Abstract
Free full text

Interaction of Serum Response Factor (SRF) with the Elk-1 B Box Inhibits RhoA-Actin Signaling to SRF and Potentiates Transcriptional Activation by Elk-1
Abstract
Serum response factor (SRF) is a transcription factor which regulates many immediate-early genes. Rho GTPases regulate SRF activity through changes in actin dynamics, but some SRF target genes, such as c-fos, are insensitive to this pathway. At the c-fos promoter, SRF recruits members of the ternary complex factor (TCF) family of Ets domain proteins through interactions with the TCF B-box region. Analysis of c-fos promoter mutations demonstrates that the TCF and ATF/AP1 sites adjoining the SRF binding site inhibit activation of the promoter by RhoA-actin signaling. The presence of the TCF binding site is sufficient for inhibition, and experiments with an altered-specificity Elk-1 derivative demonstrate that inhibition can be mediated by the Elk-1 TCF. Using Elk-1 fusion proteins that can bind DNA autonomously, we show that inhibition of RhoA-actin signaling requires physical interaction between the Elk-1 B box and SRF. These results account for the insensitivity of c-fos to RhoA-actin signaling. Interaction of the B box with SRF also potentiates transcriptional activation by the Elk-1 C-terminal activation domain. Combinatorial interactions between SRF and TCF proteins are thus likely to play an important role in determining the relative sensitivity of SRF target genes to Ras- and Rho-controlled signal transduction pathways.
Serum response factor (SRF) is a MADS-box transcription factor which controls both growth factor-responsive immediate-early genes and many muscle-specific genes (1, 35). The activity of SRF is regulated by a novel signal transduction pathway involving by Rho-family GTPases. Activated Rho GTPases suffice to activate SRF in the absence of extracellular stimuli, and functional Rho itself is required for SRF activation by serum mitogens such as LPA (17). Signal-stimulated SRF activation is inhibited by drugs that prevent actin polymerization, such as latrunculins, and conversely, SRF can be activated in the absence of external signals by drugs and proteins that promote F-actin accumulation (8, 11, 24, 32, 34). These findings suggest that SRF activity is sensitive to depletion of the G-actin pool (32).
At many of its target genes, SRF forms a ternary complex with members of a family of Ets domain proteins known as ternary complex factors (TCFs), whose activity is controlled by mitogen-activated protein kinase (MAPK) signaling pathways (36). Each of the three TCFs, Elk-1, SAP-1, and Net, contains a conserved 20-amino acid B box which mediates interaction with the SRF DNA-binding domain (4, 15, 31), and mutagenesis and structural studies have identified residues essential for ternary complex formation (13, 22, 23). A novel feature of the complex is a flexible linker between the N-terminal Ets domain and the B box which allows the TCFs to bind cooperatively with SRF to DNA sites separated by a variety of spacings (38). Phosphorylation of the TCFs at multiple sites within their conserved C-terminal regulatory domains can promote both transcriptional activation (9, 19, 25) and ternary complex formation (9, 10, 39), although at the c-fos promoter, TCF binding does not appear to be regulated in vivo (14). The c-fos TCF binding site is required for activation of the promoter by stimuli which act predominantly through the ERK signaling pathway, such as the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) and receptor tyrosine kinases (12, 16, 21, 27).
The availability of specific inhibitors of the signaling pathways controlling the TCFs and SRF allows the contribution of these pathways to the activation of SRF target genes to be assessed directly. Intriguingly, RhoA-actin and ERK signaling appear to act in a mutually exclusive manner at several SRF-controlled immediate-early genes: the c-fos and egr-1 genes are sensitive to ERK signaling but insensitive to the RhoA-actin pathway, while the converse holds true for the srf and vinculin genes (11). In this study, we show that it is TCF binding to the c-fos promoter which inhibits its activation via RhoA-actin signaling and that this requires contact between the B box and SRF. We also demonstrate that interaction of the B box with SRF potentiates transcriptional activation by the Elk-1 TCF.
MATERIALS AND METHODS
Plasmids.
The c-fos promoter mutants are derivatives of pF711, which contains the entire human c-fos gene including 711 bp of 5′ flanking sequences (37). Mutants ΔSIF, ΔTCF, ΔSIFΔTCF, and 3D.AFos have been described previously (16, 18). The ΔAP1/ATF, 2xTCF, and ΔAP1/ATFΔTCF mutations were introduced by two-step PCR with pF711 or ΔTCF as a template and replacement of the p711 small EcoRI-BssHI fragment with the mutant. MLV.FlagElk (3); NL.Elk (15); Gal-ElkΔ33 (published as GAL4-ΔRI 25); and MLV.lacZ, MLVα118, and RNase protection probe templates pSP6Fos5′ and SP6α132 (17, 18) were described previously. PCR was used to introduce into Gal-ElkΔ33 the ElkNA mutation, in which each of the nine C-terminal regulatory region S/T-P motifs is mutated to AP (a gift from R. Thomas), and the previously described Y159A and L158P mutations (22).
Transfections and gene expression assays.
NIH3T3 cells in 60-mm-diameter dishes were transiently transfected with LipofectAMINE (Life Technologies, Inc.) according to the manufacturer's recommendations. For RNase protection assays, cells were transfected with 0.5 μg of c-fos promoter mutant, 0.3 μg of MLVα118, and expression plasmids reaching a 2-μg total with the addition of MLVplink or pUC19 as appropriate. For luciferase assays, cells in a six-well plate were transfected with 10 ng of the 5xGal.LUC reporter gene, 0.3 μg of MLVlacZ, and 50 ng of GAL4.Elk derivative reaching 1 μg of total DNA with the addition of pUC19. Cells were maintained in medium containing 0.3% fetal calf serum (FCS) for 24 h and then stimulated with 15% serum or 85 nM TPA for 30 min (RNase protection assays) or 7 h (luciferase assays). The inhibitors U0126 (Promega) and latrunculin B (Calbiochem) were added at 10 and 0.5 μM, respectively, 30 min before stimulation. RNA preparation and RNase protection assays were done as described previously (17, 18, 37); quantitation was carried out by phosphorimaging using ImageQuant software (Molecular Dynamics), with normalization to the cotransfected α-globin reference signal (11). Luciferase assays were performed by standard techniques, with normalization to a cotransfected β-galactosidase control.
