Abstract
Free full text

Ubiquitin-dependent lysosomal degradation of the HNE-modified proteins in lens epithelial cells
Abstract
4-Hydroxynonenal (HNE), a highly reactive lipid peroxidation product, may adversely modify proteins. Accumulation of HNE-modified proteins may be responsible for pathological lesions associated with oxidative stress. The objective of this work was to determine how HNE-modified proteins are removed from cells. The data showed that αB-crystallin modified by HNE was ubiquitinated at a faster rate than that of native αB-crystallin in a cell-free system. However, its susceptibility to proteasome-dependent degradation in the cell-free system did not increase. When delivered into cultured lens epithelial cells, HNE-modified αB-crystallin was degraded at a faster rate than that of unmodified αB-crystallin. Inhibition of the lysosomal activity stabilized HNE-modified αB-crystallin, but inhibition of the proteasome activity alone had little effect. To determine if other HNE-modified proteins are also degraded in a ubiquitin-dependent lysosomal pathway, lens epithelial cells were treated with HNE and the removal of HNE-modified proteins in the cells was monitored. The levels of HNE-modified proteins in the cell decreased rapidly upon removal of HNE from the medium. Depletion of ATP or the presence of MG132, a proteasome/lysosome inhibitor, resulted in stabilization of HNE-modified proteins. However, proteasome-specific inhibitors, lactacystin-β-lactone and epoxomicin, could not stabilize HNE-modified proteins in the cells. In contrast, chloroquine, a lysosome inhibitor, stabilized HNE-modified proteins. The enrichment of HNE-modified proteins in the fraction of ubiquitin conjugates suggests that HNE-modified proteins are preferentially ubiquitinated. Taken together, these findings show that HNE-modified proteins are degraded via a novel ubiquitin and lysosomal-dependent but proteasome-independent pathway.
Oxidative stress is known to cause various forms of damage to most types of cells, including protein oxidation, DNA breaks, and lipid peroxidation (1-3, 4, 5). The 4-hydroxynonenal (HNE), generated by peroxidation of polyunsaturated fatty acids, such as arachidonic and linoleic acids, is the most reactive and cytotoxic product of lipid peroxidation (6, 7). HNE modification is also frequently used as a marker of lipid peroxidation and oxidative stress in vivo (7). Lysine, histidine, and cysteine residues in proteins and nucleophilic groups in DNA are all potential targets for HNE (8). Higher levels of HNE-modified proteins have been detected in animal and human tissues under certain pathological conditions, suggesting that accumulation of HNE-modified proteins is involved in the pathophysiology of degenerative diseases (9, 10) and cellular aging (11). It has also been shown that both exogenously added HNE and endogenously generated HNE can form adducts with various intracellular proteins (12, 13). Proteins modified by HNE may alter their structures and functions, and accumulation of such modified proteins may be cytotoxic. Thus, HNE-modified proteins must be removed to avoid disruption of the cellular functions.
The extent of accumulation of oxidatively modified proteins depends on the rate of production and the rate of degradation by proteases (1). It is known that, in most types of cells, intracellular proteolytic enzymes selectively degrade oxidized proteins (14, 15). Proteolytic capabilities are therefore considered to be secondary defense systems that can delay the accumulation of adversely modified proteins (1, 16, 17). The majority of intracellular proteins are degraded either by the lysosomal pathway or by the ubiquitin-proteasome pathway (UPP). The lysosome is responsible for the degradation of membrane or extracellular proteins that enter cells by endocytosis (18). The UPP is a major proteolytic pathway in eukaryotic cells, and it is responsible for conditional degradation of intracellular short-lived regulatory proteins or abnormal cytosolic and nuclear proteins (19-23). In the UPP, proteins targeted for proteasomal degradation are first ligated to multiple ubiquitin molecules. Polyubiquitination targets proteins for degradation by the 26S proteasome in an ATP-dependent manner (23, 24). Since both the formation of ubiquitin conjugates and subsequent degradation of the conjugates require ATP, a hallmark of the UPP is the ATP dependence. In some cases, ubiquitination of proteins, particularly membrane receptors, facilitate their internalization and degradation by the endosome-lysosome pathway (25).
We demonstrated previously that the UPP is involved in degradation of oxidized proteins in cell-free systems and within intact cells (26). In this study, we evaluated the role of the UPP in degradation of HNE-modified proteins. We found that the HNE-modified proteins were ubiquitinated at a faster rate than their nonmodified counterpart, but the susceptibility of HNE-modified proteins to proteasome-dependent degradation was not significantly different from that of unmodified proteins. Although depletion of ATP resulted in the accumulation of HNE-modified proteins in cells, proteasome-specific inhibitors had little effect on the degradation of HNE-modified proteins. In contrast, inhibition of the lysosomal activity stabilized the HNE-modified proteins. These data indicate that the HNE modified proteins are degraded by a novel ubiquitin-dependent lysosomal pathway.
MATERIALS AND METHODS
Materials
Na125I and 125I-protein A were supplied by PerkinElmer Life Science (Boston, MA). Nitrocellulose membrane and protein molecular mass standards were purchased from Bio-Rad (Richmond, CA). BioPORTER protein delivery reagent was purchased from Gene Therapy Systems (San Diego, CA). Carbobenzoxyl-leucinyl-leucinyl-leucinal-H (MG-132) and ubiquitin aldehyde (Ubal) were purchased from Bostonbiochem (Cambridge, MA). HNE was purchased from Cayman Chemical Company (Ann Arbor, MI). The antibodies to HNE-modified proteins were produced in Dr. L. Szweda's laboratory. Rabbit reticulocytes were purchased from PelFreez Biologicals (Rogers, AR). Ni-NTA magnetic agarose beads were purchased from Qiagen (Valencia, CA). Unless specified, all other reagents were purchased from Sigma (St. Louis, MO) and were the highest grade available.
