Z12297 | 6.6 | 3.8 | NC | |
    IFP35 IFP35 | AA013783 | 7.5 | NC | −5.6 | NC |
    IFN-activated 202 A protein IFN-activated 202 A protein | M31418 | 9.7 | 6.9 | NC | |
    G-protein-like LRG-47 G-protein-like LRG-47 | U19119 | 10.5 | 12.1 | NC | |
    Mx Mx | M12279 | 13.9 | 4.7 | −6.4 | |
    GTPase (Ran) GTPase (Ran) | L32751 | 14.9 | NC | NC | 13.9 |
    Similar to IFN-inducible protein 1-8D Similar to IFN-inducible protein 1-8D | AA002526 | 15.2 | 15 | NC | NC |
    Similar to IFN-γ GTP-binding protein Similar to IFN-γ GTP-binding protein | U53219 | 20.6 | 10.3 | −3 | |
    Similar to ISG12 (human) Similar to ISG12 (human) | AA145371 | 20.6 | 49.2 | 2.1 | NC |
    GTP-binding protein GTP-binding protein | L38444 | 22.4 | 12.4 | −2.2 | |
    GARG 39 GARG 39 | U43085 | 22.9 | NC | 3.1 | |
    GBP-2 GBP-2 | AA288442 | 23.2 | 5.5 | −3.8 | |
    Similar to IFN-inducible protein 1-8U Similar to IFN-inducible protein 1-8U | W12941 | 25.4 | 7.5 | −1.5 | 2.4 |
    GBP3 GBP3 | U44731 | 25.7 | 9.2 | −2.3 | |
    IRF-7 IRF-7 | U73037 | 26.8 | 5.3 | −2.6 | |
    GARG 49/irg2 GARG 49/irg2 | L32974 | 27 | 11.4 | −1.5 | NC |
    GARG-16 GARG-16 | U43084 | 29.1 | 7.2 | NC | NC |
    2′5′ oligonucleotide A synthetase 2′5′ oligonucleotide A synthetase | M33863 | 30.1 | 3.5 | −3 | |
    IFN-inducible GTPase IFN-inducible GTPase | AA415898 | 36.1 | 14.9 | −4.1 | |
    Similar to Best5 protein (rat) Similar to Best5 protein (rat) | AA175784 | 47.4 | 16 | −4.2 | NC |
    IFN-induced 15-kDa protein (ISG15) IFN-induced 15-kDa protein (ISG15) | X56602 | 121.2 | 38.2 | −3.2 | |
| Lipocalins | |||||
    Lipocalin 2 (24p3) Lipocalin 2 (24p3) | X81627 | 37.3 | 3 | 7.1 | NC |
    Apolipoprotein D Apolipoprotein D | L39123 | 39.8 | NC | NC | 26.1 |
| Nervous system related | |||||
    P400 Protein P400 Protein | X15373 | −5.1 | NC | 46.1 | 27.4 |
    Similar to S182 proteim (m-Numb) Similar to S182 proteim (m-Numb) | U70674 | 2 | 9.6 | 2.4 | |
    Oligodendrocyte-specific protein (OSP) Oligodendrocyte-specific protein (OSP) | U19582 | 2.9 | −12 | 12 | 30.7 |
| Protease inhibitors | |||||
    Tissue inhibitor of metalloproteinase-1 Tissue inhibitor of metalloproteinase-1 | V00755 | 8.3 | 3.4 | −1.3 | |
    Spi2 proteinase inhibitor Spi2 proteinase inhibitor | M64086 | 14.5 | 10.5 | −2.7 | NC |
| Protein degradation | |||||
    Similar to S182 protein Similar to S182 protein | U70674 | 2 | 9.6 | 2.4 | |
    LAMP-2 LAMP-2 | J05287 | 5.1 | 1.7 | NC | 3.8 |
    Cathepsin S Cathepsin S | AA255186 | 6.6 | 2.6 | 6.9 | 7.5 |
    Similar to ubiquitin-activating enzyme E1 Similar to ubiquitin-activating enzyme E1 | AA170444 | 10.5 | −2.3 | NC | |
    Lmp10 Lmp10 | D85561 | 7.2 | 2.1 | −2.2 | 2.4 |
    CTLA-2 alpha CTLA-2 alpha | X15591 | 8.4 | 4.7 | −2.9 | −1.6 |
    Cathepsin C Cathepsin C | U89269 | 9.2 | 2.2 | NC | 5.3 |
    E2 ubiquitin-conjugating enzyme E2 ubiquitin-conjugating enzyme | AA273574 | 17.6 | 10.7 | −3.6 | NC |
    Lmp2 Lmp2 | D14566 | 18.2 | 7.9 | −10.2 | NC |
    EST, similar to lysophospholipase EST, similar to lysophospholipase | AA170668 | 23 | 7 | −2.5 | |
    Ubiquitin-specific protease 18 Ubiquitin-specific protease 18 | AA286391 | 35.2 | 21 | −2.5 | |
    Lmp7 Lmp7 | U22031 | 60.2 | 17.6 | NC | NC |
| Transcriptional regulators | |||||
    Regulatory protein T-lymphocyte 1 (rpt-1) Regulatory protein T-lymphocyte 1 (rpt-1) | J03776 | 5.4 | 4.3 | 2.4 | |
    Rpo2lc1 Rpo2lc1 | J03750 | 7.2 | NC | NC | 7 |
    Estrogen-responsive finger protein (Elp) Estrogen-responsive finger protein (Elp) | D63902 | 9 | 4.5 | 2.8 | 1.3 |
    c-fos c-fos | V00727 | 13 | NC | 2.3 | |
    Stat-1 Stat-1 | U06924 | 13.4 | 11 | NC | NC |
    C/EBP delta C/EBP delta | X61800 | 18.1 | 7.4 | NC | |
| Miscellaneous | |||||
