Abstract
Free full text

A ligand for the aryl hydrocarbon receptor isolated from lung
Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-inducible transcription factor that is best known because it mediates the actions of polycyclic and halogenated aromatic hydrocarbon environmental toxicants such as 3-methylcholanthrene and 2,3,7,8-tetrachlorodibenzo-p-dioxin. We report here the successful identification of an endogenous ligand for this receptor; ≈20 μg was isolated in pure form from 35 kg of porcine lung. Its structure was deduced as 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester from extensive physical measurements and quantum mechanical calculations. In a reporter gene assay, this ligand activates the AHR with a potency five times greater than that of β-naphthoflavone, a prototypical synthetic AHR ligand. 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester competes with 2,3,7,8-[3H]tetrachlorodibenzo-p-dioxin for binding to human, murine, and fish AHRs, thus showing that AHR activation is caused by direct receptor binding, and that recognition of this endogenous ligand is conserved from early vertebrates (fish) to humans.
The Ah locus was first defined in inbred mice as a strain difference in response to treatment with polycyclic aromatic hydrocarbons such as 3-methylcholanthrene (1). The evidence for the presence of a receptor encoded by the locus was then provided by characterizing the strain-specific and stereo-specific binding sites in mouse liver cytoplasm (2). The coding sequence for the receptor, a soluble protein named aryl hydrocarbon receptor (AHR), was eventually identified and characterized (3). The receptor is found in many vertebrates including fish. The AHR is a ligand-inducible transcription factor, a member of a basic helix–loop–helix/Per-Arnt-Sim (bHLH/PAS) family. On binding to xenobiotic ligands, the AHR has been shown to regulate the expression of a variety of genes including those encoding for cytochrome P450 enzymes. In addition, receptor activation has been linked to alterations in cell proliferation, apoptosis, adipose differentiation, tumor promotion, immune function, vitamin A status, development, and reproductive functions (4–9). The generation of AHR-deficient mice also points to possible physiological functions of the receptor in liver, heart, ovary, and vascular and immune systems (7, 10–12).
The AHR is best known because polycyclic aromatic hydrocarbons, such as 3-methylcholanthrene and benzo[α]pyrene, and halogenated aromatic hydrocarbons, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), serve as ligands (1, 13). Although the ability of AHR to bind a variety of xenobiotic ligands is of great interest, it is clear that the AHR did not evolve to respond to manufactured chemicals. It is reasonable to suspect that endogenous ligands must exist for the AHR. Recently, two human urinary products were isolated that bind to the AHR (14). Whether those products are physiological AHR ligands or not is undetermined because the identified compounds are indigo, a commonly used fabric dye, and indirubin, an isomer of indigo. For the past few years we have labored under the assumption that endogenous ligands exist for the AHR in tissues of higher vertebrates. We have now succeeded in isolating from porcine lung tissue a ligand for AHR and have unequivocally identified it as 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE) by UV and IR spectrophotometry, MS, extensive NMR studies, and quantum mechanical calculations.
Materials and Methods
UV Spectroscopy.
About 3 μg of the ligand in 200 μl of methanol/water (60:40, vol/vol) was injected into a cyano HPLC column (Adsorbosphere CN-AQ 5U, i.d. = 4.6 mm, Alltech Associates) equilibrated with 60% methanol in water at a flow rate of 0.5 ml/min. Compounds eluted from the column passed through the flow cell (path length, 10 mm; i.d., 0.009 inches) of a Waters 996 photodiode array detector. The UV absorbance of the mobile phase was automatically set to zero. UV absorption spectra were recorded with MILLENNIUM 3.2 software (Waters) at a rate of one spectrum per s. The spectrum acquired at the moment of the highest UV absorption is shown.
Electron Impact MS.
A new glass probe was inserted into a MS50TC Ultrahigh Resolution Mass Spectrometer (Kratos Analytical Instruments), with electron energy of 70 eV and ion source temperature of 150°C. The instrument and probe were at first operated in this mode until a satisfactory background was achieved. A blank spectrum of the residue of column effluent eluting at the retention time of the putative ligand was recorded. The sample (≈200 ng) was then introduced on the same probe and the spectrum was taken. Any signal common to both the blank and the sample spectrum was subtracted generating the spectrum reported. A KRATOS DS-55 data acquisition system was used to record all spectroscopic data.
