Abstract
Free full text

Cyclophilin A and TRIM5α Independently Regulate Human Immunodeficiency Virus Type 1 Infectivity in Human Cells
Abstract
Cyclophilin A (CypA), a cytoplasmic, human immunodeficiency virus type 1 (HIV-1) CA-binding protein, acts after virion membrane fusion with human cells to increase HIV-1 infectivity. HIV-1 CA is similarly greeted by CypA soon after entry into rhesus macaque or African green monkey cells, where, paradoxically, the interaction decreases HIV-1 infectivity by facilitating TRIM5α-mediated restriction. These observations conjure a model in which CA recognition by the human TRIM5α orthologue is precluded by CypA. Consistent with the model, selection of a human cell line for decreased restriction of the TRIM5α-sensitive, N-tropic murine leukemia virus (N-MLV) rendered HIV-1 transduction of these cells independent of CypA. Additionally, HIV-1 virus-like particles (VLPs) saturate N-MLV restriction activity, particularly when the CA-CypA interaction is disrupted. Here the effects of CypA and TRIM5α on HIV-1 restriction were examined directly. RNA interference was used to show that endogenous human TRIM5α does indeed restrict HIV-1, but the magnitude of this antiviral activity was not altered by disruption of the CA-CypA interaction or by elimination of CypA protein. Conversely, the stimulatory effect of CypA on HIV-1 infectivity was completely independent of human TRIM5α. Together with previous reports, these data suggest that CypA protects HIV-1 from an unknown antiviral activity in human cells. Additionally, target cell permissivity increased after loading with heterologous VLPs, consistent with a common saturable target that is epistatic to both TRIM5α and the putative CypA-regulated restriction factor.
Primate cells possess an activity that blocks retroviral replication at a step after viral entry but prior to completion of reverse transcription (29, 30). In rhesus macaques and owl monkeys this activity potently restricts human immunodeficiency virus type 1 (HIV-1) but largely permits infection by simian immunodeficiency virus SIVMAC239 (9, 16, 37). Analysis of chimeric viruses generated from HIV-1 and SIVMAC239 sequences revealed viral determinants for susceptibility to this restriction activity within CA (15, 18, 41, 50).
Independent screens, one with rhesus macaque cells and the other with owl monkey cells, identified the respective TRIM5 orthologues as necessary for HIV-1 restriction in these species (48, 54). TRIM5 is a member of the tripartite motif (TRIM) protein family, defined by a cluster of RING-finger, B-box, and coiled-coil domains (40, 44). The native function of TRIM5 is unknown, but the ability of this protein to serve as a target for E2 ubiquitylation in a RING-finger-dependent manner (63) suggests that it acts as an E3 ubiquitin ligase in vivo.
The alpha isoform of TRIM5 is required for retroviral restriction in most primate species that have been studied (27, 32, 42, 54, 64). Species-specific variation in the carboxy-terminal SPRY domain accounts to a large extent for the variation in retroviral restriction specificity among orthologues (46, 52, 55, 65). For example, TRIM5α encoded by some African green monkeys (TRIM5αAGM) and by rhesus macaques (TRIM5αrh) restricts HIV-1, N-tropic murine leukemia virus (N-MLV), and certain strains of simian immunodeficiency virus (27, 32, 54, 64). Human TRIM5α (TRIM5αhu) potently inhibits N-MLV but, when overexpressed, only moderately restricts HIV-1 and simian immunodeficiency virus (27, 32, 42, 54, 64).
An unusual TRIM5 isoform accounts for the potent HIV-1 restriction activity in owl monkey cells (39, 48). All TRIM5 alleles from this genus that have been examined possess a complete cyclophilin A (CypA) cDNA inserted into intron 7 (39, 45, 48). In the resulting protein, the SPRY domain unique to the alpha isoform has been replaced by CypA. This is particularly intriguing since free CypA binds to the exposed proline-rich loop of HIV-1 CA (21, 22, 35) and promotes HIV-1 infectivity in human cells (12, 13, 21, 58). The CypA-CA interaction is required in the target cell, as opposed to the producer cell (26, 31, 51, 61), but the mechanism by which CypA promotes HIV-1 infection of human cells is unknown. The discovery of the owl monkey TRIM5 gene fusion with CypA raised suspicion that these two proteins might be functionally interdependent.
Retroviral restriction is overcome by infection at high multiplicity or by flooding target cells with nonreplicating virus-like particles (VLPs) (4, 9, 10, 16, 17, 19, 25, 43, 57, 60). These observations have been explained by postulating the existence of a saturable factor necessary for restriction. Cross-saturation experiments in human cells suggest that, in the absence of CypA, HIV-1 is particularly sensitive to the same factor that restricts N-MLV (61). Since N-MLV restriction in human cells requires TRIM5α (27, 32, 42, 64), CypA binding to HIV-1 CA might shield incoming HIV-1 cores from restriction by TRIM5αhu. This hypothesis is supported by the observation that HIV-1 infection is CypA-independent in a human cell clone that was selected for loss of N-MLV restriction activity (47). Additionally, HIV-1 restriction by rhesus macaque or African green monkey TRIM5α requires CypA (6).
RNA interference knockdown, single-cycle HIV-1 infectivity assays, and several tools that disrupt the CA-CypA interaction were used here to clarify the relationship between CypA and TRIM5αhu. We conclude that CypA promotes HIV-1 infectivity via effects on a host factor distinct from TRIM5αhu.