    Prolactin Prolactin | X02892 | NC | 6 | 8.8 | |
    Parotid secretory protein (PSP) Parotid secretory protein (PSP) | X01697 | −19.2 | −19.6 | ||
    Salivary protein 1 Salivary protein 1 | X00349 | −17.7 | −18.9 | ||
    MyD116 MyD116 | X51829 | 5.2 | NC | −8.7 | NC |
    TDAG51 TDAG51 | U44088 | 5.3 | NC | NC | 4.7 |
    Prealbumin Prealbumin | D89076 | 5.4 | NC | 1.8 | 9.9 |
    Prostaglandin E synthase Prostaglandin E synthase | W97022 | 5.4 | NC | −2.3 | |
    N1-acetyltransferase SSAT N1-acetyltransferase SSAT | L10244 | 7.1 | NC | NC | 6.1 |
    Thymidylate kinase family lipopolysaccharide-inducible protein (TYKi) Thymidylate kinase family lipopolysaccharide-inducible protein (TYKi) | L32973 | 8 | 1.9 | −2.4 | NC |
    Tumor necrosis factor alpha-induced protein 2 (B94) Tumor necrosis factor alpha-induced protein 2 (B94) | L24118 | 8.1 | −7.6 | NC | |
    Onzin Onzin | AA638539 | 12.5 | 9 | 1.4 | |
    Cyclophilin C-associated protein Cyclophilin C-associated protein | X67809 | 41.4 | 27.3 | NC | |
    Serum amyloid A3 Serum amyloid A3 | X03479 | 56.1 | −54.9 | ||
| Unknown | |||||
    EST EST | AA277082 | 5 | NC | NC | |
    EST EST | AA033381 | 5.1 | 5.1 | ||
    EST EST | C80656 | 5.2 | NC | −6.7 | NC |
    EST EST | AA255253 | 5.3 | 6.4 | NC | |
    EST EST | C78546 | 5.5 | 1.9 | NC | 2.3 |
    EST EST | AA177851 | 7.8 | 4.5 | −2.9 | |
    EST EST | AA163876 | 8.6 | 5.1 | −2.3 | |
Given the potential importance of the developmental downregulation of proapoptotic molecules such as caspase-3 and TRAF4 and the developmental upregulation of the neuroprotective chemokine, fractalkine, in developmental changes in susceptibility to Sindbis virus-induced apoptosis, we sought to confirm the observed changes in GeneChip expression by using independent methods to measure gene expression and with RNA samples derived from independent groups of mice. Using an RNase protection assay, we evaluated expression of caspase-3 and other caspases in the brains of 1-day-old, 10-day-old, and 4-week-old brains at days 1, 2, and 3, after dsTE12Q or mock infection (Fig. (Fig.3A).3A). After correcting for differences in RNA loading by normalizing signal intensities to those observed with the control probes for L32 and GAPDH, we found that only caspase-3 mRNA expression was significantly altered in the CNS during development (14.4-fold decrease in 4-week-old versus 1-day-old mouse brains). We confirmed by RT-PCR (Fig. (Fig.3B)3B) that TRAF4 mRNA expression was decreased in the brains of both mock-infected and Sindbis virus-infected 4-week-old mice compared to the brains of 1-day-old mice. In addition, RT-PCR analysis confirmed a developmental increase in CNS fractalkine mRNA expression in 4-week-old mock- and dsTE12Q-infected brains. Furthermore, immunohistochemical analyses with an anti-fractalkine antibody demonstrated that fractalkine immunoreactivity was more intense in the brains of 4-week-old compared to 1-week-old mice (see Fig. Fig.3C3C for representative photomicrographs). The fractalkine immunostaining appeared to localize predominantly to neurons, a finding that is consistent with previous reports (47, 61, 68).
The GeneChip expression data, combined with the use of independent methods to evaluate developmental regulation of CNS expression apoptosis-related molecules, demonstrates that two proapoptotic molecules—caspase-3 and TRAF4—show increased expression in neonatal mouse brain and one neuroprotective chemokine, fractalkine, shows increased expression in weanling mouse brain. These molecules are “candidate” regulators of age-dependent susceptibility to virus-induced neuronal apoptosis.
The host inflammatory gene response is more robust in the brains of Sindbis virus-infected neonatal mice than weanling mice.