Fourier Transform (FT)-IR Spectroscopy.
One microgram of the ligand in methanol was spotted onto a Teflon STI IR Card (Thermo Spectra-Tech, Shelton, CT). The sample was applied to the card in such a way that the area through which the smallest diameter IR beam passes was kept at a minimum to maximize the sample path length with limited supply of material. The solvent was evaporated and the card was placed in the sample chamber of an Infinity 60 AR FT-IR Spectrometer (Thermo-Mattson, Madison, WI). The spectrum was acquired with WINFIRST 3.00 software (resolution, 2; scan number, 200; Iris opening, 1%; window, KBr). A blank spectrum was generated by using the residue from the HPLC effluent as described for MS.
One-Dimensional NMR Spectroscopy.
Two micrograms of the ligand in specified solvents was placed in either a 2.5-mm NMR sample tube (Wilmad–Labglass, Buena, NY) or a 5-mm Shigemi tube (Shigemi, Allison Park, PA). The magnetic susceptibility of the Shigemi tube was adjusted to that of the solvent. The standard 1H NMR, single-frequency decoupling, and single frequency nuclear Overhauser effect difference spectra were obtained on Bruker DMX-750 and DMX-500 spectrometers (Bruker, Billerica, MA). The spectrum of a blank produced as described for MS was also recorded.
Two-Dimensional NMR Spectroscopy.
Twelve micrograms of the ligand in deuterated methanol was placed in a 5-mm Shigemi tube (Shigemi). The two-dimensional 1H{13C} heteronuclear single quantum correlation (HSQC) and heteronuclear multiple-bond correlation (HMBC) experiments were performed on the DMX-500 equipped with a 5-mm triple-resonance, single-axis gradient Cryoprobe (Bruker). In the case of the HMBC experiment, the direct peaks were not suppressed and a refocusing 180° pulse was applied to both 1H and 13C resonances at the midpoint of the antiphase buildup period to refocus static field inhomogeneities.
Quantum Mechanical Calculations.
Proposed structures were constructed by using the GAUSSVIEW program (Gaussian, Carnegie, PA). These models were subjected to full, unconstrained geometry optimization at the B3LYP/6–31g* level of theory by using the JAGUAR quantum chemical program (Schrodinger, Portland, OR). The absence of imaginary frequencies assured that the models were in a local ground state. Calculated frequencies were scaled by using the Modified Scaled Quantum Mechanical Force Fields (SQM) method on the optimized structures (15). Isotropic chemical shifts were calculated from optimized models by using the GAUSSIAN suite of quantum chemical programs. The Gauge Including Atomic Orbital method at the B3LYP/6–311++g(2d,2p) level of theory as implemented in GAUSSIAN 98 was used for the calculations. Calculated chemical shifts were calibrated versus experimental chemical shifts of some model compounds and the theoretical values were scaled empirically (16–18) by using the following linear equations: experimental shift (1H, ppm) = calculated shift × 0.95 + 0.26; experimental shift (13C, ppm) = calculated shift × 0.95 + 0.59. The R2 values for the linear regressions were 0.999 for both 1H and 13C fits. Expected deviations of the calibrated theoretical chemical shifts from experimental values are ≈0.15 ppm (16) for 1H and ≈3 ppm for 13C (18). The theoretical proton and carbon shifts of tetramethysilane were set to zero.
Coupling constants were calculated by the single-finite-perturbation method (19). The magnitude of the perturbation was 0.02 atomic units, which is within the linear region of response, and the calculation was performed at the B3LYP/6–311+g* level of theory. In this method, only the Fermi contact contribution to the scalar coupling is available.
Identities of Protons and Carbons in Structure Analysis.
For the convenience of identification of atoms in the compound, protons and carbons are named after their chemical shifts. For example, a proton with a chemical shift of 9.25 ppm will be named as 9.25 proton and, similarly, a carbon of 141.05 ppm as 141 carbon.
Cell Culture and Reporter Gene Assay.