MATERIALS AND METHODS
Plasmids.
pNL4-3 contains an infectious HIV-1 provirus (2). pNL4-3/G89V encodes a mutant CA that disrupts the CypA binding site (51). pNL4-3GFP, or HIV-1GFP, is pNL4-3 with an env-inactivating mutation and an enhanced green fluorescent protein (GFP) gene replacing nef (28).
p8.9NΔSB expresses HIV-1 gag and pol from the cytomegalovirus immediate-early (CMVie) promoter and bears inactivating mutations in the RNA packaging region, env, and nonessential accessory genes (7). p8.9NΔSB/A92E is p8.9NΔSB encoding the CA mutant A92E (7). CSGWΔN is an HIV-1 vector expressing enhanced green fluorescent protein under the control of the spleen focus-forming virus long terminal repeat (9). It was modified from the parent CSGW vector by blunting a NotI restriction site. pMDG encodes the vesicular stomatitis virus glycoprotein (VSV-G) (8).
pCIG3-N and -B are MLV-derived constructs expressing N- or B-tropic CA, respectively (8). pMIG is a murine stem cell virus (MSCV)-based expression vector containing an internal ribosome entry site-GFP cassette (62). pCL-Eco contains psi-minus Moloney MLV expressed from the CMVie promoter (38). pSUPER.retro.puro (pSRP; OligoEngine) is an MSCV-derived vector expressing short hairpin RNAs (shRNAs) from the H1 polymerase III promoter (14). pSRP-TRIM5hu expresses an shRNA targeting human TRIM5 mRNA (target sequence, 5′-GCTCAGGGAGGTCAAGTTG-3′) (54). pSRP-CypA expresses an shRNA that targets human CypA mRNA. It was made by subcloning the shRNA expression cassette from pMH-CypA147 (48). The control plasmid, pSRP-Luc, encodes an shRNA targeting luciferase (48).
pshRNA.lenti.puro (pshRLP) is an HIV-1-based vector derived from pCSGWΔN that contains an H1-shRNA cassette as in pSUPER (14) and a puromycin resistance gene driven by the phosphoglycerate kinase (PGK) promoter. To engineer pshRLP-TRIM5hu and pshRLP-Luc, the following oligonucleotides were used for PCR, with pSRP-TRIM5hu or pSRP-Luc as templates: 5′-GCTCTAGAGCGGCCGCGCTCCTTTCGGTCGGGCGCT-3′ and 5′-AGGAATTCTTAATTAAGATCGATCTCTCGAGGTCGAC-3′. The PCR products were subcloned into CSGWΔN using EcoRI and NotI sites.
Cells and drugs.
Adherent cells (293T, TE671, HeLa, and Mus dunni tail fibroblasts) were maintained in Dulbecco's modified Eagle medium. A human T-cell line, CEM-SS, was maintained in RPMI medium. All media were supplemented with 10% fetal bovine serum and antibiotics. CsA (Bedford Laboratories) was prepared in dimethyl sulfoxide at 10 mg/ml and diluted in tissue culture medium to 2.5 μM for each experiment. Dextran sulfate (5 mg/ml; Sigma) was prepared in H2O and was used at 100 μg/ml as described previously (8).
Viruses.
Vectors and viruses were produced by transfecting 106 293T cells/well in 6-well plates using Lipofectamine 2000 (Invitrogen). NL4-3GFP vectors were produced by cotransfecting cells with 0.3 μg of pMDG and 3.7 μg of either pNL4-3GFP or pNL4-3GFPG89V. HIV-1GFP wild-type and HIV-1GFP A92E vectors were produced using 2 μg of CSGWΔN, 0.3 μg of pMDG, and 1.7 μg of p8.9NΔSB, either wild type or A92E mutant. Full-length infectious HIV-1 viruses were produced by transfecting 293T cells with 4 μg of pNL4-3 or pNL4-3/G89V. To produce MSCV-based shRNA vectors, 293T cells were cotransfected with 1.7 μg of pCL-Eco, 0.3 μg of pMDG, and 2 μg of either pSRP-CypA, pSRP-TRIM5, or pSRP-Luc. To produce HIV-1-based shRNA vectors, 293T cells were cotransfected with 1.7 μg of p8.9NΔSB, 0.3 μg of pMDG, and 2 μg of either pshRLP-TRIM5 or pshRLP-Luc. For production of N- or B-tropic MLVGFP vectors, cells were cotransfected with 2 μg of pMIG, 0.3 μg of pMDG, and 1.7 μg of either pCIG3-N or pCIG3-B. For production of HIV-1 VLPs, 4.5 × 106 293T cells/10-cm-diameter plate were cotransfected with 18 μg of p8.9NΔSB and 2 μg of pMDG using Ca2PO4.
All virus- and VLP-containing supernatants were harvested 48 h posttransfection, spun at 300 × g for 5 min to remove cell debris, and filtered through a 0.45-μm-pore-size filter (Pall Acrodisc). For infectious NL4-3 viruses and VLPs, clarified supernatant was layered onto a 25% sucrose cushion and accelerated at 100,000 × g for 2 h in an SW41 rotor. The pelleted virions were resuspended in RPMI medium with 10% fetal bovine serum and stored at −80°C. Virions were normalized prior to infection by measuring reverse transcriptase (RT) activity as previously described (24).
Infection.