We found that dsTE12Q infection in 1-day-old mice altered CNS gene expression to a greater extent than dsTE12Q infection in 4-week-old mice (Table (Table2).2). The expression of very few genes decreased by fivefold or more in response to dsTE12Q infection in either neonatal mouse brains (n = 3) or weanling mouse brains (n = 1). However, expression levels of 93 genes increased by ≥5-fold in virus-infected neonatal mouse brains, and expression levels of 45 genes increased by ≥5-fold in virus-infected weanling mouse brains. Of the genes with increased expression in neonatal mouse brains, more than half (n = 51) had significantly greater levels of expression in direct comparisons of infected neonatal compared to infected weanling mouse brains. These results indicate that dsTE12Q infection in neonatal mouse brains results both in an increased number of host genes with altered CNS expression, as well as greater magnitudes of increased gene expression compared to dsTE12Q infection in weanling mouse brain.
The list of host genes that are upregulated in dsTE12Q-infected 1-day-old and 4-week-old infected brains (Table (Table2)2) overlaps substantially with our previous findings comparing CNS gene expression in 10-day-old mice infected with a lethal and nonlethal strain of Sindbis virus (25). The major categories of genes with altered expression in response to Sindbis virus infection include genes encoding molecules involved in antigen presentation and protein degradation, complement proteins, chemokines, lipocalins, and IFN-regulated proteins. The most marked increases (>20-fold) in gene expression were observed in dsTE12Q-infected neonatal brains in several MCH class I molecules (22.4- to 66.3-fold), lymphocyte antigen LY-6.2 (64.9-fold), the lipocalins lipocalin 2 (37.3-fold) and apolipoprotein (39.8-fold), the protein degradation molecules ubiquitin specific protease 18 (35.2-fold) and Lmp7 (60.2-fold), cyclophilin C-associated protein (41.4-fold), and the chemokines MCP-1 (24.6-fold) and RANTES (35.4-fold). In addition, expression was increased by more than 20-fold of 14 genes that are either induced by IFN and/or involved in IFN signaling, including IFN-inducible proteins involved in GTP binding or GTPase activity, glucocorticoid attenuated response genes, the IFN regulatory factor IRF-7, the well-characterized antiviral molecule 2′-5′ oligoadenylate synthetase, and IFN-inducible genes of an as-of-yet uncharacterized function. This latter group includes an IFN-inducible ubiquitin-like 15-kDA protein, ISG15, which was the gene with the greatest magnitude of induction in our study (121.2-fold) and also the IFN-inducible gene with the greatest magnitude of induction in GeneChip expression analyses of cytomegalovirus infection in fibroblast cells (150-fold) (77).
For almost all of the genes classified in categories with established roles in the host response to viral infection (i.e., molecules involved in antigen presentation and protein degradation, complement proteins, chemokines, and IFN pathway-related proteins), greater increases were observed in the brains of dsTE12Q-infected neonatal compared to weanling mice (Table (Table2).2). Although it is possible that such genes may play a role in host pathology and contribute to increased disease severity in younger mice, it is also possible that they are induced to a greater extent in neonatal mouse brains as part of a host protective response to a greater viral burden. Therefore, their pattern of expression in neonatal versus weanling mouse brains does not provide significant insight into determining potential candidate regulators of age-dependent susceptibility to Sindbis virus infection.
However, among the IFN-inducible genes, there was one notable exception to the general pattern of higher gene expression in neonatal mouse brains. Only one gene, an EST similar to human ISG12 (herein referred to as mouse ISG12), increased by a greater magnitude in the brains of 4-week-old mouse brains (49.2-fold) compared to 1-week old mouse brains (20.5-fold). To validate this finding obtained in our GeneChip expression analysis, we performed both Northern blot analysis of the pooled RNA samples used for GeneChip expression analysis and RT-PCR analysis of RNA samples from individual mouse brains harvested at serial time points after an independent infection (Fig. (Fig.4).4). Northern blot analysis demonstrated undetectable levels of mouse ISG12 expression in both mock-infected 1-day-old and 4-week-old mice, and an ~2-fold-increase in mouse ISG12 expression in the brains of dsTE12Q-infected 4-week-old compared to dsTE12Q 1-day-old mice (Fig. (Fig.4A).4A). Using RT-PCR analysis, low levels of mouse ISG12 were detected in mock-infected brains, but the level of induction of mouse ISG12 expression was also greater in 4-week-old compared to 1-day-old infected brains (Fig. (Fig.4B).4B). Thus, GeneChip expression analysis, Northern blot analysis, and RT-PCR analysis all confirm a unique pattern of ISG12 expression characterized by a greater magnitude of induction in the brains of dsTE12Q-infected 4-week-old mice that have decreased viral loads than in the brains of infected neonatal mice that have increased viral loads. This pattern suggests the possibility that increased upregulation of mouse ISG12 in response to Sindbis virus infection plays a protective role in weanling mice.
Enforced neuronal expression of mouse ISG12 results in a significant delay in Sindbis virus-induced death in neonatal mice.