Preparation of 125I-labeled proteins
Proteins were radioiodinated essentially as described previously (27). In brief, the protein (200 μg) was mixed with Na125I (200 μCi). The iodination reaction was started by addition of chloramine T, and the vials were shaken for 2 min at room temperature. Then the reaction was terminated by the addition of sodium metabisulfite and sodium iodide. Free 125I and small protein fragments were removed from the protein solution by Centricon-10 microconcentrators (Millipore Corporation, Bedford, MA).
Expression and purification of Ubc4
The cloned cDNA (2E isoform of mammalian Ubc4-1) was expressed in E. coli using the T7 promoter in the pET11d-expression vector (Novagen Inc., Madison, WI). The plasmid was generously provided by Dr. Simon Wing and was transformed into BL21 E. coli (Novagen Inc.). The expression and purification of Ubc4-1 were performed as described previously (28).
Modification of α-crystallin with HNE
The mixture of αA- and βB-crystallin was purified from bovine lenses as described previously (29). The recombinant αB-crystallin was expressed and purified as described previously (30). Native α-crystallins (2.5 mg/ml) were treated with 0-200 μM of HNE in phosphate-buffered saline (PBS), pH 7.4, at 37°C for 2 h. To determine the effects of HNE modification on the susceptibility to degradation, recombinant αB-crystallin was first iodinated as described above and the iodinated αB-crystallin was incubated with or without 100 μM HNE at 37°C for 2 h. Free HNE and free 125I were removed by centrifugation with Centricon-10 microconcentrators.
Proteolysis assays
Rabbit reticulocyte lysate was prepared as described previously by Ciechanover et al. (31). 125I-labeled αB-crystallins treated with or without 100 μM HNE were used as substrates for the degradation assay. ATP-dependent degradation was performed in 25 μl assays that contained 15 μl of rabbit reticulocyte lysate and 10 μl of buffer [50 mM Tris-HCl (pH 7.8) containing 5 mM MgCl2, 2 mM DTT, 2 mM ATP, 10 mM creatine phosphate, 4.5 μg creatine phosphokinase, 2 μg ubiquitin, and 2 μg 125I-labeled substrate]. To determine the ATP-independent degradation, ATP, creatine phosphate, and creatine phosphokinase were replaced with 30 mM 2-deoxyglucose. All these assays were supplemented with 0.5 μg recombinant Ubc4. MG132 was added to selected tubes at a final concentration of 80 μM to determine the involvement of the proteasome. The degradation was initiated by the addition of 3-5 χ 104 cpm of 125I-labeled α-crystallins (2 μg). The reaction was carried out at 37°C for 2 h and then stopped by the addition of 400 μl of 1% (w/v) bovine serum albumin, immediately followed by 100 μl of 100% (w/v) trichloroacetic acid (TCA). After standing on ice for 10 min, the samples were centrifuged 14,000 rpm at 4°C for 10 min. Aliquots of supernatant (400 μl) were counted to determine the TCA-soluble radioactivity. The amount of radioactivity in the pellet was also determined. The extent of degradation was determined as the percentage of 125I released as TCA-soluble fragments. Each assay was performed in duplicate.
Ubiquitin conjugation assays
Ubiquitin-protein conjugates were formed by incubation of 125I-labeled αB-crystallins with proteasome-free fraction II prepared from rabbit reticulocytes. Fraction II was prepared as described by Ciechanover et al. (31), and the proteasome was removed from fraction II by centrifuging at 100,000 g for 5 h (32). All ubiquitination assays were done in 25 μl containing 15 μl of proteasome-free Fraction II and 10 μl of reaction buffer (50 mM Tris, pH 7.8, 5 mM MgCl2, 2 mM DTT, 2 mM ATP, 10 mM creatine phosphate, 4.5 μg creatine phosphokinase, 2 μg of ubiquitin). For negative control, ubiquitni was omitted from the assay. The reaction was initiated by the addition of 3-5 χ 106 cpm of 125I-labeled substrates and terminated by addition of 25 μl of 2x Laemmli buffer (33) after incubation at 37°C for 1 h. Proteins in the reaction mixture were separated by SDS-PAGE and transferred to nitrocellulose. After autoradiography, the membrane was probed for ubiquitin conjugates with antibodies to ubiquitin. The immunoreactive bands were visualized using the SuperSignal Chemiluminescent Kit (Pierce, Rockford, IL). The relative amounts of ubiquitin-protein conjugates were quantified by scanning densitometry.