Cell extracts and mobility shift assays.
For gel mobility shift assays and immunoblotting, plasmids expressing Elk-1 derivatives were transfected as described above (1 μg per 6-cm-diameter dish); serum stimulation was for 15 min. For Elk-1 and NL.Elk, extracts were prepared as described previously (25). For Gal-ElkΔ33 fusion proteins, cells were lysed in a solution containing 20 mM HEPES (pH 7.9), 10% glycerol, 0.4 M KCl, 0.4% Triton X-100, 0.2 mM EDTA, 0.5 mM dithiothreitol, 5 mM NaF, 0.1 μg of okadaic acid/ml, and protease inhibitors. Binding reactions for Elk-1 and NL.Elk contained 0.4 μg of extract in 10 μl of binding buffer {10 mM Tris-HCl [pH 7.9], 50 mM NaCl, 1 mM EDTA, 0.5 mM dithiothreitol, 50 ng of ovalbumin/ml, 50 ng/of poly(dI-dC)·poly(dI-dC)μl, and SRFs from residues 133 to 265 [SRF(133-265)]} with 0.5 ng of the c-fos promoter mutant probe and were incubated for 15 min at room temperature. For Gal-ElkΔ33 binding assays, 4 μg of whole-cell extracts was used in binding buffer without EDTA. Probes were generated by PCR as described previously (38) with primers p10 (5′ CGCACTGCACCCTCGGTGTTGGCTGC 3′) and p11 (5′ ATGGCTCCCCCCAGGGCTACAGGGAAAG 3′). Complexes were resolved in a 5% 37.5:1 acrylamide-bis-acrylamide-0.5× Tris-borate-EDTA gel.
RESULTS
c-fos promoter mutations.
The c-fos upstream regulatory region contains an SRF site flanked by an Ets motif and an ATF/AP1 site, with a STAT binding site some 30 bp upstream (Fig. (Fig.1A).1A). To investigate how this sequence context can inhibit activation of SRF by the RhoA-actin pathway, we investigated the regulatory properties of c-fos promoter mutants in which the STAT binding site is replaced with a consensus Gal4 operator, the TCF Ets motif is replaced with a half-operator for LexA, and the SRF site is replaced with an optimized binding site for the SRF-related Mcm1 protein as previously reported (16) (Fig. (Fig.1A,1A, ΔSIF, ΔTCF, and ΔSRF). The ATF/AP1 site was disrupted either by a quadruple transversion (29) or by the introduction of a second optimal Ets motif to create an SRF site flanked by TCF sites (Fig. (Fig.1A,1A, ΔATF/AP1 and 2xTCF). The ΔATF/AP1 and ΔTCF mutations were also combined to generate a promoter lacking both the TCF and ATF/AP1 sites (Fig. (Fig.1A,1A, ΔTCFΔATF/AP1).
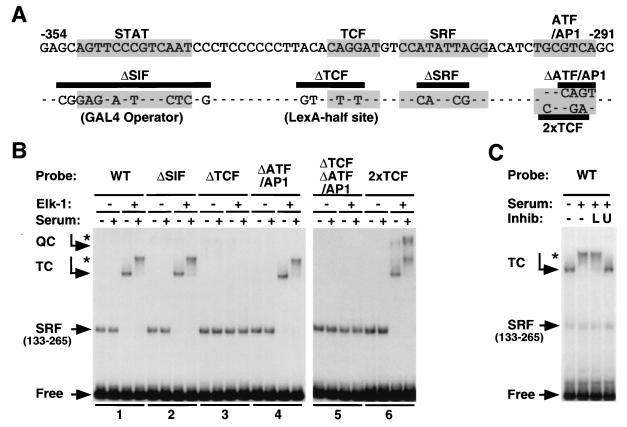
c-fos 5′ regulatory region. (A) Structure of the c-fos promoter. The sequence of the c-fos upstream regulatory region from −291 to −354 (according to reference 35) is shown. The consensus binding sites for STAT factors, TCFs, SRF, and ATF/AP1 factors are shaded. Below the sequence, the different promoter mutations are shown, with dashes indicating identity to the wild-type sequence; the ΔSIF and ΔTCF mutations generate the GAL4 operator and LexA half-site, respectively. The different promoter mutants contain the underlined clusters of mutations either singly or in combination, i.e., ΔTCFΔATF/AP1 lacks both the TCF and ATF/AP1 sites but retains an intact STAT binding site. (B) Promoter mutants. The effect of mutations on ternary complex formation is shown. Cells were transfected with MLV.plink or MLV.FlagElk. Whole-cell extracts were assayed for ternary complex activity with added SRF(133-265) by using the indicated c-fos promoter probes of equal specific activity. WT, wild type. (C) Effect of inhibitors on ternary complex formation. Cells were transfected with MLV.FlagElk and stimulated with serum for 15 min following pretreatment with 0.5 μM latrunculin B (L) or 10 μM UO126 (U).
To examine ternary complex formation on the different promoter mutants, we produced Elk-1 by overexpression in NIH3T3 cells and used it in gel mobility shift assays. All the mutants containing an intact Ets motif formed a ternary complex with the SRF DNA-binding domain, the minimal SRF region sufficient for ternary complex formation (25). An additional slower mobility complex was formed with the double Ets motif mutant 2xTCF, which presumably represents a quaternary complex containing two molecules of Elk-1 (Fig. (Fig.1B).1B). The mobility of the SRF-Elk-1 ternary complexes was further reduced upon serum stimulation, reflecting phosphorylation of the Elk-1 C terminus (Fig. (Fig.1C).1C). In agreement with functional studies, serum-induced modification of the ternary complex was prevented by blockade of Raf-ERK signaling with the specific MEK inhibitor U0126 (Fig. (Fig.1C)1C) (6). In contrast, the ternary complex was unaffected by treatment with latrunculin B, which sequesters G-actin and inhibits signaling to SRF but which does not inhibit ERK signaling (Fig. (Fig.1C)1C) (7, 11). Thus, Elk-1 modification is unaffected by inhibition of the RhoA-actin signaling pathway to SRF.