To evaluate whether increased expression of mouse ISG12 exerts protective effects in Sindbis virus infection, we used the Sindbis virus vector system to study the effects of enforced neuronal mouse ISG12 expression in neonatal mouse brain on the natural history of lethal CNS Sindbis virus infection. First, we confirmed that endogenous ISG12 is a neuronally expressed gene by performing RT-PCR to detect ISG12 mRNA in primary cultured rat DRG neurons (data not shown). Next, we performed 5′ and 3′ RACE with a mouse brain cDNA library to obtain the sequence of the full-length mouse ISG12 cDNA; the predicted amino acid sequence of mouse ISG12 is shown in Fig. Fig.5A5A (GenBank accession no. AY090098). Mouse ISG12 encodes a 91-amino-acid, highly hydrophobic protein with a predicted molecular mass of 8 kDa. BLAST alignment reveals 65% identity and 79% similarity with human ISG12 (formerly known as p27 or IFN-inducible 11.5-kDa protein) (77) and 62% identity and 79% similarity with the recently identified rat homolog, IFN-regulated gene, IRG1 (39).
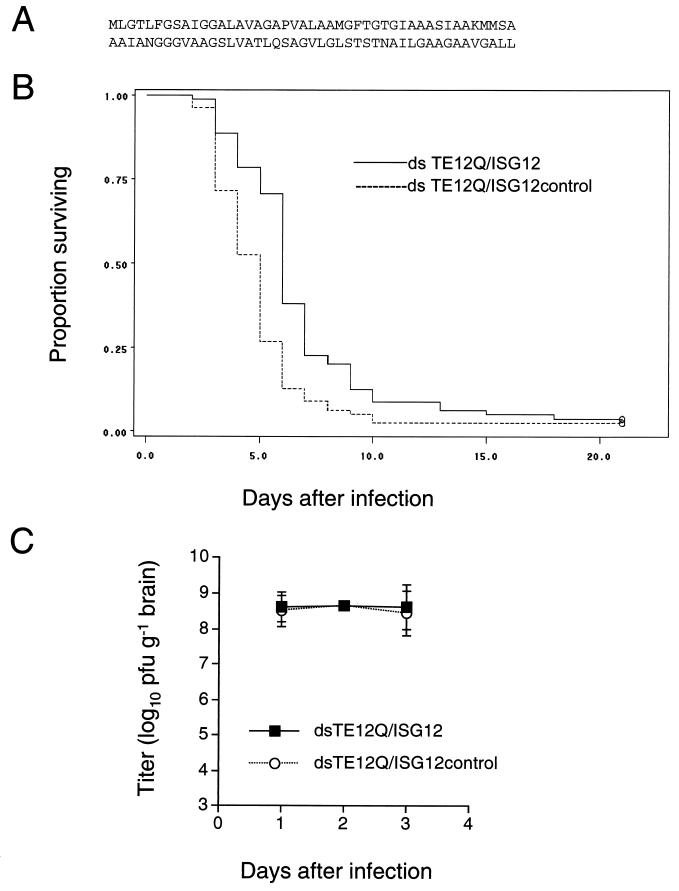
Deduced amino acid sequence of mouse ISG12 (GenBank accession no. AY090098) (A), Kaplan-Meier survival curves of 1-day-old mice infected intracerebrally with 1,000 PFU of dsTE12Q/ISG12 or dsTE12Q/ISG12control (B), and viral growth curves in mouse brains infected with dsTE12Q/ISG12 or dsTE12Q/ISGcontrol. The results in panel B represent combined survival curves for eight independent litters with 10 to 12 mice per litter. Similar differences in survival curves were observed in each independent litter. Significance of survival difference between dsTE12Q/ISG12- and dsTE12Q/ISG12 control-infected mice is P = 0.0001 (log-rank sum test). The results in panel C represent the geometric mean viral titers of five to six mice per time point per virus group.
We then cloned flag epitope-tagged mouse ISG12 and a control construct of ISG12 lacking the start codon into dsTE12Q to generate the recombinant chimeric viruses, dsTE12Q/ISG12 and dsTE12Q/ISG12control. Western blot analysis of BHK cell lysates infected with dsTE12Q/ISG12 with an anti-flag antibody confirmed expression of a flag epitope-tagged 8-kDa protein and immunohistochemical staining of mouse brains with an anti-flag antibody confirmed flag-ISG12 expression in similar regions of the brain that stained positive for Sindbis virus antigen (data not shown). After intracerebral infection of 1-day-old mice with dsTE12Q/ISG12, the average survival time was ca. 7 days (6.96 ± 0.434), whereas the average survival time was decreased to ca. 5 days (5.09 ± 0.348) after infection with the control virus, dsTE12Q/ISG12control. Kaplan-Meier analysis of the survival probabilities of dsTE12Q/ISG12 versus dsTE12Q/ISG12control-infected mice demonstrated a significant protective effect of ISG12 expression (P = 0.0001, log-rank sum test) (Fig. (Fig.5B).5B). The mechanism by which enforced neuronal ISG12 expression delays Sindbis virus-induced death is unknown, since we observed no significant differences in viral growth curves (Fig. (Fig.5C),5C), Sindbis virus antigen staining, or TUNEL apoptotic nuclei labeling in the brains of dsTE12Q/IS12 compared to dsTE12Q/IS12control-infected mice (data not shown). Nonetheless, the ability of ISG12 overexpression to significantly delay death in neonatal Sindbis virus-infected mice suggests that higher endogenous levels of induction of ISG12 expression in the brains of Sindbis virus-infected weanling mice may contribute to age-dependent resistance to lethal CNS disease.