Adherent cells were plated in 48-well plates (2 × 104 cells/well). Cells in suspension were seeded in 48-well plates at 8 × 104 cells/well. Virion stocks were added to the cells in a final volume of 250 μl/well. GFP expression in infected cells was analyzed 48 h postinfection by flow cytometry (see below). For infections with full-length virus, dextran sulfate was added 16 h postinfection (100 μg/ml) to prevent spread of the virus (8). Forty-eight hours postinfection, cells were fixed in 4% formaldehyde-phosphate-buffered saline and assayed for intracellular p24 using a monoclonal anti-p24 antibody (183-H12-5C; NIH AIDS Research & Reference Reagent Program).
To generate TE671 and HeLa cells with stable expression from pSRP vectors, cells were seeded into 24-well plates at 3 × 104 cells/well, covered with 500 μl of clarified 293T supernatant, and spinoculated for 70 min at 1,200 × g. Forty-eight hours posttransduction, cells were subjected to puromycin selection with quantum increases in drug concentration (1, 5, 10, 15, 20, 30, 40, and 50 ng/μl) over a period of 10 days.
To generate CEM-SS cells with stable expression from pshRLP vectors, cells were seeded into 24-well plates at 105 cells/well and infected with 50 μl of clarified 293T supernatant in a total volume of 500 μl. All cells were subjected to puromycin selection 48 h posttransduction as described above.
Flow cytometry.
FACSCalibur and Cellquest Pro software (Becton Dickinson) were used to record GFP fluorescence in the FL1 channel. A quantity of 104 live cells was analyzed per sample.
Western blotting.
Cells were normalized by number, and lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with antibodies to CypA (rabbit polyclonal; Biomol) and β-actin (mouse monoclonal; Sigma). Coomassie blue staining of the membranes confirmed that loading of samples had been normalized.
RESULTS
Endogenous TRIM5αhu restricts HIV-1.
To test the importance of TRIM5αhu for HIV-1 infectivity, shRNA was used to downregulate endogenous TRIM5αhu. TE671 cells were transduced with an MSCV-based retroviral vector delivering a pSUPER-shRNA cassette that targets human TRIM5 (14). A construct delivering an shRNA specific for luciferase (48) was used as a control. Pools of transduced cells were selected for maintenance of the shRNA-encoding transgenes by exploiting the PGK-puromycin resistance cassette that is part of the retroviral vector.
A functional assay was utilized to determine whether TRIM5αhu knockdown was successful. Since TE671 cells restrict N-MLV more than 100-fold relative to B-MLV, in a TRIM5αhu-dependent fashion (27, 32, 42, 64), TE671 cells knocked down for TRIM5 (TE671-TR5-shRNA cells) were challenged with VSV-G-pseudotyped B- or N-tropic MLVGFP. The two viral stocks were normalized based on RT activity as well as the titer on nonrestrictive Mus dunni tail fibroblasts, as previously described (8, 47, 59). In TE671-Luc-shRNA cells, the N-MLV titer was more than 100-fold lower than that of B-MLV (Fig. (Fig.1A).1A). In TE671-TR5-shRNA cells, the N-MLV titer was almost identical to that of B-MLV (Fig. (Fig.1B),1B), indicating that TRIM5αhu-dependent restriction activity was eliminated by the shRNA.
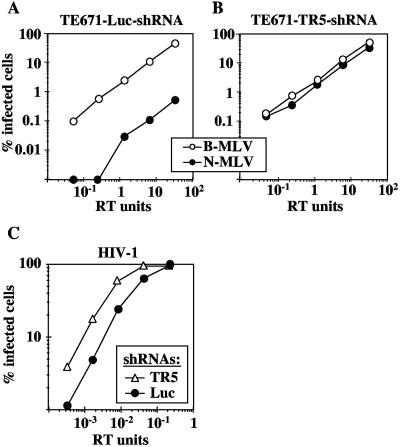
Knockdown of endogenous TRIM5αhu enhances HIV-1 infectivity. TE671 cells were transduced with an MLV-based vector delivering an shRNA expression construct specific for human TRIM5 (TR5-shRNA) or luciferase (Luc-shRNA). VSV-G-pseudotyped, N- and B-tropic MLVGFP vectors were normalized by RT and by infectivity on nonrestrictive Mus dunni tail fibroblasts and used to infect (A) TE671-Luc-shRNA cells or (B) TE671-TR5-shRNA cells. (C) VSV-G-pseudotyped HIV-1GFP virions were used to infect TE671-Luc-shRNA and TE671-TR5-shRNA cells. The percentage of GFP-positive (infected) cells was determined by flow cytometry. Shown are representative results of a single experiment. Identical results were obtained on three separate occasions using independently produced viral stocks.
The effect of TRIM5αhu knockdown on HIV-1 infectivity was examined using VSV-G-pseudotyped NL4-3GFP (HIV-1GFP). The titer of HIV-1GFP was threefold higher on TE671-TR5-shRNA cells than on control TE671-Luc-shRNA cells (Fig. (Fig.1C),1C), demonstrating that endogenous TRIM5αhu inhibits HIV-1 infectivity. This result is consistent with previous reports that TRIM5αhu overexpression decreases HIV-1 infectivity (27, 42, 52, 55).
Inhibition of HIV-1 infectivity by factors that disrupt the CA-CypA interaction is TRIM5αhu independent.