TCF and AP1/ATF sites inhibit c-fos activation by RhoA-actin signaling.
To evaluate the contributions of RhoA-actin and MEK-ERK signaling to c-fos transcription, activation of the c-fos promoter mutants was measured in cells pretreated with the pathway-specific inhibitors latrunculin B and U0126. Each mutant was transfected into NIH3T3 cells, and transcriptional activation following serum stimulation was quantified relative to that of a cotransfected human α-globin reference plasmid. Representative results are shown in Fig. Fig.2A2A and summarized in Fig. Fig.2B.2B. As with the endogenous c-fos gene, induction of a transfected wild-type c-fos gene was strongly inhibited upon blockade of ERK activation by U0126 but only slightly affected upon inhibition of RhoA-actin signaling by latruculin B (Fig. (Fig.2A,2A, lanes 1 [11]). Serum induction of a transfected synthetic SRF reporter gene, 3D.AFos, was substantially inhibited by latrunculin B treatment but essentially unaffected by U0126, as previously observed with an integrated version of this reporter (Fig. (Fig.2A,2A, compare lanes 1 and 7). Signaling to both the c-fos gene and an SRF reporter is thus faithfully reproduced by using transfected templates.
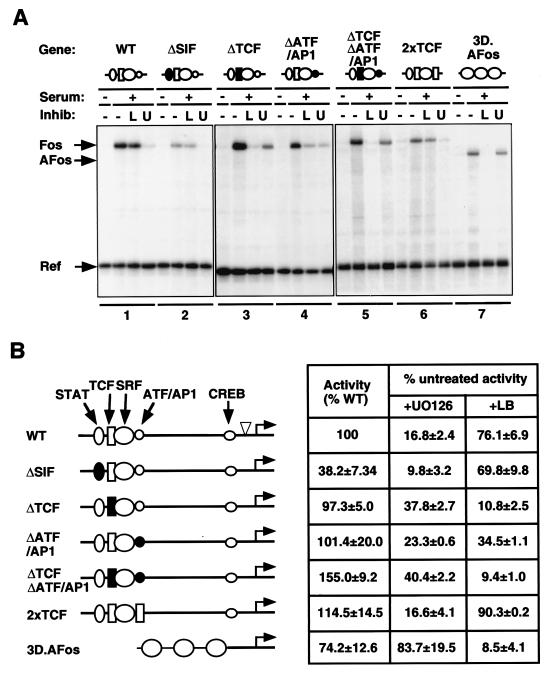
Promoter elements restricting the RhoA-actin signaling pathway to the c-fos promoter. (A) Representative experimental data. Cells were transfected with the indicated c-fos promoter mutants together with the α-globin reference gene. In this and subsequent figures, the c-fos regulatory region is shown schematically at the top with symbols as follows: ellipse, STAT site; rectangle, TCF binding site; circle, SRF binding site; small circle, AP1/ATF site. Open and filled symbols denote intact and mutated sites, respectively. Cells were serum-stimulated following pretreatment with 0.5 μM latrunculin B (L) or 10 μM U0126 (U) as indicated; RNA was prepared 30 min later for analysis by RNase protection assay. WT, wild type. (B) Data summary. Human c-fos transcript levels were quantified relative to those of the α-globin reference plasmid using the PhosphorImager. The first column shows the RNA level relative to that of the intact gene ± the standard error of the mean. The second and third columns show the activity remaining upon inhibitor treatment, taking the untreated value as 100% for each mutant, ± the standard error of the mean.
Disruption of the STAT binding site did not affect signaling pathway utilization, although it significantly reduced promoter activity (Fig. (Fig.2A,2A, compare lanes 1 and 2, and B) as previously reported (16). Mutation of the TCF binding site did not impair serum induction, in agreement with that found in previous reports (12, 16), but resulted in a substantially increased sensitivity to latrunculin B compared with that of the intact promoter (Fig. (Fig.1A,1A, compare lanes 1 and 3, and 2B). The ΔTCF mutant was also less sensitive to inhibition by U0126 than the intact promoter but remained more sensitive to this inhibitor than the SRF reporter gene 3D.AFos, suggesting that activation of the c-fos promoter requires other ERK-sensitive elements (Fig. (Fig.2A,2A, lanes 1 and 3, and B). Compared to that of the wild-type promoter, serum induction of the ΔATF/AP1 mutant was also more sensitive to latrunculin B and less sensitive to U0126 (Fig. (Fig.2A,2A, compare lanes 1 and 4). However, the effect of the ΔATF/AP1 mutation was less marked than that of the ΔTCF mutation (Fig. (Fig.2A,2A, compare lanes 3 and 4, and B). Combination of the ΔATF/AP1 and ΔTCF mutations had no greater effect on signal pathway utilization than did disruption of the TCF site alone (Fig. (Fig.2A,2A, compare lanes 5 and 3). Finally, replacement of the ATF/AP1 site with a second TCF site in the 2xTCF mutant generated a promoter that behaved in a manner similar to that in the wild-type gene, demonstrating that the ATF/AP1 site is not required for the inhibition of RhoA-actin signaling to SRF (Fig. (Fig.2A,2A, compare lanes 6 and 1, and B). These data establish that the TCF binding site is a major determinant of the insensitivity of the c-fos promoter to the RhoA-actin signaling pathway. However, the substantial remaining dependence of all the mutants on ERK signaling suggests that the c-fos promoter contains other ERK-sensitive elements that are required for its full activation (see Discussion).
Elk-1 binding inhibits RhoA-actin signaling to SRF.
To test the relevance of TCF proteins to the inhibition of RhoA signaling to SRF, we exploited the fact that the c-fos ΔTCF mutation introduces a LexA half-operator next to the SRF binding site. Replacement of the Elk-1 Ets domain with the N-terminal DNA-binding domain of the LexA repressor generates an altered-specificity Elk-1 derivative, NL.Elk, which can form an SRF-dependent ternary complex with the ΔTCF mutant DNA in vitro (15) (Fig. (Fig.3A3A and B). The NL.Elk protein also forms a functional ternary complex with the ΔTCF promoter in vivo, since its expression can restore TPA inducibility to the c-fos ΔTCF mutant, which is defective in this response (18). We therefore tested whether the expression of NL.Elk affected signaling pathway utilization by the c-fos ΔTCF promoter mutant. Expression of NL.Elk did not affect the serum-induced activity of the ΔTCF mutant but rendered transcription substantially resistant to inhibition by latrunculin B (Fig. (Fig.3C).3C). Control experiments showed that NL.Elk expression did not affect the serum inducibility of the wild-type c-fos promoter (Fig. (Fig.3C).3C). These data show that recruitment of Elk-1 to the c-fos promoter interferes with RhoA-actin signaling to SRF.