DISCUSSION
In this study, we used GeneChip microarray technology to analyze gene expression patterns in the brains of neonatal mice that are susceptible to fatal Sindbis virus infection and weanling mice that develop asymptomatic infection. We found that numerous genes are developmentally regulated in mouse brains, including three genes, caspase-3, TRAF4, and fractalkine that influence neuronal susceptibility to apoptosis. Moreover, in response to infection the brains of neonatal mice have greater alterations in host inflammatory gene expression than the brains of weanling mice, in parallel with increased viral growth, increased apoptosis, and increased neurovirulence. Only one gene, an IFN-inducible gene of unknown function, mouse ISG12, was induced by Sindbis virus infection to greater levels in the brains of weanling mice than neonatal mice. Enforced expression of this gene in neonatal mouse brain resulted in a significant delay in Sindbis virus-induced death, suggesting that this may be a novel host protective factor that is induced at higher levels in older mice and a candidate regulator of age-dependent resistance to Sindbis virus infection.
Several previous reports have shown a correlation between neuronal maturation and decreased susceptibility to virus infection and virus-induced apopotosis (reviewed in references 8 and 34). In the present study, we identify specific genes that have differential expression in the brains of neonatal mice that are highly susceptible to lethal infection and weanling mice that are resistant to lethal infection. While further studies will be required to evaluate the role of developmental regulation of specific genes in determining age-dependent susceptibility to Sindbis virus infection, two categories of genes emerge from our study as interesting candidates.
The first category includes genes that are upregulated with CNS development and also selectively upregulated in response to Sindbis virus infection in neonatal but not weanling mice, since such a pattern of expression could be consistent with that predicted for a gene involved in host protection against CNS viral infection. The only gene within this category in our study was apolipoprotein D, a lipocalin that showed 26.1-fold-increased expression in weanling versus neonatal mock-infected brains, 39.8-fold-increased expression in Sindbis virus versus mock-infected neonatal mouse brains, and no change in expression in Sindbis virus versus mock-infected weanling mouse brains. Although the precise physiologic function of apolipoprotein D is unknown, several studies have described increased expression of apolipoprotein D mRNA and/or protein expression at sites of nerve regeneration after structural or excitotoxic injury (44, 51, 56, 67), as well as in stressed cortical neurons in Alzheimer's disease patients (2, 66). Consequently, lipid metabolism involving apoplipoprotein D is postulated to play a role in neuronal repair in response to different stress stimuli. Similar to our observations in mouse brain, electron microscopic immunocytochemical analyses have also demonstrated increased postnatal expression of apolipoprotein D in 10-day-old and 20-day-old rat brain compared to neonatal rat brain (52). Viewed together with our GeneChip expression data, these observations raise the possibility that apolipoprotein D also plays a protective role in neurons in response to viral infection and that the postnatal increase in basal levels of apolipoprotein D expression could contribute to the resistance of older mice to virus-induced injury.
Given the well-established role of apoptosis in the pathogenesis of Sindbis virus and other neurotropic virus infections (reviewed in references 8 and 34), developmentally regulated genes that influence neuronal susceptibility to apoptosis constitute a second category of candidate molecular determinants of age-dependent resistance to lethal Sindbis virus infection. Most notably, we found a marked developmental downregulation of the brain mRNA expression of caspase-3, a downstream caspase in both the “extrinsic” and “intrinsic” cell death pathways (reviewed in reference 55), which plays an important role in brain apoptosis (30; reviewed in reference 72). Our findings are consistent with other reports describing a reduction in caspase-3 levels with mouse (3) and rat (45, 63, 75) brain maturation. Decreased levels of caspase-3 expression with increased postnatal age may contribute not only to age-related resistance to Sindbis virus-induced neuronal apoptosis but also to age-related resistance to apoptosis induced by other neurotropic viruses and by other neurotoxic stimuli, such as hypoxia-ischemia and excitotoxic injury (4, 10, 21, 26, 54). The previous finding that the cowpox-encoded caspase inhibitor, CrmA, reduces mortality in Sindbis virus-infected neonatal mice (46) is also consistent with the hypothesis that caspases are important determinants of lethality in neonatal infected mice. However, caspase-3 itself is not absolutely required for neonatal lethality, since we have found that neonatal caspase-3 knockout mice also succumb to fatal disease (data not shown). This observation could reflect the existence of alternate, caspase-3-independent pathways of neuronal death such as those reported in the spinal cords of older mice infected with a neuroadapted strain of Sindbis virus (18) and do not necessarily rule out a role for downregulation of caspase-3 mRNA expression in age-related resistance to Sindbis virus-induced apoptosis.