The cell line H1L1.1C2 (20) was maintained in a culture medium (DMEM + 10% FBS) with G418 at a concentration of 400 μg/ml and incubated at 37°C in 6% CO2. A 96-well, sterile cell culture plate with about 30,000 cells per well in 200 μl of culture medium without G418 was prepared and incubated for at least 12 h before dosing with AHR ligands in DMSO (0.5% in culture medium) for the reporter gene assay. The cells were incubated for an additional 4 h after dosing. The cells were washed twice with PBS (pH 6.9) and lysed by adding 50 μl of Cell Culture Lysis Reagent (Promega). Lysate from each well (10 μl) was transferred into a corresponding well of a Microlite 2 plate (Dynex Technologies, Chantilly, VA). A MLX Microtiter Plate Luminometer (Dynex Technologies) was programmed to inject 50 μl of a luciferase substrate solution (Promega) into each well, and the light intensities in relative light units generated were recorded (for 5 s after a 1-s delay) and averaged.
Receptor Binding Studies.
Aryl hydrocarbon receptors (human, murine, killfish, and zebrafish) were expressed by in vitro transcription and translation by using the TnT Quick-coupled Reticulocyte Lysate Systems reaction (Promega). Receptor ligand binding was studied by velocity sedimentation on sucrose gradients (21). The transcription and translation reactions were diluted 8-fold, divided into 100-μl aliquots containing 1/4 reaction each, and incubated overnight at 4°C with [3H]TCDD (2 nM) ± ITE at the concentrations shown. Incubation mixtures were fractionated (150 μl per fraction) on gradients of 10–30% sucrose. For competition binding studies, transcription and translation expressed murine AHR was incubated with [3H]TCDD (2 nM) in the absence or presence of increasing concentrations of unlabeled ITE, β-naphthoflavone (BNF), or TCDD. Bound ligand was separated from that free in solution by adsorption to hydroxylapatite (22) and washing by using a filter manifold (23).
Results
Extraction and Purification of the Ligand.
In preliminary work with rats, lung seemed to have the highest concentration of AHR ligand activity. By using 35 kg of porcine lung tissue as a source of ligand, we isolated 20 μg of ligand (flow chart shown in Fig. Fig.11a). Material (20 μg) was estimated on the basis of the reporter gene assay using BNF as the standard. This estimate assumes that the isolated ligand has the same activity as BNF, which proved to be a good approximation. After the final purification, an HPLC analytical column revealed a single substance (Fig. (Fig.11b).
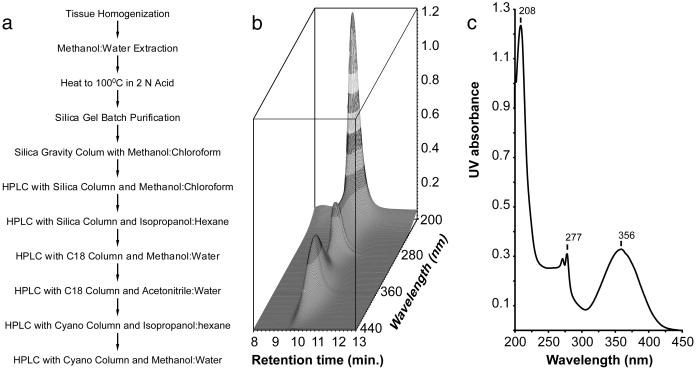
AHR ligand purification and UV spectrum. (a) Flow chart showing the steps taken in purifying the ligand. (b) The photodiode array representation of an HPLC chromatogram of the purified AHR ligand (3 μg). x axis, retention time in minutes; y axis, wavelengths monitored in nanometers; z axis, UV absorbance. (c) UV spectrum of the AHR ligand. The spectrum was taken at an HPLC retention time of ≈10 min, at which moment the UV absorbance reached the maximum. x axis, wavelengths in nm; y axis, UV absorbance.
Chemical Identification of the Ligand.
The UV absorption spectrum of the ligand revealed a maximum at 356 and another double maximum at 277 nm (Fig. (Fig.11c). The absorption spectrum suggests a highly conjugated structure. High-resolution electron impact MS revealed a molecular ion of 286.0399 giving an empirical formula of C14H10N2O3S (Fig. (Fig.22a; Table Table1).1). The presence of sulfur is evidenced by the appearance of an isotope cluster that shows the characteristic abundance of M + 2 ion heavily contributed by 34S. With only 10 protons, a highly condensed multiringed compound was expected. The mass fragmentation analysis also revealed a base fragment at 144.0453 or C9H6NO. Clearly the sulfur-containing moiety was lost during the fragmentation to its most stable ion.