TCF Elk-1 inhibits RhoA-actin signaling to SRF in the c-fos promoter. (A) Structure of the altered-specificity NL.Elk protein. Black boxes represent the Ets domain (ETS), B box (B), and C-terminal regulatory region (C); the white box represents the LexA DNA-binding domain. (B) Ternary complex formation by NL.ELK. Gel mobility shift assays were performed by using extracts from cells transfected with the MLV.plink vector or NL.ELK expression plasmid. Binding reactions included recombinant SRF(133-265) and probe from the c-fos ΔTCF mutant. (C) Effect of overexpression of NL.Elk on activation of c-fos ΔTCF. Cells were transfected with either wild-type (WT) or ΔTCF mutant c-fos genes, α-globin reference, and either vector or NL.ELK expression plasmids and stimulated as described in the legend to Fig. Fig.22.
B-box mutations do not affect transcriptional activation and DNA binding by Gal4-Elk-1 fusion proteins.
The experiments described in the preceding sections show that the Elk-1 TCF can prevent activation of the c-fos promoter via RhoA-actin signaling to SRF. However, they do not identify the Elk-1 sequences involved. Moreover, since recruitment of the NL.Elk protein to the promoter is absolutely dependent on its interaction with SRF, it cannot be used to determine whether the inhibition of RhoA-actin signaling to SRF requires physical contact between Elk-1 and SRF or merely the presence of Elk-1 on the promoter. To address these issues, we exploited an Elk-1 derivative, Gal-ElkΔ33, whose interaction with DNA can occur independently of SRF binding. In Gal-ElkΔ33, the N-terminal 33 residues of the Elk-1 Ets domain are replaced with the intact Gal4 DNA-binding domain (Fig. (Fig.4A)4A) (25). Previous biochemical and structural studies of ternary complex formation indicate that the linker between the TCF Ets domain and B box is flexible (4, 13, 15, 38), and we reasoned that if Gal-ElkΔ33 were bound to DNA in the vicinity of SRF, interactions between SRF and its B box should still be able to occur. We also investigated mutant derivatives of Gal-ElkΔ33 lacking all the C-terminal phosphorylation sites (NA mutants) or containing the B-box point mutations L158P or Y159A, which abolish interaction with SRF (22) (Fig. (Fig.4A4A).
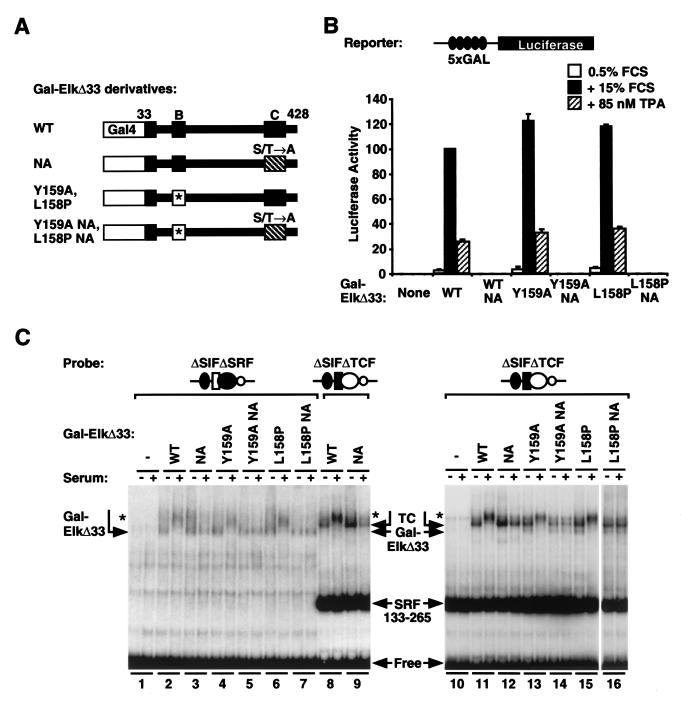
Gal-ElkΔ33 fusion protein. (A) Structure of the protein and its derivatives. Elk-1 sequences are shown in black as in Fig. Fig.2A,2A, with the Gal4 DNA-binding domain represented by a white box. The mutant B box is indicated in white with an asterisk, and the Ala substitutions at the nine C-terminal MAPK phosphorylation sites are indicated by stripes. WT, wild type. (B) Reporter assay of Gal-ElkΔ33 proteins. Cells were transfected with the 5xGal.LUC reporter plasmid and either the vector or Gal-ElkΔ33 expression plasmids, together with the control plasmid MLV.LacZ, and maintained in 0.5% FCS for 24 h. Following stimulation with 15% FCS or 85 nM TPA, reporter activity was measured. Luciferase activity, normalized to control β-galactosidase activity, is expressed relative to that of serum-induced GAL4.Elk Δ33 as 100%. Data are given as averages from three independent experiments ± standard errors of the means, except for the NA mutants (n = 2). (C) DNA binding properties of the Gal-ElkΔ33 fusion proteins. Whole-cell extracts were prepared from serum-starved or -stimulated cells expressing Gal-ElkΔ33 fusion proteins as indicated. Lanes 1 to 6, binding assays performed in the absence of the SRF core DNA-binding domain by using the ΔSIFΔSRF probe; lanes 7 to 16, binding assays performed with the SRF core DNA-binding domain by using the ΔSIFΔTCF probe. Arrows indicate the Gal-ElkΔ33 complex, the Gal-ElkΔ33/SRF complex, and SRF(133-265); phosphorylated forms are indicated by asterisks.