To investigate whether HIV-1 infectivity is more susceptible to TRIM5αhu-mediated restriction when the CypA-CA interaction is disrupted, TE671-TR5-shRNA cells and TE671-Luc-shRNA cells were infected with HIV-1GFP in the presence of CsA, a competitive inhibitor of the CA-CypA interaction (35). The magnitude of reduction of HIV-1 infectivity by CsA in TE671-TR5-shRNA cells was identical to the magnitude of reduction by the drug in the control TE671-Luc-shRNA cells (Fig. 2A and B). Similarly, the titer of HIV-1GFP G89V, a CA mutant that disrupts interaction with CypA (67), was equally decreased in TE671-TR5-shRNA and TE671-Luc-shRNA cells (Fig. 2C and D). These results show that the reduction in HIV-1 infectivity that results from CsA or the G89V mutation does not result from increased susceptibility to TRIM5αhu-mediated restriction.
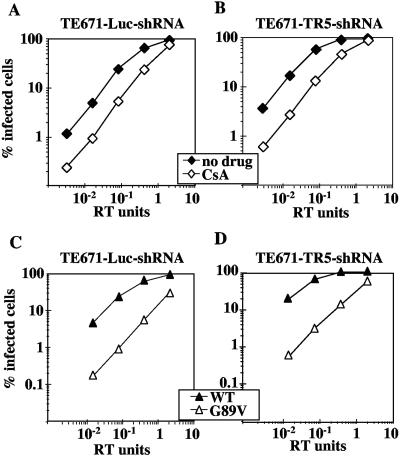
Reduction of HIV-1 infectivity by CsA or CA mutant G89V is not mediated by TRIM5αhu. VSV-G-pseudotyped, RT-normalized, HIV-1GFP wild-type and G89V mutant virions were used to infect TE671-Luc-shRNA (A and C) and TE671-TR5-shRNA (B and D) cells in the presence or absence of CsA, as indicated. The percentage of GFP-positive (infected) cells was determined by flow cytometry. Shown are representative results of a single experiment. Identical results were obtained on three separate occasions using independently produced viral stocks.
Reduction of HIV-1 infectivity by CypA knockdown is independent of TRIM5αhu.
To verify that the CsA-mediated decrease in HIV-1 titer resulted from the inhibitory effect of CsA on CypA, TE671 cells were transduced with a construct delivering an shRNA specific for CypA, as previously described (48, 51). After selection of a pool of transduced cells with puromycin, CypA was undetectable by Western blotting (Fig. (Fig.3A).3A). CypA downregulation resulted in a three- to fivefold decrease in HIV-1 infectivity compared to control cells (Fig. (Fig.3B).3B). The magnitude of reduction of HIV-1 infectivity in the context of the CypA knockdown was almost identical to the magnitude of reduction of HIV-1 by CsA in control TE671-Luc-shRNA cells (Fig. (Fig.3B),3B), indicating that CsA reduces HIV-1 infectivity via effects on CypA. As expected, since the G89V mutation disrupts the CypA binding site in CA, HIV-1 G89V infectivity was decreased no further by CypA knockdown (Fig. (Fig.3C3C).
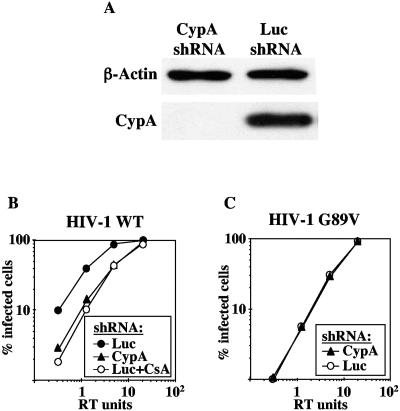
Knockdown of CypA reproduces the effect of CsA and G89V on HIV-1 infectivity. TE671 cells were transduced with MLV-based vectors delivering shRNA expression constructs specific for CypA (CypA-shRNA) or luciferase (Luc-shRNA). (A) Cells were normalized by number, and CypA knockdown was demonstrated by Western blotting for CypA and β-actin. VSV-G-pseudotyped, RT-normalized, HIV-1GFP wild-type (B) or G89V mutant (C) vectors were used to infect TE671-CypA-shRNA or TE671-Luc-shRNA cells in drug-free media or in the presence of 2.5 μM CsA, as indicated. The percentage of GFP-positive (infected) cells was determined by flow cytometry. Shown are representative results of a single experiment. Identical results were obtained on three separate occasions.
To directly test the interdependence of TRIM5αhu and CypA in mediating HIV-1 restriction, we generated cell lines in which both TRIM5αhu and CypA were downregulated using shRNA. To accomplish this, the singly transduced TE671-TR5-shRNA and TE671-Luc-shRNA cells (Fig. (Fig.1)1) were subjected to a second round of transduction either with the shRNA specific for CypA or with the control construct targeting luciferase. CypA knockdown was confirmed by Western blotting (Fig. (Fig.4A).4A). Both TE671-TR5/CypA-shRNA and TE671-TR5/Luc-shRNA cell lines still failed to restrict N-MLV, confirming that the TRIM5αhu knockdown was stable (data not shown).