Degradation of HNE-modified proteins in lens epithelial cells
Human lens epithelial cells (HLEC, SRA 01/04 cell line) were established at Dr. V. Reddy's laboratory. The HLEC were cultured in Dulbeco's Modified Eagle's Medium (DMEM) containing 10% (v/v) fetal bovine serum, penicillin (100 units/ml), streptomycin (100 μg/ml), and amphotericin (250 ng/ml), at 37°C in a humidified incubator with 5% CO2. 125I-labeled αBcrystallins were delivered into HLEC using BioPORTER protein delivery reagent according to the instructions of the manufacturer. An equal amount 125I-labeled unmodified or HNE-modified αB-crystallins (5x104 cpm, ~1 μg αB-crystallins) were mixed with BioPORTER protein delivery reagent and added to each dish of HLEC (35 mm dish). After incubation for 2 h, the medium was removed and the cells were washed with PBS twice to remove any 125I-αB-crystallins that did not enter the cells. Then the cells were incubated for another 4 h in the absence or presence of MG132 (5 μM), clasto-lactacystin-β-lactone (10 μM), or chloroquine (100 μM). Then the cells were collected and proteins were resolved by SDS-PAGE. Levels of 125I-labeled αB-crystallins remained in the cells were determined by autoradiography.
To determine the removal of intracellular HNE-modified proteins, subconfluent cultures were washed once gently with PBS and then incubated at 37°C for 1 h with 5 μM HNE in PBS containing 5 mM glucose, 0.3 mM CaCl2, and 0.62 mM MgCl2. Control cells were incubated in the same conditions and for the same period of time but without HNE. After treatment with HNE, cells were washed once with PBS and cultured in normal medium for 0-8 h. The cells were collected at the indicted time points with a cell scraper. The cell pellets were lysed in 10 mM Tris-HCl (pH 7.4), 1% Nonidet P-40, 0.1% SDS, 20 mM N-ethylmaleimide, and 2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride on ice for 30 min with occasional vortexing. After centrifugation (14,000 rpm, 15 min, 4°C), protein concentrations in the supernatants were determined with the modified method of Lowry, using the protein assay kit from Sigma according to the instructions of the manufacturer. Supernatants of the cells were resolved on 12% SDS gels and transferred to nitrocellulose membranes. After being blocked with 5% fat-free milk in TST (50 mM Tris-HCl, pH 7.4, 150 mM NaCl and 0.02% Tween 20), the membrane was probed with an antibody specific to HNE-modified proteins, followed by goat anti-rabbit IgG conjugated to horseradish peroxidase. The immunoreactive bands were visualized using the SuperSignal Chemiluminescent Kit (Pierce). The ATP depletion was achieved by incubation with 8 μM antimycin A and 20 mM 2-deoxyglucose (34).
Determination of HNE-modified proteins in the fraction of ubiquitin conjugates
To facilitate the isolation of ubiquitin conjugates, a recombinant adenovirus encoding an RGS(His)6-tagged ubiquitin was constructed (Shang et al., unpublished observations). Cultured HLEC were infected with an equivalent amount of the recombinant adenovirus or the control adenovirus for 48 h prior HNE treatment. After removal of HNE from the medium for 1 h, cells were collected and lysed in a buffer containing 8 M urea and 10 mM imidazole. The lysates were mixed with Ni-NTA magnetic agarose beads (Qiagen) for 1 h. This mixture was then layered onto a 50 μl column of Ni-NTA resin. After extensive wash with buffered 8 M urea containing 20-50 μM imidazole, the bound proteins were then eluted from the column using buffered 8 M urea containing 250 mM imidazole. Protein concentrations in the eluate were determined and resolved in SDS-PAGE. Levels of HNE-modified proteins in the eluate were determined by Western blot. An equal amount of proteins that bound to Ni-NTA resin nonspecifically (proteins from cells that do not express his-tagged proteins) was used as negative control.
RESULTS
Ubiquitination and degradation of HNE-modified protein in cell-free systems
Western blot analysis using polyclonal antibodies that specifically recognize HNE-modified proteins showed that α-crystallins were modified by HNE in a dose-dependent manner (Fig. 1). The molecular masses of the majority of these HNE-modified products were ~20 kDa, which corresponds to the molecular masses of the α-crystallin monomers. With increasing concentrations of HNE, a band at molecular mass ~40 kDa showed strong reaction with antibodies to HNE adducts. This 40 kDa HNE-reactive band comigrated with the dimer of α-crystallin. At higher HNE concentrations, some lower mass HNE adducts were also observed (Fig. 1). These lower molecular masses adducts are most likely the products of fragmentation of HNE-modified α-crystallins. Whereas the ratio of αA/αB was ~3:1 in the α-crystallin preparation (Fig. 1, lower panel), the levels of HNE modified αB-crystallin were ~3-fold higher than those of HNE-modified αA-crystallin (Fig. 1, upper panel). These data indicate that αB-crystallin is more susceptible than αA-crystallin to being modified by HNE. To test the susceptibility of HNE-modified α-crystallins to ubiquitination and degradation, we chose to use recombinant αB-crystallin treated with 100 μM HNE for 2 h. This level of HNE resulted in modification of the majority of αB-crystallin but left most of the lysine residues available for ubiquitination.
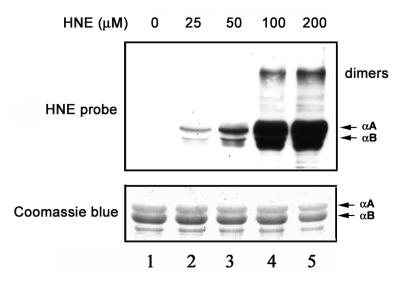
Formation of HNE-protein adducts. Lane 1 is α-crystallin not treated with HNE. Lanes 2-5 are α-crystallins exposed to 25, 50, 100, and 200 μM HNE, respectively, for 2 h at 37°C. Lower panel) Protein profiles of α-crystallins isolated from bovine lenses after incubation with HNE (Coomassie Blue staining). Upper panel) modification of α-crystallins by HNE as determined by Western blotting with antibodies to HNE-modified proteins.