We first tested the properties of Gal-ElkΔ33 in a transient transfection assay using a reporter gene controlled by five tandem copies of the Gal4 operator. Upon activation of the ERK pathway by serum or TPA stimulation, activity of the reporter was substantially increased in cells expressing Gal-ElkΔ33. Activation required the C-terminal phosphorylation sites but was not affected by either B-box mutation (Fig. (Fig.4B).4B). To examine DNA binding by the Gal-ElkΔ33 derivatives, we performed gel mobility shift assays with a probe prepared from the c-fos promoter mutant ΔSIFΔSRF, which contains a single Gal4 operator and no SRF binding site (Fig. (Fig.1A).1A). Extracts from cells expressing Gal-ElkΔ33 generated a low-mobility complex whose mobility was further decreased upon serum stimulation; a similar complex formed by Gal-ElkΔ33NA, which lacks the C-terminal phosphorylation sites, remained unaffected by serum stimulation, as expected (Fig. (Fig.4C,4C, lanes 2 and 3). The B-box mutant derivatives of Gal-ElkΔ33 and Gal-ElkΔ33NA behaved similar to those of the corresponding intact proteins (Fig. (Fig.4C,4C, compare lanes 4 to 7 with 2 and 3). Taken together, these results suggest that the Gal-ElkΔ33 proteins can bind DNA irrespective of serum stimulation and that mutation of the B box affects neither DNA binding nor transcriptional activation.
We examined the ability of Gal-ElkΔ33 to bind DNA in the presence of SRF by using a probe prepared from the c-fos ΔSIFΔTCF promoter mutant which contains an intact SRF binding site in addition to the Gal4 operator. In the absence of added SRF, extracts containing Gal-ElkΔ33 generated complexes of mobility identical to that generated on the ΔSIFΔSRF probe (data not shown). In the presence of the excess recombinant SRF core DNA-binding domain SRF(133-265), however, Gal-ElkΔ33 extracts generated a complex of slightly lower mobility than those containing Gal-ElkΔ33 alone (Fig. (Fig.4C,4C, compare lanes 2 to 7 with 8 and 9). Under these assay conditions, the majority of Gal-ElkΔ33 is present in complexes which also contain bound SRF(133-265). As expected, the mobility of these complexes was further reduced upon serum stimulation, provided that the C-terminal Elk-1 phosphorylation sites were intact (Fig. (Fig.4C,4C, lanes 8 and 9 and 11 and 12). The Gal-ElkΔ33 derivatives containing the B-box mutations L158P or Y159A bound DNA with equal efficiency in this assay (Fig. (Fig.4C,4C, lanes 13 to 16). However, in this case, complexes characteristic of the Gal4-ElkΔ33 mutants bound alone were also weakly detectable, suggesting that the intact B box favors the binding of Gal4-ElkΔ33 to an SRF(133-265)-bound DNA probe (Fig. (Fig.4C,4C, compare lanes 13 to 16 with 11 and 12). Similar results were obtained when the interaction of the Gal-ElkΔ33 derivatives with endogenous cellular SRF was analyzed (data not shown). Taken together, these data show that the Gal-ElkΔ33 protein binds autonomously to DNA and suggest that the B box remains available for interaction with SRF in this context.
Contact with the B box inhibits RhoA-actin signaling to SRF.
To test the effect of Gal-ElkΔ33 proteins on RhoA-actin signaling to SRF at the c-fos promoter, we used the promoter mutant ΔSIFΔTCF. In this gene, the SRF binding site remains intact while the STAT and TCF sites are respectively replaced by a Gal4 operator and a LexA half-site (Fig. (Fig.1A).1A). In transfection assays, the ΔSIFΔTCF mutant exhibits reduced serum inducibility and is not inducible by TPA (16). Consistent with the results of the promoter mutant studies, the serum induction of the ΔSIFΔTCF promoter was substantially inhibited by latrunculin B, presumably owing to the absence of the TCF binding site (Fig. (Fig.5A,5A, lanes 1). Expression of the NL.Elk protein did not affect the serum inducibility of ΔSIFΔTCF but rendered it resistant to inhibition by latrunculin B (Fig. (Fig.5A,5A, compare lanes 1 and 2). The ΔSIFΔTCF promoter thus behaves similarly to the ΔTCF mutant and can therefore be used to study the effect of Gal4-Elk-1 fusion proteins on signaling to SRF.
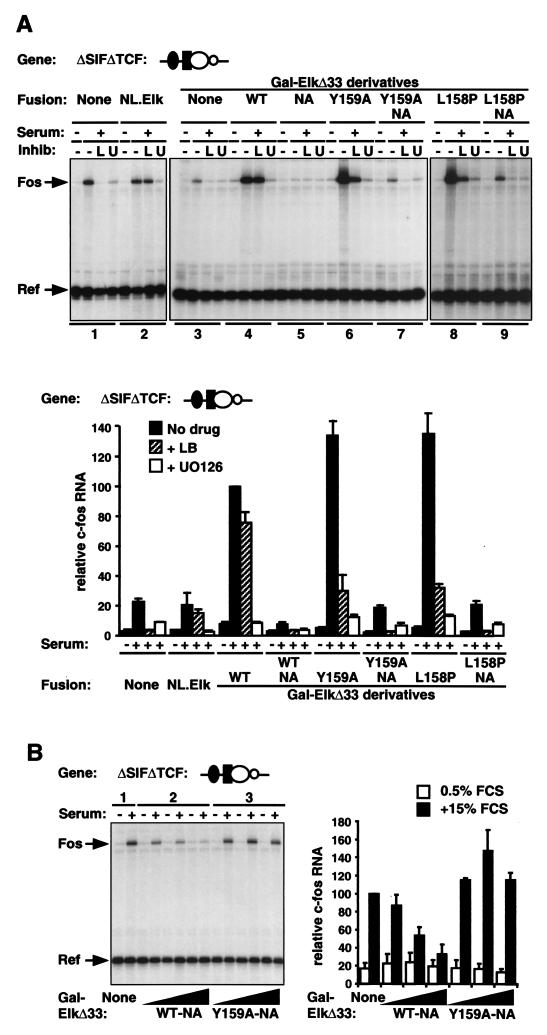
Activation of the c-fos promoter by Gal-ElkΔ33 derivatives. (A) Representative experimental data. Cells were transfected with the ΔSIFΔTCF reporter gene and either the vector or Gal-ElkΔ33 expression plasmid (50 ng), together with the α-globin reference plasmid. Cells were serum stimulated with or without pretreatment with latrunculin B (L) or U0126 (U), and RNA was prepared for analysis by RNase protection 30 min later. Human c-fos transcript levels were quantified relative to those of the α-globin reference plasmid by using the PhosphorImager, taking serum-stimulated activity in the presence of intact Gal-ElkΔ33 as 100%. WT, wild type. Data are presented as means ± half-ranges from two independent experiments. (B) Inhibition of RhoA-actin signaling to SRF by the B box. Cells were transfected with the c-fos ΔSIFΔTCF mutant and vector or the indicated Gal-ElkΔ33 plasmids (5, 50, or 500 ng) with a reference as described for panel A. Following serum stimulation, RNA was analyzed by RNase protection. Representative data are shown to the left, and the data from two independent experiments (means ± half-ranges) are summarized to the right.