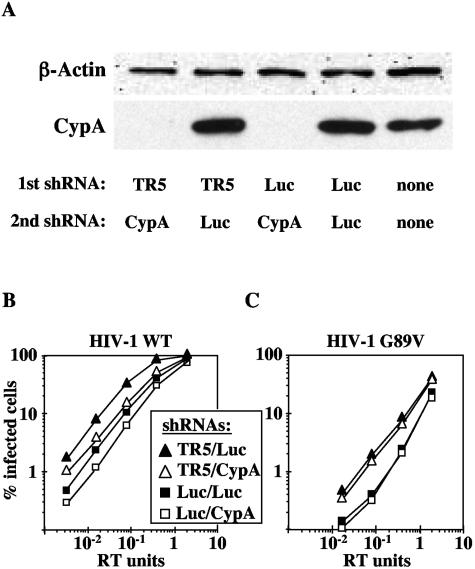
Reduction of HIV-1 infectivity by CypA knockdown is independent of TRIM5αhu. TE671 cells were transduced with an MLV-based vector delivering an shRNA expression construct specific for human TRIM5 (TR5-shRNA) or luciferase (Luc-shRNA). Each pool of cells was then transduced a second time with shRNA vectors specific for CypA or luciferase. (A) CypA knockdown in the indicated pools of cells was demonstrated by Western blotting for CypA and β-actin. VSV-G-pseudotyped HIV-1 wild-type (WT) (B) or G89V mutant (C) virions were used to infect the indicated pools of cells. The percentage of GFP-positive (infected) cells was determined by flow cytometry. Shown are representative results of a single experiment. Identical results were obtained on three separate occasions.
As before (Fig. (Fig.11 and and3),3), cells in which TRIM5αhu was downregulated were more permissive for HIV-1 infection than control cells, and cells in which CypA was knocked down were less permissive for HIV-1 infection (Fig. (Fig.4B).4B). The magnitude of the infectivity decrease due to CypA knockdown was the same in TE671-TR5/CypA-shRNA and TE671-Luc/CypA-shRNA cells, compared to the corresponding control cell lines TE671-TR5/Luc-shRNA and TE671-Luc/Luc-shRNA (Fig. (Fig.4B).4B). Similarly, the magnitude of the increase in infectivity due to disruption of TRIM5αhu was the same whether CypA was expressed or not, or whether HIV-1 wild type or G89V was used for infection (Fig. 4B and C). As expected, infectivity of HIV-1 G89V remained unaltered by CypA knockdown (Fig. (Fig.4C).4C). These results demonstrate that TRIM5αhu and CypA independently modulate HIV-1 infectivity. In other words, disruption of TRIM5αhu does not rescue the reduction in HIV-1 infectivity caused by CypA knockdown.
CypA and TRIM5αhu independently modulate HIV-1 infectivity in human CD4+ T cells.
To determine whether the results described above were peculiar to the TE671 rhabdomyosarcoma cell line, the CD4+ T-cell line CEM-SS was transduced with an HIV-1-based vector delivering a pSUPER-shRNA cassette (14) specific for human TRIM5 (54). A construct delivering an shRNA specific for luciferase (48) was used as a control. Pools of transduced cells were then selected in puromycin.
As described above for the TE671 cells, TRIM5αhu knockdown in CEM-SS was confirmed by challenging transduced cells with N- and B-tropic MLVGFP (Fig. 5A and B). HIV-1GFP infectivity was reduced to the same extent by CsA in CEM-SS-TR5-shRNA as it was in CEM-SS-Luc-shRNA (Fig. 5C and D). Similarly, the titer of HIV-1GFP G89V was decreased in both TRIM5αhu knockdown and control cells (Fig. 5C and D). These results confirm that CypA acts independently of TRIM5αhu to modulate HIV-1 infectivity in human CD4+ T cells.
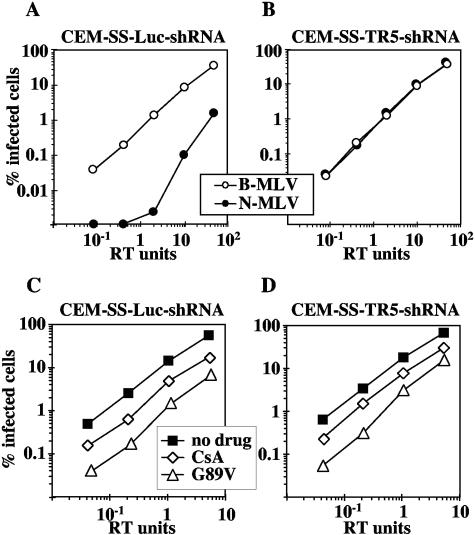
Reduction of HIV-1 infectivity by CsA or the G89V mutant is independent of TRIM5αhu in human T cells. CEM-SS cells were transduced with an HIV-based vector delivering an shRNA expression construct specific for human TRIM5 or luciferase as a control. VSV-G-pseudotyped, N- and B-tropic MLVGFP virions were used to infect CEM-SS-Luc-shRNA cells (A) or CEM-SS-TR5-shRNA cells (B). VSV-G-pseudotyped, HIV-1GFP wild-type, or G89V mutant vectors were used to infect CEM-SS-Luc-shRNA cells (C) and CEM-SS-TR5-shRNA cells (D) in drug-free medium or in the presence of 2.5 μM CsA, as indicated. The percentage of GFP-positive (infected) cells was determined by flow cytometry. Shown are representative results of a single experiment. Identical results were obtained on three separate occasions using independently produced viral stocks.
Effects of CypA and TRIM5αhu on HIV-1 infectivity are independent of the route by which virions enter target cells.