To compare the susceptibilities of untreated αB-crystallins and HNE-modified αB-crystallins to being ubiquitinated, the untreated and HNE-modified αB-crystallins were incubated with ATP and Ubc4-supplemented proteasome-free fraction II in the presence or absence of 20 μM ubiquitin. Autoradiography shows that in the absence of ubiquitin, the αB-crystallins migrated as a single band (Fig. 2A, lanes 3 and 4). After incubation in the presence of ubiquitin, ~5% αB-crystallin migrated at a slower rate (Fig. 2A, lanes 1 and 2). This slower migrating band colocalized with a ubiquitinated protein (Fig. 2A, lanes 5 and 6), indicating that the slower migrating band was the mono-ubiquitinated form of αB-crystallin. Longer exposure of the film revealed that <1% of total αB-crystallin was polyubiquitinated (data not shown). Levels of ubiquitinated HNE-modified αB-crystallin were ~2-fold greater than those for untreated αB-crystallin (Fig. 2A, compare lane 2 with 1 and lane 6 with 5), indicating that HNE-modified αB-crystallin is a preferred substrate for ubiquitination.
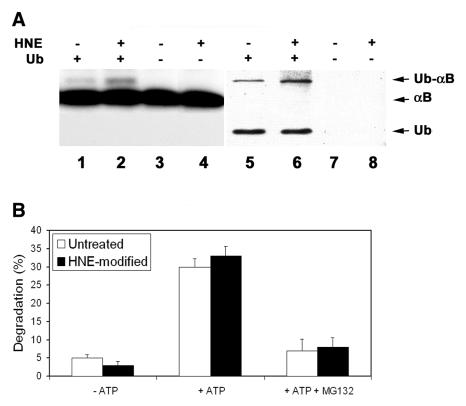
Ubiquitination and degradation of HNE-modified αB-crystallin. 125I-labeled recombinant αB-crystallin was incubated with or without 100 μM HNE for 2 h at 37°C . After removal of unreacted HNE, 125I-labeled αB-crystallins were incubated in proteasome-free fraction II prepared from rabbit reticulocyte lysate in the presence or absence of ubiquitin. A) Ubiquitination of HNE-modified αB-crystallin: Lanes 1, 3, 5, and 7 are unmodified αB-crystallin and lanes 2, 4, 6, and 8 are HNE-modified αB-crystallins. Lanes 1, 2, 5 and 6 show formation of ubiquitin conjugates in the presence of ubiquitin and lanes 3, 4, 7, and 8 are negative control after incubation without ubiquitin. Formation of ubiquitin-125I-labeled αB-crystallin conjugates was assessed by autoradiography following SDS-PAGE and transferring to nitrocellulose membrane (lanes 1-4), and the ubiquitin conjugates were confirmed by Western blotting with antibody to ubiquitin (lanes 5-8). B) degradation of HNE-modified αB-crystallin. Percent degradation was calculated from acid-soluble radioactivity recovered in supernatants after 2 h of incubation with reticulocyte lysate at 37°C. Values are means ± SD of 4 independent determinations; each was done in duplicate.
The susceptibility of the HNE-modified αB-crystallin to proteasome-dependent degradation was subsequently determined. During the 2 h of incubation with reticulocyte lysate, the ATP-independent degradation of untreated αB-crystallin and HNE-modified αB-crystallin was ~5 and 3%, respectively (Fig. 2B). If ATP was added to the degradation system, the degradation rates for untreated αB-crystallin and HNE-modified αB-crystallin were 30 and 33% during the same period of incubation (Fig. 2B). Furthermore, the presence of MG132 blocked the majority of ATP-dependent degradation of untreated α-crystallins and HNE-modified αB-crystallins (Fig. 2B), suggesting that the ATP-dependent degradation observed under these conditions was mainly proteasome dependent. It should be pointed out that although MG132 inhibits both the proteasome and the lysosome activity in cells, the lysosome enzymes are not active at the pH used in this assay. Therefore, the effects of MG132 under this experimental setting reflect the proteasome activity but not the lysosome activity. The above data indicate that both untreated and HNE-modified αB-crystallins are degraded by the ubiquitin-proteasome pathway. However, the difference of proteasome-dependent degradation between untreated and HNE-modified αB-crystallins was not statistically significant.