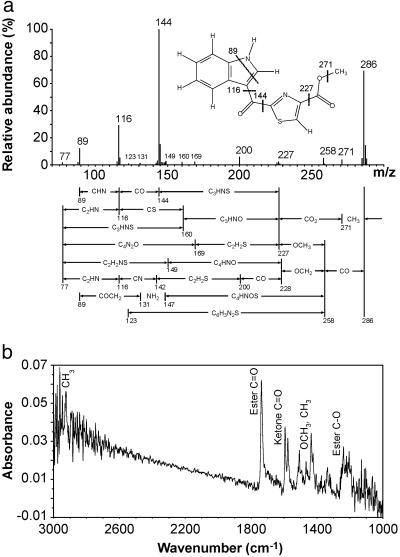
Electron impact mass and Fourier transform-IR spectra. (a) High-resolution electron impact mass spectrum of the AHR ligand. x axis, the ratio of mass to charge, m/z; y axis, relative abundance of ions. The ligand structure showing the formation of the major fragments is presented. Proposed fragmentation pathways are as mapped under the spectrum. (b) Fourier transform-IR spectrum of the AHR ligand. x axis, the frequencies of IR beam expressed as wavenumber (cm−1). The region from 1,000 to 3,000 cm−1 is shown here. y axis, the absorbance. Signals with clear attribution are as labeled.
Table 1.
Principal high-resolution electron impact mass fragments of the AHR ligand
ligand
| Mass | Abundance, % | Composition | Deviation |
|---|---|---|---|
| 288.0414 | 4.22 | C14H10N2O334S | 14.93 |
| 287.0441 | 14.21 | 13C112C13H10N2O3S | −2.79 |
| 286.0399 | 69.19 | C14H10N2O3S | −4.89 |
| 285.0361 | 4.64 | C14H9N2O3S | 9.23 |
| 145.0495 | 12.50 | 13C112C8H6NO | 9.80 |
| 144.0453 | 100.00 | C9H6NO | 2.22 |
| 143.0383 | 2.89 | C9H5NO | 8.04 |
| 117.0550 | 5.13 | 13C112C7H6N | 11.28 |
| 116.0503 | 28.98 | C8H6N | 1.90 |
| 115.0435 | 2.02 | C8H5N | 10.87 |
 89.0393 89.0393 | 12.00 | C7H5 | 1.68 |
 88.0401 88.0401 | 1.80 | C7H4 | 99.74 |
Other fragments will be reported in detail elsewhere.
Our attention next shifted to NMR spectroscopy in an attempt to characterize the nature of the protons and their connectivity relationships to carbons in the molecule (Fig. (Fig.3).3). Three protons are found at 3.99 ppm as a singlet and are methyl protons. Of particular importance is the presence of a carboxylic methyl ester group. Besides the high-resolution MS and the fragmentation analysis revealed in Fig. Fig.22a and Table Table1,1, the presence of a methyl ester group was revealed by the elimination of biological activity when the ligand was treated with 0.1 M KOH. The activity was largely restored by methylation with diazomethane (data not shown), strongly suggesting the presence of an ester group. The presence of a methyl ester functionality is also revealed from the IR spectrum with bands at 1,737 cm−1 (ester C O) and around 1,205 cm−1 (ester C—O) (Fig. (Fig.22b). Furthermore, both HSQC and HMBC data support the presence of a methyl ester (Fig. (Fig.33b).
O) and around 1,205 cm−1 (ester C—O) (Fig. (Fig.22b). Furthermore, both HSQC and HMBC data support the presence of a methyl ester (Fig. (Fig.33b).