Previous studies indicated that pseudotyping with VSV-G renders HIV-1 insensitive to CsA (3). VSV-G and the HIV-1 env-encoded glycoproteins (gp120/gp41) are believed to promote virion entry into target cells via two distinct mechanisms. VSV-G enters cells via an endocytic pathway, while the HIV-1 env glycoproteins mediate direct fusion into the target cell cytoplasm (5, 36, 53). To determine whether the functional interdependence of CypA and TRIM5αhu is altered by the entry mechanism, CEM-SS-TR5-shRNA and CEM-SS-Luc-shRNA cells were challenged with full-length, infectious HIV-1NL4-3. In the presence of CsA, or the G89V mutation, HIV-1 infectivity was decreased to the same extent in CEM-SS-TR5-shRNA cells as it was in control CEM-SS-Luc-shRNA cells (Fig. (Fig.6).6). These findings were confirmed in TE671 cells transfected with CD4 (data not shown). Thus, CypA and TRIM5αhu act independently to regulate HIV-1 infectivity, irrespective of the route of entry utilized by the virus.
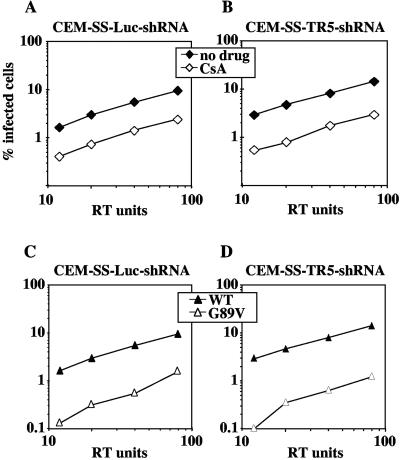
Promotion of HIV-1 infectivity by CypA is independent of TRIM5αhu, regardless of the route of viral entry. HIV-1NL4-3 virions were used to infect CEM-SS-Luc-shRNA (A) and CEM-SS-TR5-shRNA cells (B), in the presence or in absence of CsA, as indicated. HIV-1NL4-3 virions, either wild type or G89V mutant, as indicated, were used to infect CEM-SS-Luc-shRNA (C) and CEM-SS-TR5-shRNA cells (D). The percentage of infected cells was determined by flow cytometry after immunostaining for CA. Shown are representative results of a single experiment. Identical results were obtained on three separate occasions using independently produced viral stocks.
Disruption of the CypA-CA interaction enhances the efficiency by which HIV-1 VLPs saturate restriction.
Prior to the discovery that TRIM5 restricts retroviruses in primate cells (48, 54), and prior to the widespread use of RNA interference in mammalian systems, HIV-1 VLPs were shown to enhance N-MLV infectivity, if the CA-CypA interaction was disrupted using CsA or the CA G89V mutation (61). Given that TRIM5αhu exhibits a mild inhibitory effect on wild-type HIV-1 (Fig. (Fig.1),1), one would expect wild-type HIV-1 VLPs to also increase the N-MLV titer, though small phenotypes in such technically difficult saturation experiments might be difficult to detect. The effect of HIV-1 VLPs on N-MLV titer was reexamined here in the context of CypA and TRIM5 knockdown cells.
TE671-CypA-shRNA cells or control TE671-Luc-shRNA cells were infected with a constant amount of N-MLVGFP in the presence of increasing amounts of HIV-1 VLPs, either wild type or G89V. Consistent with previous observations (61), HIV-1 G89V VLPs enhanced N-MLV titer in both TE671-CypA-shRNA and control TE671-Luc-shRNA cells (Fig. (Fig.7A).7A). In TE671-Luc-shRNA cells, wild-type HIV-1 VLPs increased N-MLV titer approximately threefold, though these effects were only evident at the highest quantities of VLPs administered (Fig. (Fig.7B).7B). The same VLPs were much more efficient at abrogating restriction to N-MLV in TE671-CypA-shRNA cells, indicating the importance for the saturation mechanism of a factor that selectively recognizes HIV-1 CA in the absence of CypA (Fig. (Fig.7B7B).
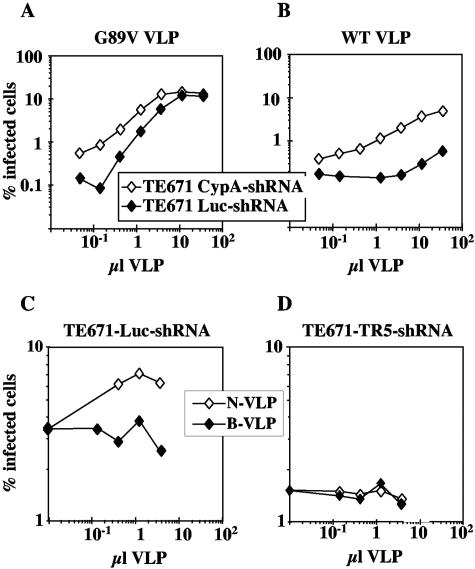
Abrogation of N-MLV restriction in TE671 cells by HIV-1 VLPs is enhanced by CypA knockdown. TE671-CypA-shRNA or TE671-Luc-shRNA cells were infected with constant amounts of N-MLVGFP in the presence of increasing quantities of HIV-1 G89V VLPs (A) or HIV-1 WT VLPs (B). TE671-Luc-shRNA (C) or TE671-TR5-shRNA (D) cells were infected with constant amounts of HIV-1GFP G89V in the presence of increasing quantities of N- or B-MLV VLPs. The percentage of GFP-positive (infected) cells was determined by flow cytometry. Identical results were obtained on three separate occasions using independent VLP stocks.