Degradation of HNE-modified proteins in HLEC
The increased susceptibility of HNE-modified proteins to being ubiquitinated without a significant alteration in their susceptibility to being degraded by the proteasome in the cell-free systems suggests that HNE-modified proteins may be degraded by a proteolytic pathway other than the proteasome pathway. To further determine how the HNE-modified proteins were removed from intact cells, 125I-labeled, untreated, or HNE-modified αB-crystallins were delivered into cultured HLEC using the BioPORTER protein delivery reagent. Both untreated and HNE-modified αB-crystallins were efficiently delivered into HLEC (~50% of the 125I-labeled crystallins), and the delivery efficiencies between untreated and HNE-modified αB-crystallins were comparable (data not shown). In the absence of protease inhibitors, both untreated and HNE-modified αB-crystallins were degraded rapidly, and it appears that HNE-modified αB-crystallin was degraded faster than unmodified B-crystallin (Fig. 3, compare lane 2 with 1). This result is consistent with our previous observations that native α-crystallins are rapidly degraded by the ubiquitin-proteasome pathway. In the presence of MG132, which inhibits both the proteasome and the lysosome, both untreated and HNE-modified αB-crystallins were stabilized (Fig. 3, lanes 3 and 4). To determine if the proteasome is involved in degradation of HNE-modified αB-crystallin in the cells, we tested the effect of a specific proteasome inhibitor (clasto-lactacystin-β-lactone) on levels of αB-crystallins in the cells. In contrast to MG132, clasto-lactacystin-β-lactone partially stabilized untreated αB-crystallin in the cells, but it did not stabilize the HNE-modified αB-crystallin (Fig. 3, compare lanes 5 and 6 with lanes 1 and 2, respectively). In the presence of lactacystin-β-lactone, the levels of untreated αB-crystallin that remained in the cells were much higher than those of HNE-modified αB-crystallin (Fig. 3, compare lane 5 with 6). This greater difference is due to the preferential stabilization of untreated αB-crystallin by the proteasome inhibitor. The inability of clasto-lactacystin-β-lactone, in contrast to MG132, to stabilize HNE-modified αB-crystallin suggests that HNE-modified αB-crystallin was degraded by the lysosome pathway, since MG132 not only inhibits the proteasome, it also inhibits cathepsins in the lysosome. To confirm the involvement of the lysosome in degradation of HNE-modified α -crystallin, we determined the effect of chloroquine (a lysosome inhibitor) on the stability of HNE-modified αB-crystallin in the cells. Indeed, HNE-modified αB-crystallins were partially stabilized in the presence of chloroquine (Fig. 3, lanes 7 and 8) and the levels of untreated and HNE-modified αB-crystallins were comparable under these conditions, indicating that HNE-modified αB-crystallin was preferentially degraded by the lysosomal pathway. If clasto-lactacystin-β-lactone and chloroquine were added together to the cells, both untreated and HNE-modified αB-crystallins were stabilized and levels of crystallins remaining in the cells were similar to those observed in the presence of MG132 (Fig. 3, compare lanes 9 and 10 with lanes 3 and 4). Longer exposure of the film revealed a small fraction (<1%) of 125I-labeled αB-crystallin migrated at a slower rate. Based on the molecular weight, the slower migrating moiety appears to be mono-ubiquitinated αB-crystallin (Fig. 3, upper panel). This putative mono-ubiquitinated αB-crystallin was more readily detectable in the presence of MG132 (Fig. 3, upper panel, lanes 3 and 4) or in the presence of both lactacystin-β-lactone and chloroquine (Fig. 3, lanes 9 and 10). Taken together, these data indicate that untreated αB-crystallin is a good substrate for the ubiquitin-proteasome pathway, but HNE-modified αB-crystallin is a preferred substrate for the lysosome pathway.
Effects of different protease inhibitors on the stability of HNE-modified αB-crystallin delivered into lens cells. αB-crystallin was iodinated and modified by HNE as described in Fig. 2. 125I-αB-crystallins were delivered into cultured human lens epithelial cells using the BioPORTER protein delivery reagent. Levels of 125I-αB-crystallins (middle panel) and the putative mono-ubiquitnated 125I-αB-crystallins (upper panel) were determined after incubation in the absence or presence of indicated inhibitors for 4 h. Autoradiograms were scanned, and levels of 125I-αB-crystallins in the cells were quantified with a densitometer (lower panel). Data in lower panel are means ± SD of 3 independent experiments.
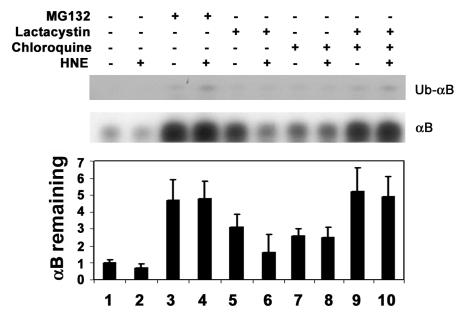
To determine if the ubiquitin-dependent lysosomal degradation of HNE-modified αB-crystallin pertains to other HNE-modified proteins, HLEC were treated with HNE and the effects of proteasome and lysosome inhibitors on the removal of HNE-modified proteins were determined. After 1 h of treatment with 5 μM HNE, several of the cellular proteins formed adducts with HNE (Fig. (Fig.44--6,6, lane 2). Upon removal of HNE from the medium, levels of the HNE-modified proteins decreased. After 24 h of recovery in HNE-free medium, the levels of HNE-modified proteins returned almost to the levels observed in untreated cells, indicating that HNE-modified proteins are rapidly removed from the cells (Fig. 4, compare lanes 3, 5, and 7 with lane 2). To explore which of proteolytic pathways are involved in the removal of HNE-modified proteins, we first evaluated the effect of MG132, which inhibits both the proteasomal and the lysosomal pathways, on the removal of the HNE-modified proteins. In contrast to the rapid removal of HNE-modified proteins in the absence protease inhibitors, the HNE-modified proteins were significantly stabilized in cells treated with MG132 (Fig. 4, compare lanes 4, 6, and 8 with lanes 3, 5, and 7, respectively), suggesting that the HNE-modified proteins in the cells are removed via proteolytic degradation. To determine if the proteasome pathway was involved, we determined the effects of proteasome specific inhibitors on the removal of HNE-modified proteins from cells. Both clastolactacystin-β-lactone and epoxomicin treatment increased the level of ubiquitin conjugates (data not shown), indicating that the proteasome activity was inhibited. However, neither clastolactacystin-β-lactone nor epoxomicin slowed the removal of HNE-modified proteins from the cells (Fig. 5, compare lanes 5 and 6 with lane 3). These data indicate that the proteasome did not play a major role in the removal of HNE-modified proteins from the lens cells. In contrast, chloroquine inhibited the removal of HNE-modified proteins (Fig 5, compare lane 7 with lane 3). These data indicate that the majority of HNE-modified proteins in intact cells are degraded by the lysosomal pathway, rather than by the proteasome. The effect of these protease inhibitors on the removal of HNE-modified proteins was not due to cytotoxicity, because cells were viable as judged by Trypan blue exclusion assay (data not shown). There were no detectable changes in cell morphology under these conditions.