Expression of Gal-ElkΔ33 increased the serum inducibility of ΔSIFΔTCF; moreover, activation was substantially resistant to latrunculin B, indicating its relative independence from RhoA-actin signaling (Fig. (Fig.5A,5A, compare lanes 3 and 4). Induction was completely abolished by the MEK inhibitor U0126, consistent with a role for the Elk-1 C-terminal regulatory domain (Fig. (Fig.5A).5A). These results suggest that, in the presence of Gal-ElkΔ33, RhoA-actin signaling to SRF does not contribute to serum-induced activation of ΔSIFΔTCF. Expression of Gal-ElkΔ33 NA, which lacks the Elk-1 C-terminal phosphorylation sites, failed to potentiate the serum-induced activation of ΔSIFΔTCF and instead reduced activity to below the basal level (Fig. (Fig.5A,5A, lanes 3 to 5, and B, lanes 1 and 2). Thus, inactive Gal-ElkΔ33 on neighboring DNA interferes with serum-induced signaling to SRF.
We investigated the role of the Elk-1 B box in these phenomena by using the Gal-ElkΔ33 L158P and Y159A mutants. Both proteins potentiated the serum inducibility of ΔSIFΔTCF to a similar extent as did intact Gal-ElkΔ33, consistent with the observation that the B-box mutations do not affect DNA binding (Fig. (Fig.4C).4C). However, in contrast to serum induction by intact Gal-ElkΔ33, induction by the mutants was substantially sensitive to latrunculin B, although it also remained sensitive to U0126 (Fig. (Fig.5A,5A, compare lanes 4 and 5 with 6 to 9) (see Discussion). As with the intact Gal-ElkΔ33, maximal activation by the Gal-ElkΔ33 B-box mutants was completely dependent on the integrity of the Elk-1 C-terminal phosphorylation sites (Fig. (Fig.5A,5A, lanes 7 and 9). Expression of the Gal-ElkΔ33 B-box mutants lacking the C-terminal phosphorylation sites did not reduce the serum-induced activity of the ΔSIFΔTCF promoter below its basal level, in contrast to the inhibition seen when the B box was intact (Fig. (Fig.5A,5A, compare lanes 5, 7, and 9, and B). These results show that physical interaction between the B box and SRF, rather than mere binding of the Elk-1 TCF to the c-fos promoter, is required to inhibit RhoA-actin signaling to SRF.
SRF potentiates Elk-1 transcriptional activation through the B box.
The results presented above show that mutation of the B box in Gal-ElkΔ33 reduces the latrunculin B-resistant component of the serum response (Fig. (Fig.5A,5A, compare shaded bars). One potential explanation for this observation is that the activity of the Elk-1 C-terminal domain in Gal-ElkΔ33 is potentiated by the interaction of the B box with SRF; if this were the case, disruption of the interaction would lower Elk-1 transcriptional activity. To address this issue, we investigated the activity of the Elk-1 C-terminal domain in the absence of signaling to SRF. To do this, we examined activation of the reporter by TPA, a stimulus that can activate the Elk-1 C-terminal domain but does not activate transiently transfected SRF reporter genes (17).
TPA treatment did not significantly activate the ΔSIFΔTCF promoter (Fig. (Fig.6A,6A, left), in agreement with the findings of a previous study (16). In the presence of Gal-ElkΔ33, however, TPA induced transcription to a level comparable to that of serum (Fig. (Fig.6A,6A, compare left and right panels). TPA induction was resistant to latrunculin B (Fig. (Fig.6A,6A, left panel) and was also both dependent on the Elk-1 C-terminal phosphorylation sites and inhibited by U0126 (data not shown). Thus, as expected, RhoA-actin signaling to SRF does not contribute to TPA-induced promoter activation. We next tested the Gal-ElkΔ33 B-box mutants Y159A and L158P. TPA-stimulated activation of the ΔSIFΔTCF promoter by these proteins was substantially less efficient than that observed with intact Gal-ElkΔ33 and was again completely resistant to latrunculin B (Fig. (Fig.6A,6A, left panel). The effect of the B-box mutations on the TPA-induced transcription of ΔSIFΔTCF was strikingly similar to their effect on the latrunculin B-independent component of the serum response, as measured in parallel experiments (Fig. (Fig.6A,6A, compare left and right panels).