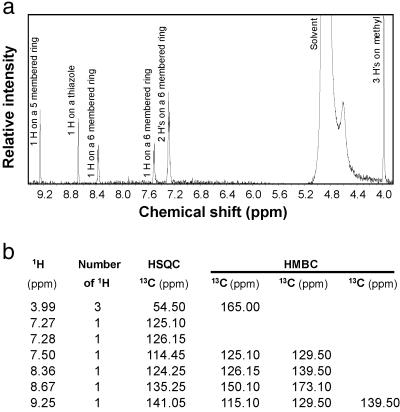
Proton NMR spectrum and proton-carbon-13 correlations. (a) 1H NMR spectrum of the AHR ligand in d-methanol. The peak labeled Solvent is contributed from NMR solvent. No signal existed in the solvent masked area confirmed by examining a spectrum taken in d-chloroform (data not shown). Signals contributed from numbers and locations of the protons are as labeled. (b) Table of 1H-13C correlation of the AHR ligand from both HSQC and HMBC spectra. The protons and carbon-13s with detectable connections are listed in the same row. The carbon-13s listed under a column headed HSQC are the ones directly connected (through one bond) to the corresponding proton(s) (in the same row), whereas the ones under the columns headed HMBC are connected to the corresponding proton(s) through 2–3 bonds.
Four protons (7.27, 7.28, 7.50, and 8.36 protons) must be attached to a six-membered aromatic ring judging from their chemical shifts, results of decoupling and nuclear Overhauser effect experiments (data not shown), and both HSQC and HMBC data (Fig. (Fig.33b). This ring, in turn, is fused with a five-membered aromatic ring on which the 9.25 proton is attached (from HMBC data). The five-membered ring must be heterocyclic, containing either a nitrogen or an oxygen because the 144 base fragment in the mass spectrum is composed of C9H6NO. However, the oxygen cannot be a component of the five-membered ring because the chemical shifts of the two carbons directly connected to the oxygen would have larger shifts than observed in the HSQC and HMBC spectra. A nitrogen, therefore, must be the component in the five-membered ring resulting in an indole structure. The NMR signals of the proton (11.33 ppm) attached to the indole nitrogen and the nuclear Overhauser effect response of its neighboring (7.50 and 9.25) protons to it were visible when d-acetone instead of d-methanol was used as NMR solvent (data not shown). To satisfy the composition of C9H6NO of the 144 base fragment in the mass spectrum, one of the positions on the five-membered ring must be substituted with either a C O or C—O moiety. Considering the fact that both the 9.25 and 8.36 protons (Fig. (Fig.3)3) are highly deshielded, a group must be located between the 9.25 and 8.36 protons. Thus, the 9.25 proton must be located between the nitrogen and the substituted carbon, further confirming the indole structure. The C—O or C
O or C—O moiety. Considering the fact that both the 9.25 and 8.36 protons (Fig. (Fig.3)3) are highly deshielded, a group must be located between the 9.25 and 8.36 protons. Thus, the 9.25 proton must be located between the nitrogen and the substituted carbon, further confirming the indole structure. The C—O or C O moiety could be a C—O in a six-membered ring, which would not deshield its neighboring protons, or an attached ketone, which will highly deshield its neighboring (9.25 and 8.36) protons. The presence of a ketone on a carbon attached to the indole nucleus is revealed by the IR spectrum providing a band at 1,593 cm−1 (Fig. (Fig.22b), characteristic of ketones on a carbon in the vicinity of an indole as revealed by model compounds such as 3-benzoylindole [see Integrated Spectral Data Base System (SDBS) for Organic Compounds, National Institute of Advanced Industrial Science and Technology, Tsukuba, Ibaraki, Japan; www.aist.go.jp/riodb/sdbs/; ref. 24], (cyclohex-1-en-1-yl)-3-indolylmethanone, and 3-(E-2-methylbut-2-enoyl)indole (24). Those same models also displayed a unique feature of forming, by losing other fragments, the same stable 144 (C9H6NO) mass fragment composed of indole and the attached ketone. This ketone is attached to another five-membered heteroaromatic ring (either thiazole or isothiazole).
O moiety could be a C—O in a six-membered ring, which would not deshield its neighboring protons, or an attached ketone, which will highly deshield its neighboring (9.25 and 8.36) protons. The presence of a ketone on a carbon attached to the indole nucleus is revealed by the IR spectrum providing a band at 1,593 cm−1 (Fig. (Fig.22b), characteristic of ketones on a carbon in the vicinity of an indole as revealed by model compounds such as 3-benzoylindole [see Integrated Spectral Data Base System (SDBS) for Organic Compounds, National Institute of Advanced Industrial Science and Technology, Tsukuba, Ibaraki, Japan; www.aist.go.jp/riodb/sdbs/; ref. 24], (cyclohex-1-en-1-yl)-3-indolylmethanone, and 3-(E-2-methylbut-2-enoyl)indole (24). Those same models also displayed a unique feature of forming, by losing other fragments, the same stable 144 (C9H6NO) mass fragment composed of indole and the attached ketone. This ketone is attached to another five-membered heteroaromatic ring (either thiazole or isothiazole).