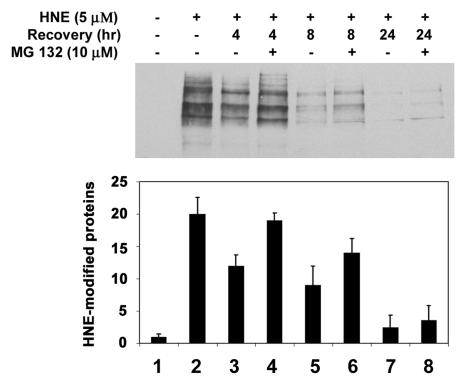
Removal of HNE-modified proteins in human lens epithelial cells. Cells were treated with 5 μM HNE for 1 h and allowed to recover in a medium with or without 40 μM MG132 for 0-24 h. Cells were collected, and an equal amount of protein (30 μg) from cell lysate was analyzed by Western blotting with antibodies to HNE-conjugated proteins. Upper panel ) Typical Western blotting result. Lower panel) Densitometry quantification of HNE-modified proteins in the cells. Data are lower panel are means ± SD of 3 independent experiments.
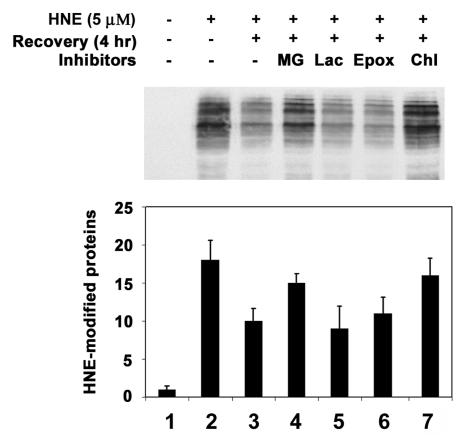
Effects of protease inhibitor on removal of HNE-modified proteins from lens epithelial cells. Cells were treated with 5 μM HNE for 1 h and allowed to recover in a medium contains different protease inhibitors for 4 h. Cells were then collected and an equal amount of protein (30 μg) from the cell lysate was analyzed by Western blotting with antibodies to HNE-conjugated proteins. Lane 1, untreated cells; lane 2; cells treated with HNE for 1 h; lane 3, recovered without any inhibitor; lane 4, recovered in the presence of MG132; lane 5, recovered in the presence of clasto-lactacystin-β-lactone; lane 6, recovered in the presence of epoxomicin; lane 7, recovered in the presence of chloroquine. Upper panel) Typical Western blotting result. Lower panel) Densitometry quantification of HNE-modified proteins in the cells. Data in the lower panel are means ± SD of 3 independent experiments.
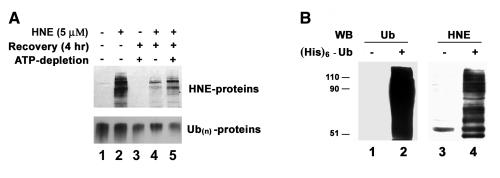
HNE-modified proteins are ubiquitinated in lens cells. A) ATP-dependent removal of HNE-modified proteins. Human lens epithelial cells were treated with 5 μM HNE for 1 h and allowed to recover in a normal medium or in a medium containing antimycin A and 2-deoxyglucose for 4 h. The cells were collected and an equal amount of protein (30 μg) from cell lysate was analyzed by Western blotting with antibodies to HNE-conjugated proteins (upper panel) or antibodies to ubiquitin (lower panel). B) Ubiquitination of HNE-modified proteins in cells. Cells were infected with a control virus or with a recombinant adenovirus encoding (His)6-tagged ubiquitin for 48 h, and then cells were treated with HNE for 1 h. Ubiquitin conjugates were isolated from the cell lysate by Ni-NTA resin and resolved by SDS-PAGE. Lanes 1 and 2 were probed with antibodies to ubiquitin, and lanes 3 and 4 were identical samples as lanes 1 and 2 but probed with antibodies to HNE-conjugated proteins.