A VLP titration experiment was also performed in the context of TRIM5αhu knockdown. Here cells were challenged with a fixed amount of HIV-1GFP G89V in the presence of increasing quantities of either N-MLV or B-MLV VLPs. Compared with B-MLV VLPs, N-MLV VLPs specifically increased HIV-1GFP G89V titer in TE671-Luc-shRNA cells (Fig. (Fig.7C).7C). No infectivity increase was observed in TE671-TR5-shRNA cells (Fig. (Fig.7D).7D). These results indicate that TRIM5αhu is required for saturation of restriction machinery by N-MLV VLPs.
HIV-1 CA mutant A92E is not hypersensitive to TRIM5αhu.
HIV-1 CA mutant A92E was selected by serial passage of HIV-1NL4-3 in the presence of a nonimmunosuppressive analogue of CsA (1, 26, 51, 66). In some cell lines, A92E renders HIV-1 CsA resistant. In HeLa cells, A92E is CsA dependent and its infectivity is stimulated by removal of target cell CypA (1, 26, 51, 66). The inhibitory effect of CypA on A92E replication in HeLa cells is reminiscent of the situation in Old World primate cells, where CypA is required for TRIM5αhu-mediated restriction of HIV-1 (6).
To directly examine the mechanism underlying the CsA dependence of A92E replication, shRNA was used to downregulate endogenous TRIM5αhu in HeLa cells. N-tropic and B-tropic MLVGFP virions were used to confirm TRIM5αhu knockdown (Fig. 8A and B), as described above. Cells were then challenged with HIV-1GFP wild type or A92E mutant, in the presence or absence of CsA. HIV-1GFP A92E showed the same CsA dependence in HeLa-TR5-shRNA cells as in the control HeLa cells (Fig. 8C and D). These results indicate that the unusual phenotype of this CA mutant is not explained by increased sensitivity to TRIM5αhu-mediated restriction.
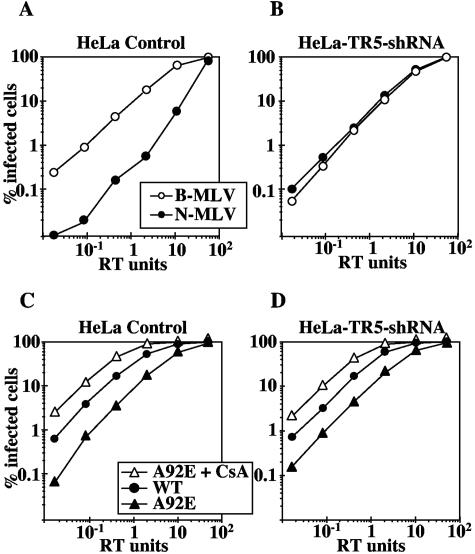
CsA dependence of HIV-1 CA mutant A92E is TRIM5αhu independent. HeLa cells were transduced with an MLV-based vector delivering an shRNA expression construct specific for human TRIM5. VSV-G-pseudotyped, N- and B-tropic MLVGFP virions were used to infect control HeLa cells (A) or HeLa-TR5-shRNA cells (B). VSV-G-pseudotyped, HIV-1GFP WT or A92E mutant virions were used to infect control HeLa cells (C) or HeLa-TR5-shRNA (D) cells, in the presence or absence of CsA, as indicated. The percentage of infected cells was determined by flow cytometry. Shown are representative results of a single experiment. Identical results were obtained on three separate occasions using independently produced viral stocks.
DISCUSSION
Endogenous TRIM5αhu restricts HIV-1.
In this report, several issues were addressed concerning CypA and TRIM5αhu. Previous studies had suggested that HIV-1 is susceptible to restriction by TRIM5αhu, but only when interaction between CA and CypA is disrupted (61). Other studies had shown that supranormal levels of TRIM5αhu inhibit the titer of wild-type HIV-1, even in the presence of CypA (27, 42, 52, 55). Here, endogenous levels of TRIM5αhu were shown to restrict HIV-1 in cells with normal CypA expression (Fig. (Fig.1).1). This innate anti-HIV-1 activity of TRIM5αhu holds significant potential for understanding the epidemiology, clinical course, and pathogenesis of AIDS. Polymorphisms in the gene encoding TRIM5αhu, or differences in TRIM5αhu expression level, might determine an individual's susceptibility to the acquisition of HIV-1 infection. Alternatively, TRIM5αhu might be added to the long list of HIV-1 replication barriers that contribute to the duration of clinical latency.
Effects of CypA on HIV-1 infectivity are independent of endogenous TRIM5αhu.
Endogenous TRIM5αhu and CypA both influence HIV-1 infectivity, but, in contrast to findings in Old World primates, where HIV-1 restriction by TRIM5α is CypA dependent (6), it was clearly demonstrated here that endogenous TRIM5αhu and CypA influence HIV-1 infectivity independently of each other (Fig. (Fig.22 and and4).4). These findings held for HIV-1 infection of adherent cells and of CD4+ T cells (Fig. (Fig.5).5). In apparent contrast to a previous study with CsA (3), the effects of the two host factors were the same whether HIV-1 entered target cells via direct fusion with the cytoplasm or via an endocytic pathway (Fig. (Fig.6).6). The discrepancy with the earlier report can be explained by the fact that the phenomena studied here concern CypA's role in postentry events (26, 51, 61) and the previous study examined the effect of CsA when the drug was administered during virion production (3). The inhibitory mechanism of the drug given during virion production, in fact, does not involve disruption of CypA or the CA-CypA interaction (51).
Evidence that CypA protects HIV-1 from a restriction factor.