Two other putative regulators of apoptosis, TRAF4 and fractalkine, were also developmentally regulated in a pattern that is consistent with increased susceptibility of younger mice to Sindbis virus-induced apoptosis. The neurotrophin receptor, p75NTR, induces NF-κB activation and apoptosis through interactions with TRAF family members, and TRAF4 binds to the type II cell death domain of p75NTR, blocking NF-κB activation and suppressing the dimerization-induced inhibition of p75NTR-mediated apoptosis (76). Therefore, if the p75 signaling pathway is involved in neuronal apoptosis triggered by Sindbis virus infection, increased expression of TRAF4 in neonatal mouse brain could contribute to increased susceptibility to virus-induced apoptosis. Interestingly, in mouse development studies, TRAF4 expression is noted predominantly during ontogenesis of the central and peripheral nervous system, preferentially in postmitotic undifferentiated neurons, and remains expressed in the adult hippocampus and olfactory bulb, which are structures that contain multipotential cells responsible for neoneurogenesis (41). In the present study and in our previous study of Sindbis virus neurotropism and Sindbis virus-induced apoptosis in neonatal mice (36), we found extensive viral antigen staining and neuronal apoptosis in the olfactory bulb, hippocampus, and other regions containing neuronal progenitor cells, such as the supraventricular zone. Thus, TRAF4 expression and susceptibility to Sindbis virus infection may be correlated in neonatal mice.
Fractalkine is a unique chemokine, distinguished structurally by the presence of a CX3C motif, as well as transmembrane-spanning and mucin-like domains, and shows atypical constitutive expression in a number of nonhematopoeitic tissues, including brain (reviewed in reference 17). Within rodent brain, fractalkine expression is mostly in neurons and highest in the olfactory bulb, cerebral cortex, hippocampus, caudate putamen and nucleus accumbens (47, 61). Fractalkine has been shown to block human immunodeficiency virus (HIV) gp120-induced apoptosis in cultured hippocampal neurons (27, 42), protect cultured neurons against the neurotoxin platelet activating factor and the HIV regulatory gene product Tat (68), and to inhibit TNF secretion and prevent neuronal death induced by lipopolysaccharide-activated microglia (78). In addition, in hippocampal neurons, fractalkine lowers the frequency of spontaneous, glutamergic excitatory postsynaptic currents, reduces voltage-dependent Ca2+ currents, and activates the transcription factor CREB (42), which is involved in protection against neuronal apoptosis induced by trophic factor deprivation (58). The neuroprotective effects of fractalkine against HIV gp120-induced apoptosis are thought to involve activation of the protein kinase, AKT, a major component of prosurvival signaling pathways, through a recently identified neuronal CX3CR1 chemokine receptor (43).
Given the neuroprotective effects of fractalkine (which is neuronally expressed and acts in both a paracrine and autocrine fashion), we speculate that increased expression of fractalkine in older mouse brain compared to neonatal mouse brain may contribute to resistance to Sindbis virus-induced neuronal apoptosis. We attempted to address this hypothesis by using the previously described Sindbis virus dsTE12Q vector system to express fractalkine in an enforced manner in neonatal infected neurons; however, such studies were not technically feasible due to the large size of the fractalkine insert and lack of neurovirulence in neonatal mice of the control chimeric virus containing a noncoding construct of fractalkine. Further studies are therefore planned to evaluate Sindbis virus pathogenesis in fractalkine and fractalkine receptor knockout mice.
In addition to identifying genes that are developmentally regulated in the brain and candidate regulators of age-dependent susceptibility to Sindbis virus infection, the present study confirms and extends our previous report describing the relationship between viral replication, virus-induced apoptosis, neurovirulence, and CNS gene expression. Previously, we compared CNS gene expression in 10-day-old mice with fatal Sindbis virus infection (strain dsTE12H) and with asymptomatic Sindbis virus infection (strain dsTE12Q) and found that fatal infection was associated with greater changes in host inflammatory response genes than asymptomatic infection (25). Similarly, in the present study, we found that fatal dsTE12Q infection in neonatal mice was associated with greater changes in host inflammatory response gene expression than asymptomatic dsTE12Q infection in weanling mice. Of note, neonatal mice infected with the dsTE12Q strain (which is virulent in 1-day-old but not 10-day-old mice) in the present study had greater levels of induction of host inflammatory gene expression than did 10-day-old mice infected with the neurovirulent strain, dsTE12H, in our previous study. This finding suggests that the magnitude of the host gene response to infection observed in the previous study relates in a general manner to neurovirulence but is not specific to the E2-55 Q→H mutation that renders Sindbis virus neurovirulent in older mice (71).
Moreover, the results of the present study suggest that the specific nature of the host gene response to virulent Sindbis virus is stereotypic, since the majority of genes with expression increased by ≥5-fold in dsTE12Q-infected neonatal mouse brains also had increased expression in dsTE12H-infected 10-day-old mouse brains. These similarities help confirm the consistency of changes in expression of a specific subset of host inflammatory response genes across different models of Sindbis virus neurovirulence and support the notion that specific microbial pathogens may produce characteristic host gene response patterns that are detectable by microarray analysis. Furthermore, they identify specific genes that are markedly upregulated in different models of neurovirulence that are likely to play an important role in the host response to Sindbis virus infection (e.g., the chemokines MCP-1 and RANTES; the IFN-inducible genes ISG15 and GARG-39, -49, and -16; the lipocalins, apoplipoprotein D and lipocalin 2; and cyclophilin C).