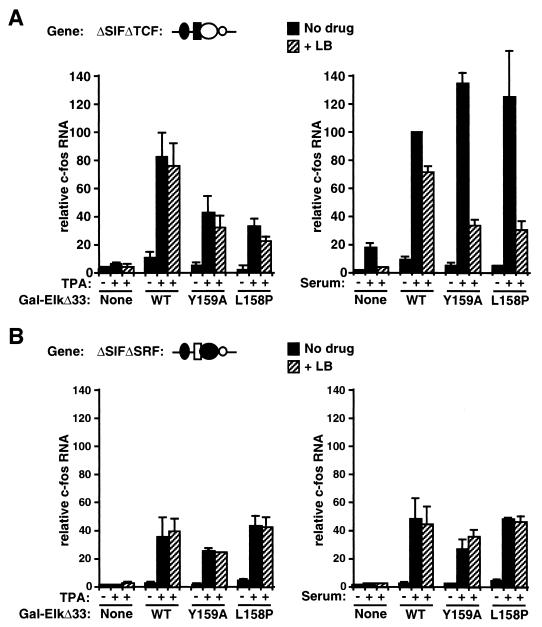
Interaction of the B box with SRF potentiates transcriptional activity of Elk-1. (A) B-box mutations reduce Gal-ElkΔ33 activity in the presence of SRF. Cells were transfected with c-fos promoter ΔSIFΔTCF, together with the α-globin reference gene and either the vector plasmid or the indicated Gal-ElkΔ33 plasmids. c-fos transcription was analyzed following stimulation with either 85 nM TPA (left panel) or 15% serum (right panel), with or without latrunculin B pretreatment (LB). Transcript levels were quantified relative to those of the α-globin reference plasmid by using the PhosphorImager, taking the serum-stimulated activity of ΔSIFΔTCF in the presence of intact Gal-ElkΔ33 as 100%. WT, wild type. Data are presented as means ± standard errors of the means from three independent experiments, except for the B-box mutants (n = 2). (B) B-box mutations do not affect Gal-ElkΔ33 activity in the absence of SRF. The experiment shown in panel A was repeated by using the c-fos promoter ΔSIFΔSRF, which does not contain the SRF binding site.
These results contrast with those obtained in the simple Gal4 reporter gene assay, where the TPA inducibility of Gal-ElkΔ33 was similar to that of its B-box mutant derivatives Y159A and L158P (Fig. (Fig.4B).4B). To test whether this difference in behavior reflects interaction with SRF, we repeated the assays with the c-fos mutant ΔSIFΔSRF, which, in contrast to ΔSIFΔTCF, cannot bind SRF. The ΔSIFΔSRF promoter is unresponsive to stimulation by serum or TPA (Fig. (Fig.6B)6B) (16). As with ΔSIFΔTCF, expression of the intact Gal-ElkΔ33 protein conferred substantial TPA inducibility on ΔSIFΔSRF and this was resistant to latrunculin B (Fig. (Fig.6B,6B, left panel). In this context, however, mutation of the Gal-ElkΔ33 B box had no effect on induction by TPA (Fig. (Fig.6B,6B, left panel). Similar results were obtained when serum was used as the stimulus (Fig. (Fig.6B,6B, right panel). These results strongly suggest that interaction of the Elk-1 B box with SRF potentiates the transcriptional activation of Gal-ElkΔ33. Taken together with the results described in the preceding sections, these experiments suggest a model in which the interaction of SRF with the Elk-1 B box both inhibits activation of SRF via the RhoA-actin pathway and potentiates transcriptional activation by the Elk-1 C-terminal domain.
DISCUSSION
In this study, we investigated the role of TCF in signaling specificity to immediate-early genes controlled by the transcription factor SRF. Using the prototypic immediate-early gene c-fos promoter and derivatives of the Elk-1 TCF as a model system, we found that interaction of SRF with the TCF Elk-1 is sufficient to prevent its serum-induced activation via the RhoA-actin signal pathway. Using Gal4-Elk-1 derivatives which can bind DNA autonomously, we obtained evidence that the interaction of the B box with SRF is required both to inhibit RhoA-actin signaling to SRF and for maximal transcriptional activation by Elk-1 (Fig. (Fig.7A).7A). Our results suggest a model in which formation of the SRF-TCF ternary complex thus both controls signaling specificity to SRF and relieves an autoinhibitory interaction that suppresses activity of the Elk-1 C-terminal activation domain (Fig. (Fig.7B7B).

Regulatory interactions between SRF and TCF. (A) Interactions between Gal-Elk fusion proteins and SRF. Left panel, interaction between SRF and the B box of Gal-ElkΔ33 blocking access of the RhoA-actin signal pathway to SRF. Only a single unit of the Gal4 dimer is shown; it remains unclear whether both the Elk-1 B boxes present in the dimeric fusion protein contact SRF. Right panel, effect of the B-box mutations upon interaction with SRF. Mutation of the B box in the fusion indicated by the asterisk impairs transcriptional activation and allows a cofactor X, which mediates RhoA-actin signaling, to interact with SRF. The depicted interaction of X, which may or may not be dimeric, with SRF is figurative. (B) Proposed interactions Elk-1 proteins and SRF. Diagrams are as described for panel A. Recruitment of Elk-1 to DNA is dependent on interaction with SRF, which relieves an autoinhibitory interaction masking the Elk-1 activation domain. Interaction with SRF also inhibits access of the putative RhoA-actin signaling cofactor X as described for panel A. At promoters lacking a TCF binding site, TCF is not recruited and activation of SRF occurs via recruitment of putative cofactor X.
The use of specific inhibitors of RhoA-actin and MEK-ERK signaling in conjunction with different c-fos promoter mutants provides strong evidence that it is the combinatorial interactions between transcription factors that prevent c-fos activation via the RhoA-actin pathway. Two transcription factor binding sites flanking the c-fos SRF binding site were implicated in the control of signaling specificity: the 5′ TCF-binding Ets motif and the 3′ ATF/AP1 site, which appeared less important. Substitution of the 3′ ATF/AP1 site with a second Ets motif did not increase RhoA-actin signaling to SRF, indicating that this site is not absolutely required to restrict signaling specificity. The sensitivity of many of our c-fos promoter derivatives to the inhibition of ERK signaling by U0126, even when RhoA-actin signaling provides the main activation stimulus to SRF, strongly suggests that optimal c-fos transcriptional activation involves the activation of factors other than the TCFs via the ERK pathway. A candidate for such a promoter element is the TATA-proximal CREB site, which is a target for MAPK signaling via the RSK- or MSK-controlled activity of CREB (2, 5). Our studies with c-fos promoter mutants demonstrate that, at least in principle, it is possible to design promoters that are equally sensitive both to RhoA-actin and to ERK signaling.