Considering the published (25) proton and carbon chemical shifts of thiazole and isothiazole and the fact that the two other positions will be substituted by two groups with deshielding power, only the 5-position (numbering from S to N) can be considered for the 8.67 proton attachment in the case of the thiazole structure. With the 2- and 4-positions attached to the ketone and the methyl ester, respectively, and vice versa, there will be two possible structures based on thiazole. Similar to the thiazole situation, there are two possible structures based on isothiazole because only the 4-position can be considered for the 8.67-proton attachment.
The 1H-13C coupling constant for the 8.67 proton to its directly attached carbon obtained from the HMBC spectrum is ≈193 Hz. We investigated the coupling constants between the position 5 proton and carbon in the thiazole and the position 4 proton and the carbon in isothiazole. Faure et al. (26) and Tseng (27) reported that the coupling constant between the position 5 proton and carbon in thiazole is ≈191 Hz and the range is from 188 to 200 with different substitutions at the other positions, while the one between the position 4 proton and carbon in isothiazole is ≈173 Hz and the range is from 168 to 171 with different substitutions at the other positions. Furthermore, we calculated the theoretical coupling constants for both thiazole- and isothiazole-based structures. The theoretical coupling constant between the 8.67 proton and its connected carbon in the thiazole structure is 185.4 Hz, whereas that in the isothiazole-based structure is 175.7 Hz. Judging from both theoretical coupling constants and experimental values in the literature, the isothiazole structures were eliminated. Of the thiazole structures, one is derived from cysteine in which the position 2 of thiazole is substituted with a ketone and position 4 a methyl ester. The other is a noncysteine-related structure with a reversed order of substitution.
Finally, the chemical shift theoretical calculations show that the cysteine-related structure best fits the experimental values. The standard deviation of the theoretical chemical shifts from the experimental shifts was 2.07 ppm for the carbon nuclei and 0.098 ppm for the proton. This deduction is strongly supported by experimental evidence reported in the literature. When an aldehyde (equivalent to a ketone in this case) is connected to the 2-position of the thiazole, the chemical shift of carbon 2 is 166 ppm and carbon 4 is 146 ppm (28). When a methyl ester is attached to the 4-position of the thiazole, the chemical shift of carbon 4 is 146 ppm and the carbon 2 (with a methyl group attached) is 167 ppm (29). We can, therefore, confidently deduce that when an aldehyde or ketone is attached to carbon 2 and the methyl ester to carbon 4 of a thiazole at the same time, similar to the arrangement of the cysteine-related structure, the chemical shifts of carbon 2 and 4 would be 166 and 146, respectively, which agrees with both calculated and measured chemical shifts for the structure. Similar analysis and calculations led to the elimination of the noncysteine-related structure. Thus, the cysteine-related structure was deduced as 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester, which we abbreviate as ITE (Fig. (Fig.44a). To be sure of the structure, chemical synthesis was accomplished (to be reported elsewhere). As shown in Fig. Fig.5,5, the synthetic material exactly comigrates with the isolated ligand on HPLC. Further, the biological response curves using the reporter gene assay are identical for synthetic and isolated ligand (Fig. (Fig.5).5).