We focused further attention on one specific IFN-inducible gene, ISG12, that had greater expression in Sindbis virus-infected weanling mouse brains than in neonatal mouse brains, despite the presence of ca. 10- to 50-fold more virus in the brains of Sindbis virus-infected neonatal mice. Our findings indicate that enforced neuronal ISG12 expression resulted in a significant delay in mortality in neonatal mice, providing the first evidence for a role of this protein in antiviral host defense. The mechanism by which mouse ISG12 delays Sindbis virus-induced death is unknown and, to our knowledge, no studies have been performed with ISG12-like proteins in other species that elucidate potential functions of these proteins. Human ISG12 was originally cloned as an estrogen-induced gene in the human breast epithelial cell line, MCF7, and originally designated p27 (57). A subsequent study demonstrated that ISG12 is strongly induced by IFN-α and only slightly induced by IFN-γ in a panel of seven different human cell lines (11). In addition to increased expression in human breast carcinoma cells, increased ISG12 mRNA expression has also been detected in cardiac and skeletal muscle cells from cardiomyopathic hamsters (5) and in the human and rat endometrium on different days of the oestrous cycle (39). Among IFN-inducible proteins, ISG12 is unique in that it is the only protein found thus far to localize to the nuclear envelope (40). However, the function of ISG12 in the nuclear envelope has not yet been explored. Our findings that ISG12 exerts protective effects during lethal Sindbis virus infection and that ISG12 is the only IFN-regulated gene with greater expression in weanling than neonatal virally infected mouse brains, suggests that further studies are warranted to define the molecular mechanisms by which this protein functions in IFN-regulated host responses to viral infections.
Acknowledgments
We thank the Microarray Facility of the Albert Einstein College of Medicine for assistance in performing Affymetrix GeneChip exression analysis and Daniel Heitjan of the Department of Biostatistics at Columbia University for assistance with data analysis.
This work was supported by NIH grants R29AI40246 and RO1 AI44157 and an Irma T. Hirshl Trust Scholar Award to B.L.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.76.22.11688-11703.2002
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc136759?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Effect of Viral Strain and Host Age on Clinical Disease and Viral Replication in Immunocompetent Mouse Models of Chikungunya Encephalomyelitis.
Viruses, 15(5):1057, 26 Apr 2023
Cited by: 5 articles | PMID: 37243143 | PMCID: PMC10220978
Genomic Evidence for the Nonpathogenic State in HIV-1-Infected Northern Pig-Tailed Macaques.
Mol Biol Evol, 40(5):msad101, 01 May 2023
Cited by: 1 article | PMID: 37134013 | PMCID: PMC10182734
Antiviral response within different cell types of the CNS.
Front Immunol, 13:1044721, 15 Nov 2022
Cited by: 3 articles | PMID: 36458002 | PMCID: PMC9706196
Review Free full text in Europe PMC
Immature Brain Cortical Neurons Have Low Transcriptional Competence to Activate Antiviral Defences and Control RNA Virus Infections.
J Innate Immun, 15(1):50-66, 23 Jun 2022
Cited by: 2 articles | PMID: 35738238 | PMCID: PMC10643910
Network meta-analysis of transcriptome expression changes in different manifestations of dengue virus infection.
BMC Genomics, 23(1):165, 27 Feb 2022
Cited by: 3 articles | PMID: 35220956 | PMCID: PMC8882220
Go to all (77) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 165 of 165)
- (3 citations) ENA - AA145371
- (2 citations) ENA - L32751
- (2 citations) ENA - X01697
- (2 citations) ENA - X14194
- (2 citations) ENA - X00349
- (2 citations) ENA - M37761
- (2 citations) ENA - AA255186
- (2 citations) ENA - D89076
- (2 citations) ENA - L10244
- (2 citations) ENA - U89269
- (2 citations) ENA - AY090098
- (2 citations) ENA - U70674
- (2 citations) ENA - L39123
- (1 citation) ENA - U18797
- (1 citation) ENA - M31314
- (1 citation) ENA - M18184
- (1 citation) ENA - L20899
- (1 citation) ENA - AA060835
- (1 citation) ENA - M13444
- (1 citation) ENA - J04953
- (1 citation) ENA - M13685
- (1 citation) ENA - M77831
- (1 citation) ENA - U05245
- (1 citation) ENA - X97281
- (1 citation) ENA - U43084
- (1 citation) ENA - U43085
- (1 citation) ENA - U55060
- (1 citation) ENA - W41070
- (1 citation) ENA - X00496
- (1 citation) ENA - M29881
- (1 citation) ENA - M74570
- (1 citation) ENA - U47329
- (1 citation) ENA - U44731
- (1 citation) ENA - AA152671