Our results demonstrate that TCF binding is likely to prevent RhoA-actin signaling to SRF at other genes possessing SRF-associated TCF binding sites. However, the identification of TCF target genes from sequence considerations alone is complicated by the extreme flexibility in spacing between the Ets and SRF motifs tolerated by the SRF-TCF complex (4, 38). Functional TCF sites have been identified in the c-fos and egr-1 genes (12, 18, 21, 26, 30), and the egr-1 gene is also insensitive to RhoA-actin signaling (11, 32). In contrast, the srf, vinculin, and actin genes, which do not contain well-defined TCF binding sites, are regulated via the SRF-linked RhoA-actin pathway (11). Together with the results reported here, these observations are consistent with the idea that TCF binding is a primary determinant of signaling specificity. In support of this, the cyr61 gene, which contains no obvious TCF site, is sensitive to RhoA-actin signaling while the PIP92 and junB genes, which contain TCF-associated SRF sites, are insensitive (K. Murai, D. Gineitis, and R. Treisman, unpublished observations). However, two observations suggest that TCF binding is not the sole determinant of signaling specificity at SRF target genes. First, the results with the ATF/AP1 c-fos promoter mutants suggest that at least ATF/AP1 family proteins can also inhibit RhoA-actin signaling when bound in the vicinity of SRF. Second, mutational analysis of the vinculin promoter suggests that the introduction of a consensus TCF binding site is insufficient to render the promoter insensitive to RhoA-actin signaling (G. Smith and R. Treisman, unpublished observations).
Our results show that inhibition of RhoA-actin signaling to SRF requires physical contact between SRF and the TCF B-box sequence, suggesting that it does not reflect the recruitment of an inhibitory factor to the promoter by TCF. The B box inserts hydrophobic side chains into a hydrophobic channel on the surface of the SRF DNA-binding domain (13) in a manner similar to that of the interaction of the yeast SRF relative Mcm1 with its accessory factor MATα2 (33). It was previously shown that SRF mutations which impair interaction with TCF also impair the TCF-independent, serum-induced activity of the protein and proposed that they disrupt the docking site for an unknown factor which mediates TCF-independent signaling to SRF (18). The present results are consistent with this model and suggest that the TCF B box physically competes for this unknown factor. Given the dimeric nature of SRF, it is puzzling that a single TCF binding site appears sufficient to inhibit signaling, but this might be expected if the RhoA-actin signaling factor were dimeric. We are presently using biochemical approaches to identify other factors that interact with this region of the SRF DNA-binding domain.
Our results suggest that the interaction between the B box of Elk-1 and SRF potentiates Elk-1 transcriptional activity: in the presence of neighboring DNA-bound SRF, the activity of the Gal4-Elk-1 fusion protein Gal-ElkΔ33, which contains the entire Elk-1 coding sequence apart from the N-terminal 33 amino acids, was substantially reduced upon mutation of the B box. It is possible that our assays underestimated the effect of the SRF interaction upon transcriptional activity, since it is unclear whether both the B boxes in the dimeric Gal-ElkΔ33 protein are capable of interaction with SRF. Previous studies have provided evidence for a variety of intramolecular interactions within the Elk-1 TCF. The unphosphorylated C-terminal regulatory domain and the B-box region can inhibit both ternary complex formation and autonomous DNA binding (4, 20, 28, 38). Physical interactions between the Ets domain and both the B box and the C-terminal domain are detectable in vitro, and the integrity of the B box is required for phosphorylation of the C-terminal domain to promote DNA binding (39). These studies have led to an autoinhibitory model for Elk-1 in which the B box and the unphosphorylated C-terminal region cooperate to inhibit DNA binding by Elk-1 (39). Our data suggest an additional autoinhibitory mechanism operates within Elk-1, in which the function of the C-terminal activation domain is inhibited in the absence of interaction with SRF. The B-box sequence is highly conserved among the TCFs, even at positions not implicated in ternary complex formation (13, 22), and we speculate that such conserved residues may mediate intramolecular interactions with the Elk-1 C-terminal domain. Elucidation of the mechanism of such interactions will require a comprehensive structural analysis of Elk-1 and its ternary complex with SRF.
Acknowledgments
We thank Dziugas Gineitis and Ross Thomas for plasmids and the members of the laboratory and Caroline Hill for thoughtful comments on the manuscript.
This work was funded by Cancer Research UK.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.22.20.7083-7092.2002
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc139817?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.22.20.7083-7092.2002
Article citations
Systematic morphological profiling of human gene and allele function via Cell Painting.
Elife, 6:e24060, 18 Mar 2017
Cited by: 69 articles | PMID: 28315521 | PMCID: PMC5386591
p49/STRAP, a Serum Response Factor Binding Protein (SRFBP1), Is Involved in the Redistribution of Cytoskeletal F-Actin Proteins during Glucose Deprivation.
J Nutr Health Aging, 21(10):1142-1150, 01 Jan 2017
Cited by: 3 articles | PMID: 29188873
Stress-dependent phosphorylation of myocardin-related transcription factor A (MRTF-A) by the p38(MAPK)/MK2 axis.
Sci Rep, 6:31219, 05 Aug 2016
Cited by: 12 articles | PMID: 27492266 | PMCID: PMC4974569
Dynamic transcription factor activity networks in response to independently altered mechanical and adhesive microenvironmental cues.
Integr Biol (Camb), 8(8):844-860, 29 Jul 2016
Cited by: 15 articles | PMID: 27470442 | PMCID: PMC4981188
Overexpression of Striated Muscle Activator of Rho Signaling (STARS) Increases C2C12 Skeletal Muscle Cell Differentiation.
Front Physiol, 7:7, 08 Feb 2016
Cited by: 15 articles | PMID: 26903873 | PMCID: PMC4745265
Go to all (38) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.
J Mol Biol, 314(3):495-506, 01 Nov 2001
Cited by: 46 articles | PMID: 11846562
Growth hormone regulates ternary complex factors and serum response factor associated with the c-fos serum response element.
J Biol Chem, 272(41):25951-25958, 01 Oct 1997
Cited by: 40 articles | PMID: 9325329
Integration of growth factor signals at the c-fos serum response element.
Philos Trans R Soc Lond B Biol Sci, 351(1339):551-559, 01 Apr 1996
Cited by: 29 articles | PMID: 8735278
Critical Protein-Protein Interactions Determine the Biological Activity of Elk-1, a Master Regulator of Stimulus-Induced Gene Transcription.
Molecules, 26(20):6125, 11 Oct 2021
Cited by: 5 articles | PMID: 34684708 | PMCID: PMC8541449
Review Free full text in Europe PMC