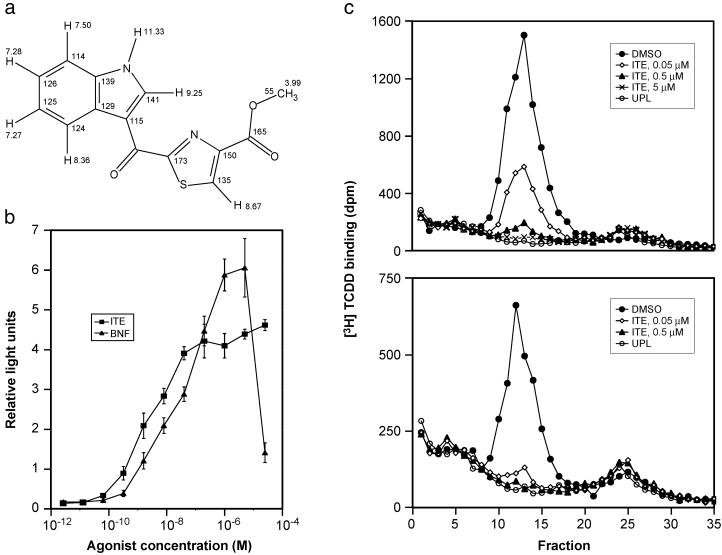
The structure and biochemical characterization of ITE. (a) The structure of the endogenous AHR ligand and its proton and carbon-13 chemical shifts (same as their identification numbers). (b) Dose–response curves for ITE and BNF. The ability of ITE and BNF to activate a DRE-reporter gene construct is shown (mean ± SD). (c) Competition of ITE for specific [3H]TCDD binding to in vitro-synthesized murine (Upper) and human (Lower) AHRs as analyzed by velocity sedimentation on sucrose density gradients. UPL refers to unprogrammed lysate (i.e., in vitro transcription and translation in the absence of AHR expression plasmid), which serves as a measure of nonspecific binding in the velocity sedimentation analysis.

Comparison of isolated vs. synthetic ITE. An HPLC chromatograph of a mixture of natural (≈20 ng) and synthetic (≈20 ng) ITE is shown. HPLC condition: C18 analytical column (4.6 × 250 mm), methanol/water (70:30 vol/vol); flow rate = 0.5 ml/min; λ = 356 nm; x axis, retention time. y axis, UV absorbance. (Insets) Dose–response curves (mean ± SD) generated by the natural (Left) and synthetic (Right) ITE, relative right units (ordinate) vs. agonist concentration.
Biochemical Characterization of the Ligand.
Fig. Fig.44b indicates that ITE is about five times more potent than BNF in reporter gene activity after 4 h of incubation. The agonist activity of ITE through the AHR was demonstrated by the ability of an AHR antagonist, 3′-methoxy-4′-nitroflavone (30), to block the reporter gene response to ITE in a dose-dependent fashion (data not shown).
Competitive binding studies show that ITE inhibits binding of [3H]TCDD to the AHR. Sucrose density gradient analyses demonstrated that ITE not only competes effectively for binding to the human and murine AHRs (Fig. (Fig.44c), but is equally effective in competing for [3H]TCDD binding to AHRs from the killifish Fundulus heteroclitus (AHR1 and AHR2) and the zebrafish Danio rerio (data not shown). Competition for [3H]TCDD (2 nM) binding to the murine AHR by using a range of ITE, BNF, and TCDD concentrations resulted in nearly equivalent binding affinities for ITE and BNF, whereas TCDD showed slightly higher binding affinity (Ki values for ITE, BNF, and TCDD of 3, 2, and 0.5 nM, respectively; data not shown). Thus, ITE is a ligand that binds directly to the AHR to induce receptor activity.
Although ITE is similar to BNF in affinity for binding to the AHR, it is more potent (≈5-fold) than BNF in transactivating the DRE-containing reporter gene construct in Hepa1c1c7 cells. The enhanced activity of ITE in transactivation could be caused by greater stability or access to the nucleus, or alternatively, ITE may induce a change in receptor conformation that is more condusive to the binding of comodulatory proteins.
Discussion
Although it is likely that more than one endogenous ligand will be found for the AHR, we have now demonstrated the existence of one of these, which we obtained from lung tissue. The ability of acid treatment of methanolic extracts to render the material chloroform soluble suggests that the compound may exist as a higher-order complex in tissue. Release of ITE from this complex can also be accomplished in the absence of methanol ruling out transesterification during the workup. A minor concern is whether our acid treatment could generate ITE from inactive components in the methanolic extract. We are unable to conceive any reaction catalyzed by 2 N acid in aqueous methanol that could yield ITE, but we cannot totally exclude it. Furthermore, the fact that ITE is so active argues for its existence before extraction.
Several groups have reported the identification of endogenous ligands for the AHR. The closest example is the indigo-related compounds isolated from human urine (14). However, because these compounds were isolated from urine, the question of whether they represent urinary excretion products from an exogenous source or were generated from endogenous compounds remains unanswered. Similarly, the lipoxin A(4) (31), bilirubin-related compounds (32), and tryptophan-related compounds (indole and tryptamine) (33) are certainly endogenous but whether they are the true physiological ligands for AHR has not yet been resolved.