- (1 citation) ENA - M33863
- (1 citation) ENA - Y00305
- (1 citation) ENA - M55637
- (1 citation) ENA - W53443
- (1 citation) ENA - J03750
- (1 citation) ENA - X64068
- (1 citation) ENA - X51829
- (1 citation) ENA - R74641
- (1 citation) ENA - X83562
- (1 citation) ENA - U73037
- (1 citation) ENA - W97022
- (1 citation) ENA - AA255253
- (1 citation) ENA - AA098307
- (1 citation) ENA - AA072252
- (1 citation) ENA - M35244
- (1 citation) ENA - AA638539
- (1 citation) ENA - X56602
- (1 citation) ENA - AA415898
- (1 citation) ENA - W15873
- (1 citation) ENA - AA170251
- (1 citation) ENA - X00246
- (1 citation) ENA - AA175784
- (1 citation) ENA - D14566
- (1 citation) ENA - AA049662
- (1 citation) ENA - X03479
- (1 citation) ENA - M83219
- (1 citation) ENA - D63902
- (1 citation) ENA - X01838
- (1 citation) ENA - M69070
- (1 citation) ENA - L48687
- (1 citation) ENA - X16133
- (1 citation) ENA - M83218
- (1 citation) ENA - L24118
- (1 citation) ENA - U11860
- (1 citation) ENA - U53219
- (1 citation) ENA - U19582
- (1 citation) ENA - M11284
- (1 citation) ENA - X02892
- (1 citation) ENA - U29396
- (1 citation) ENA - W20873
- (1 citation) ENA - AA013783
- (1 citation) ENA - AA139510
- (1 citation) ENA - M31418
- (1 citation) ENA - K00811
- (1 citation) ENA - M69069
- (1 citation) ENA - X81627
- (1 citation) ENA - V00727
- (1 citation) ENA - M63244
- (1 citation) ENA - U31908
- (1 citation) ENA - AA673405
- (1 citation) ENA - X91144
- (1 citation) ENA - U19119
- (1 citation) ENA - M19681
- (1 citation) ENA - U50631
- (1 citation) ENA - J00423
- (1 citation) ENA - J03776
- (1 citation) ENA - AA277082
- (1 citation) ENA - Z70023
- (1 citation) ENA - R75354
- (1 citation) ENA - AA065915
- (1 citation) ENA - AA175767
- (1 citation) ENA - X52264
- (1 citation) ENA - U44088
- (1 citation) ENA - C80656
- (1 citation) ENA - X61800
- (1 citation) ENA - U92565
- (1 citation) ENA - W12941
- (1 citation) ENA - J05287
- (1 citation) ENA - X67783
- (1 citation) ENA - AA163876
- (1 citation) ENA - J04596
- (1 citation) ENA - M12279
- (1 citation) ENA - U06924
- (1 citation) ENA - M13366
- (1 citation) ENA - AA033381
- (1 citation) ENA - AA145127
- (1 citation) ENA - AA170444
- (1 citation) ENA - L00606
- (1 citation) ENA - U02298
- (1 citation) ENA - U05683
- (1 citation) ENA - AA516966
- (1 citation) ENA - AA002526
- (1 citation) ENA - X15373
- (1 citation) ENA - Z30940
- (1 citation) ENA - J02623
- (1 citation) ENA - Z12297
- (1 citation) ENA - M17440
- (1 citation) ENA - M33266
- (1 citation) ENA - U10120
- (1 citation) ENA - AA059662
- (1 citation) ENA - X67809
- (1 citation) ENA - M63170
- (1 citation) ENA - AA273574
- (1 citation) ENA - D14883
- (1 citation) ENA - D86232
- (1 citation) ENA - M21531
- (1 citation) ENA - X59382
- (1 citation) ENA - AA288442
- (1 citation) ENA - R74735
- (1 citation) ENA - AA638530
- (1 citation) ENA - AA408420
- (1 citation) ENA - AA727845
- (1 citation) ENA - R75030
- (1 citation) ENA - AA170668
- (1 citation) ENA - AA286391
- (1 citation) ENA - L32973
- (1 citation) ENA - AA177851
- (1 citation) ENA - L32974
- (1 citation) ENA - W08455
- (1 citation) ENA - AA153021
- (1 citation) ENA - AA521958
- (1 citation) ENA - AA638086
- (1 citation) ENA - X15591
- (1 citation) ENA - U50712
- (1 citation) ENA - C78546
- (1 citation) ENA - X53825
- (1 citation) ENA - AB004879
- (1 citation) ENA - L22144
- (1 citation) ENA - X56304
- (1 citation) ENA - AA166452
- (1 citation) ENA - K01496
- (1 citation) ENA - X70393
- (1 citation) ENA - M64086
- (1 citation) ENA - M21952
- (1 citation) ENA - U59807
- (1 citation) ENA - D85561
- (1 citation) ENA - L38444
- (1 citation) ENA - V00755
- (1 citation) ENA - U22031
- (1 citation) ENA - AA266791
- (1 citation) ENA - X06368
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Identification of genes involved in the host response to neurovirulent alphavirus infection.
J Virol, 75(21):10431-10445, 01 Nov 2001
Cited by: 65 articles | PMID: 11581411 | PMCID: PMC114617
Sindbis Virus Can Exploit a Host Antiviral Protein To Evade Immune Surveillance.
J Virol, 90(22):10247-10258, 28 Oct 2016
Cited by: 16 articles | PMID: 27581990 | PMCID: PMC5105660
Fatal Sindbis virus infection of neonatal mice in the absence of encephalitis.
Virology, 224(1):73-83, 01 Oct 1996
Cited by: 51 articles | PMID: 8862401
Age-dependent susceptibility to fatal encephalitis: alphavirus infection of neurons.
Arch Virol Suppl, 9:31-39, 01 Jan 1994
Cited by: 37 articles | PMID: 8032263
Review
Funding
Funders who supported this work.
NIAID NIH HHS (3)
Grant ID: K23 AI079394
Grant ID: R01 AI44157
Grant ID: R29AI40246