Because the HNE-modified proteins are preferred substrates for ubiquitination in cell-free systems (Fig. 2), we determined if ubiquitination is involved in the lysosomal degradation of HNE-modified proteins. A hallmark of ubiquitination is the requirement of ATP. Levels of ubiquitin conjugates are often correlated with intracellular ATP levels. When intracellular ATP was depleted with antimycin A and 2-deoxyglucose (34), levels of endogenous ubiquitin conjugates decreased significantly (Fig. 6A, lower panel) and the removal of HNE-modified proteins was significantly slowed as compared with cells with normal levels of ATP (Fig. 6A, upper panel, compare lane 5 with lane 4), indicating the degradation of HNE-modified proteins is ATP dependent. To further determine if ubiquitination is involved in the lysosomal degradation of HNE-modified proteins, we expressed (His)6-tagged ubiquitin in the cells and isolated ubiquitin conjugates from HNE-treated cells. Figure 6B showed that the fraction of ubiquitin conjugates was enriched with of HNE-modified proteins as compared with the proteins that were nonspecifically bound to the Ni-NTA resin. These data imply that HNE-modified proteins are recognized by the ubiquitinating system and targeted to the lysosome for degradation.
DISCUSSION
An increasing body of evidence suggests that HNE, a lipid peroxidation product, is involved in many of the pathophysiological lesions associated with oxidative stress in cells and tissues (35). HNE exhibits a wide variety of biological effects including inhibition of DNA and protein synthesis (7), glutathione depletion (36), and inhibition of various enzymes (37). Cellular proteins are the primary targets of HNE modification via forming Michael adducts on lysine, histidine and cysteine residues, or through formation of Schiff bases with lysine residues (38). It appears that the efficient removal of HNE-modified proteins is essential, because accumulation of HNE-modified proteins is associated with many cellular dysfunctions (9, 10) and cellular aging (11). This study elucidated how HNE-modified proteins are removed from the cells.
We found that HNE-modified proteins were preferentially ubiquitinated in a cell-free system (Fig. 2A). However, the rate of proteasome-dependent degradation of the HNE-modified proteins was not significantly different from that of unmodified proteins in the cell-free system (Fig. 2B). Similarly, we found that HNE-modified proteins are efficiently removed from lens epithelial cells by a proteolytic mechanism (Figs. (Figs.33--5).5). Although ATP is required for this proteolytic pathway (Fig. 6A), inhibition of the cellular proteasome activity did not affect the removal of HNE-modified proteins (Figs. (Figs.33 and and5).5). In contrast, inhibition of the lysosome activity resulted in an accumulation of HNE-modified proteins in the cells (Figs. (Figs.33 and and5).5). Moreover, it suggests that HNE-modified proteins are preferentially ubiquitinated by the ubiquitin-conjugating system since ubiquitin conjugates isolated from HNE-treated cells were enriched with HNE-modified proteins (Fig. 6B). Taken together, these results indicate that, like other types of damaged proteins, HNE-modified proteins are also recognized by the ubiquitin-conjugating system and the ubiquitin-conjugating system may serve as a common mechanism to recognize various types of modified or damaged proteins, although different E2 s and E3 s may be used to ubiquitinate different types of damaged proteins. The ubiquitinated proteins can either be degraded by the proteasome (24, 39) or by the lysosome (23, 40, 41). However, the mechanism for the differential destinations of the ubiquitinated proteins remains elusive. The destination of ubiquitinated proteins could be determined by the nature of the protein substrates or by the structures of the ubiquitin conjugates. For example, proteins destined to the proteasome are often polyubiquitinated (24) and proteins destined to the lysosome are often mono-ubiquitinated (42). The putative mechanism for substrate recognition is summarized in Fig. 7.
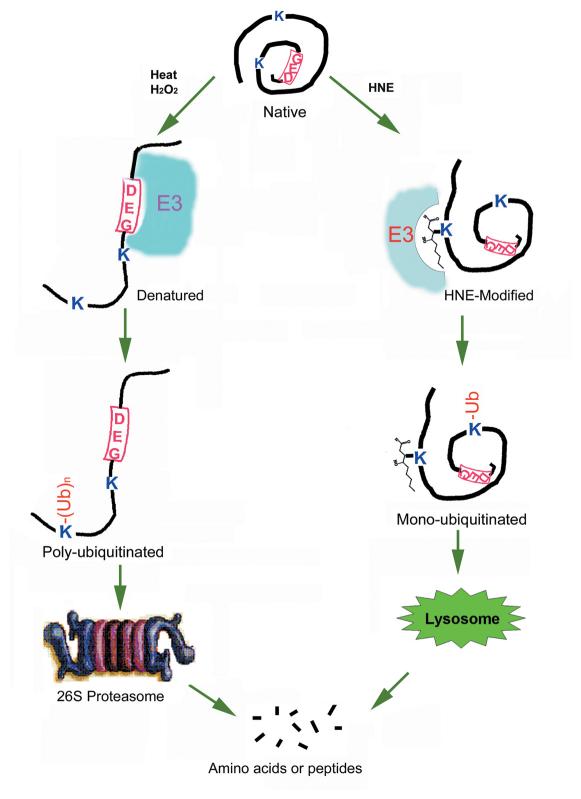
Hypothesis of ubiquitin-dependent protein quality control mechanism. We hypothesize that many proteins have internal signal for ubiquitination as indicated by “DEG” in the diagram. Ubiquitination signal is hidden in the native conformation, and therefore native proteins are not recognized by the ubiquitination machinery. However, various modifications and damage may cause protein unfolding and exposure of ubiquitination signal. Thus, the unfolded proteins will be recognized by the combination of specific E2 s and E3 s and be ubiquitinated. Specific modification, such HNE, may serve as the signal for ubiquitination. Poly-ubiquitinated proteins will be recruited to the 26S proteasome for degradation, and mono-ubiquitinated proteins may enter into the lysosome for degradation.