The origin of ITE is very likely from tryptophan and cysteine by an expected condensation reaction. Of course, this must be investigated in vitro and in vivo. Even if the two fragments arise from the two amino acids, exactly how it occurs will be of considerable interest. Furthermore, whether the production of this compound is regulated and in what way can now be approached experimentally. The biological function of ITE is difficult to ascertain at this moment. Based on the effects of xenobiotics that bind AHR, a diverse group of functions for ITE can be envisioned. With the availability of this ligand, physiological functions of the AHR can now be directly investigated.
Acknowledgments
We thank Mr. Rowland Randall, Department of Biochemistry, University of Wisconsin, for carrying out the mass spectral work; Dr. Darrell McCaslin of the Biophysics Laboratory in the Department of Biochemistry, University of Wisconsin, for the help in Fourier transform-IR measurement; and Diana G. Franks of the Biology Department, Woods Hole Oceanographic Institution, for assistance with the receptor-binding assays. We are also grateful to Dr. M. S. Denison, University of California, Davis, for the reporter gene system. This work was supported by the Wisconsin Alumni Research Foundation, the University of Wisconsin Sea Grant Institute, and the National Institutes of Health. NMR studies were carried out at the National Magnetic Resonance Facility at Madison supported by the National Institutes of Health Biomedical Technology Program, National Science Foundation Academic Infrastructure Program, National Institutes of Health Shared Instrumentation Program, National Science Foundation Biological Instrumentation Program, and U.S. Department of Agriculture. This is contribution no. 10717 from the Woods Hole Oceanographic Institution.
Abbreviations
AHR, aryl hydrocarbon receptor
TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin
ITE, 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester
BNF, β-naphthoflavone
HSQC, heteronuclear single quantum correlation
HMBC, heteronuclear multiple bond correlation
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.232562899
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc137481?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.232562899
Article citations
Telmisartan potentiates the ITE-induced aryl hydrocarbon receptor activity in human liver cell line.
Arch Toxicol, 31 Oct 2024
Cited by: 0 articles | PMID: 39480548
The role of the AHR in host-pathogen interactions.
Nat Rev Immunol, 16 Oct 2024
Cited by: 0 articles | PMID: 39415055
Review
The aryl hydrocarbon receptor agonist ITE reduces inflammation and urinary dysfunction in a mouse model of autoimmune prostatitis.
Am J Clin Exp Urol, 12(4):149-161, 25 Aug 2024
Cited by: 0 articles | PMID: 39308590 | PMCID: PMC11411176
Microparticles incorporating dual apoptotic factors to inhibit inflammatory effects in macrophages.
J Pharm Sci, 113(11):3196-3205, 08 Jun 2024
Cited by: 1 article | PMID: 38852674
Timing influences the impact of aryl hydrocarbon receptor activation on the humoral immune response to respiratory viral infection.
Toxicol Appl Pharmacol, 489:117010, 18 Jun 2024
Cited by: 0 articles | PMID: 38901696
Go to all (159) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
An endogenous aryl hydrocarbon receptor ligand enhances de novo generation of regulatory T cells in humans.
J Leukoc Biol, 105(2):291-295, 30 Nov 2018
Cited by: 6 articles | PMID: 30500079
TCDD and a putative endogenous AhR ligand, ITE, elicit the same immediate changes in gene expression in mouse lung fibroblasts.
Toxicol Sci, 114(1):90-100, 19 Nov 2009
Cited by: 34 articles | PMID: 19933214 | PMCID: PMC2819971
ITE, an endogenous aryl hydrocarbon receptor ligand, suppresses endometrial cancer cell proliferation and migration.
Toxicology, 421:1-8, 03 Apr 2019
Cited by: 8 articles | PMID: 30953668
The Aryl Hydrocarbon Receptor in Immunity: Tools and Potential.
Methods Mol Biol, 1371:239-257, 01 Jan 2016
Cited by: 22 articles | PMID: 26530806
Review
Funding
Funders who supported this work.
NIEHS NIH HHS (2)
Grant ID: R01 ES006272-13
Grant ID: R01 ES006272