Since the discovery of the HIV-1 CA/CypA interaction (35), several models have been proposed to explain the role of CypA in HIV-1 replication. The finding that HIV-1 is particularly good at saturating the TRIM5αhu-dependent restriction of N-MLV when CypA is removed (61) set the stage for the most recent model, namely, that CypA protects HIV-1 from restriction by TRIM5αhu. This model was supported by the discovery of the owl monkey fusion protein TrimCyp (48), the existence of which hinted that the functions of CypA and TRIM5 were somehow associated. In cells from rhesus macaques and African green monkeys, the TRIM5α orthologues are indeed CypA dependent (6). Ever-more-convincing evidence was provided when a TE671 clone (17H1) that had been selected for loss of N-MLV restriction was found to be permissive for HIV-1 in a CypA-independent manner (47). Though all the results described above indicate that CypA protects HIV-1 from a host restriction factor, it was disappointing to find that neither TRIM5αhu expression levels nor the TRIM5αhu sequence was altered in clone 17H1 (47) (data not shown). To accommodate these reports, and the data presented here, the model requires modification: CypA does not protect HIV-1 from TRIM5αhu but from an unknown restriction factor. Though much progress has been made over the past 13 years, the exact role of CypA in HIV-1 replication remains a mystery.
Possible mechanisms by which CypA regulates HIV-1 CA recognition by the restriction machinery.
CypA catalyzes the cis-trans isomerization of peptidyl-prolyl bonds (20, 33, 34, 56), and HIV-1 CA has been shown to be a catalytic substrate by nuclear magnetic resonance (11). In the absence of CypA, nearly 90% of the covalent bonds between glycine 89 and proline 90 are in the trans conformation (23). If restriction machinery only recognizes CA in its trans conformation, then CypA might protect HIV-1 by assisting in the formation of the restriction-resistant cis-isoform. Alternatively, CypA binding might mask CA residues or induce an allosteric change in CA that prevents recognition by the restriction machinery.
Cross-saturation and retroviral restriction pathways.
Given that restriction of N-tropic MLV requires TRIM5αhu (27, 32, 42, 64), and given that CypA and TRIM5αhu act independently to regulate HIV-1 infectivity (Fig. (Fig.4),4), how does one explain the heightened ability of N-MLV and HIV-1 to cross-saturate when CypA is removed (61) (Fig. (Fig.7)?7)? Cross-saturation implies the existence of a common factor required for restriction of both viruses. Biochemical experiments (49), as well as extensive genetic experiments (27, 32, 42, 64), indicate that TRIM5αhu is the recognition element for N-MLV. Data presented here indicate that an unknown factor must be required for HIV-1 CA recognition in the absence of CypA. How then can these findings be reconciled? The answer is suggested by the phenotype of clone 17H1 (47), which belies the presence of yet another unknown factor that is epistatic to the CA recognition components and required for restriction of both N-MLV and CypA-free HIV-1.
Acknowledgments
We thank David Sayah and Sarah Sebastian for reagents and ideas.
This work was supported by the National Institutes of Health grant RO1AI36199 to J.L.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.80.6.2855-2862.2006
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1395419?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Exploring HIV-1 Maturation: A New Frontier in Antiviral Development.
Viruses, 16(9):1423, 06 Sep 2024
Cited by: 0 articles | PMID: 39339899 | PMCID: PMC11437483
Review Free full text in Europe PMC
Capsid-dependent lentiviral restrictions.
J Virol, 98(4):e0030824, 18 Mar 2024
Cited by: 0 articles | PMID: 38497663 | PMCID: PMC11019884
Review Free full text in Europe PMC
A Tripartite Complex HIV-1 Tat-Cyclophilin A-Capsid Protein Enables Tat Encapsidation That Is Required for HIV-1 Infectivity.
J Virol, 97(4):e0027823, 11 Apr 2023
Cited by: 3 articles | PMID: 37129415 | PMCID: PMC10134889
HIV-1 capsid variability: viral exploitation and evasion of capsid-binding molecules.
Retrovirology, 18(1):32, 26 Oct 2021
Cited by: 14 articles | PMID: 34702294 | PMCID: PMC8549334
Review Free full text in Europe PMC
Cytoplasmic CPSF6 Regulates HIV-1 Capsid Trafficking and Infection in a Cyclophilin A-Dependent Manner.
mBio, 12(2):e03142-20, 23 Mar 2021
Cited by: 29 articles | PMID: 33758083 | PMCID: PMC8092277
Go to all (82) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Cyclophilin A protects HIV-1 from restriction by human TRIM5α.
Nat Microbiol, 4(12):2044-2051, 21 Oct 2019
Cited by: 89 articles | PMID: 31636416 | PMCID: PMC6879858
Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells.
J Virol, 79(1):176-183, 01 Jan 2005
Cited by: 145 articles | PMID: 15596813 | PMCID: PMC538701
The V86M mutation in HIV-1 capsid confers resistance to TRIM5α by abrogation of cyclophilin A-dependent restriction and enhancement of viral nuclear import.
Retrovirology, 10:25, 28 Feb 2013
Cited by: 20 articles | PMID: 23448277 | PMCID: PMC3598646
The control of viral infection by tripartite motif proteins and cyclophilin A.
Retrovirology, 4:40, 12 Jun 2007
Cited by: 149 articles | PMID: 17565686 | PMCID: PMC1906832
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (2)
Grant ID: R01 AI036199
Grant ID: R01AI36199