Available data regarding the degradation of HNE-modified proteins are limited. One study showed that HNE-modified proteins in homogenates of kidney were degraded by the ubiquitinproteasome pathway (43). Other studies showed that HNE-modified proteins, such as HNE-modified glucose-6-phosphate dehydrogenase, were resistant to proteasome-dependent degradation (44) and in some cases the HNE-modified proteins acted as potent noncompetitive inhibitors of the proteasome (44, 45). A recent paper showed that histones and other proteins treated with low levels of HNE (<100 μM) were better substrate for the 20S proteasome (46). However, proteins modified with higher levels of HNE (>100 μM) were resistant to degradation by the proteasome (46). The demonstration that HNE-modified proteins are degraded by the ubiquitin-dependent lysosomal pathway reconciles the apparent differences in opinion regarding which proteolytic pathways are responsible for the degradation of HNE-modified proteins.
Ubiquitin-dependent lysosomal degradation was involved in regulation of several membrane proteins, particularly receptor proteins (42, 47, 48). Ubiquitination of these membrane proteins triggers their internalization and targets them for degradation by the lysosome pathway. Similar to many other proteins that are degraded by the ubiquitin-dependent lysosomal pathway, the majority of the ubiquitinated HNE-modified proteins were mono-ubiquitinated (Figs. (Figs.33 and and6B).6B). This work showed that not only membrane receptors, but also HNE-modified membrane or intracellular proteins, are removed from the cells by the ubiquitin-dependent lysosomal pathway. Considering that both membrane receptors and HNE-modified proteins are monoubiquitinated and degraded by the lysosome, it is tempting to speculate that they are recognized by the same or similar ubiquitin ligases, such as c-Cbl.
Selective degradation of damaged proteins is essential for cellular functions. The ubiquitin conjugating system provides a mechanism to distinguish damaged or adversely modified proteins from native proteins. Identification of the enzymes that are required for the recognition and ubiquitination of various types of modified or damaged proteins will help to understand the molecular mechanisms of the cellular protein quality control system and will provide clues for prevention and/or treatment of diseases that are related to the accumulation of damaged proteins, such as cataract and other age-related degenerative diseases.
ACKNOWLEDGMENTS
This work was supported by NIH grant EY11717 (to F. Shang), EY 13250 (to A. Taylor), USDA contract 5000-052 (to A. Taylor), and a grant from Portuguese Foundation for Science and Technology (FCT) with the support of FEDER through program POCTI (to P. Pereira and C. Marques). Mark Siegal and Matthew Gallagher are thanked for help in preparation of this manuscript.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1096/fj.04-1743fje
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1382276?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
ECPUB5 Polyubiquitin Gene in Euphorbia characias: Molecular Characterization and Seasonal Expression Analysis.
Genes (Basel), 15(7):957, 21 Jul 2024
Cited by: 0 articles | PMID: 39062736 | PMCID: PMC11275293
Ubiquitination contributes to the regulation of GDP-mannose pyrophosphorylase B activity.
Front Mol Neurosci, 17:1375297, 24 Jun 2024
Cited by: 0 articles | PMID: 38979475 | PMCID: PMC11228364
Alleviating the unwanted effects of oxidative stress on Aβ clearance: a review of related concepts and strategies for the development of computational modelling.
Transl Neurodegener, 12(1):11, 13 Mar 2023
Cited by: 3 articles | PMID: 36907887 | PMCID: PMC10009979
Review Free full text in Europe PMC
Dienone Compounds: Targets and Pharmacological Responses.
J Med Chem, 63(24):15075-15093, 04 Nov 2020
Cited by: 7 articles | PMID: 33146523 | PMCID: PMC7770821
Review Free full text in Europe PMC
MPTP-induced vulnerability of dopamine neurons in A53T α-synuclein overexpressed mice with the potential involvement of DJ-1 downregulation.
Korean J Physiol Pharmacol, 21(6):625-632, 30 Oct 2017
Cited by: 13 articles | PMID: 29200905 | PMCID: PMC5709479
Go to all (69) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Heat shock factor 4 regulates lysosome activity by modulating the αB-crystallin-ATP6V1A-mTOR complex in ocular lens.
Biochim Biophys Acta Gen Subj, 1864(3):129496, 29 Nov 2019
Cited by: 12 articles | PMID: 31786107
alpha-Crystallin localizes to the leading edges of migrating lens epithelial cells.
Exp Cell Res, 306(1):203-215, 17 Mar 2005
Cited by: 47 articles | PMID: 15878345
Degradation of HNE-modified proteins--possible role of ubiquitin.
Redox Rep, 12(1):63-67, 01 Jan 2007
Cited by: 12 articles | PMID: 17263912
Cell biological roles of αB-crystallin.
Prog Biophys Mol Biol, 115(1):3-10, 25 Feb 2014
Cited by: 41 articles | PMID: 24576798
Review
Funding
Funders who supported this work.
NEI NIH HHS (9)
Grant ID: R01 EY013250
Grant ID: R29 EY011717-04
Grant ID: EY11717
Grant ID: R01 EY011717
Grant ID: R29 EY011717-03
Grant ID: R29 EY011717-05
Grant ID: EY 13250
Grant ID: R01 EY011717-06A1
Grant ID: R29 EY011717
U.S. Department of Agriculture (1)
Grant ID: 5000‐052
 and
and  *
*



