Abstract
Free full text

High-Efficiency Transfection of Primary Human and Mouse T Lymphocytes Using RNA Electroporation
Abstract
The use of nonviral gene transfer methods in primary lymphocytes has been hampered by low gene transfer efficiency and high transfection-related toxicity. In this report, high gene transfection efficiency with low transfection-related toxicity was achieved by electroporation using in vitro-transcribed mRNA. Using these methods, >90% transgene expression with >80% viable cells was observed in stimulated primary human and murine T lymphocytes transfected with GFP or mCD62L. Electroporation of unstimulated human PBMCs or murine splenocytes with GFP RNA yielded 95 and 56% GFP+ cells, respectively. Electroporation of mRNA for NY-ESO-1, MART-1, and p53 antigen-specific TCRs into human T lymphocytes redirected these lymphocytes to recognize melanoma cell lines in an MHC-restricted manner. The onset of gene expression was rapid (within 30 min) and durable (up to 7 days postelectroporation) using both GFP and TCR-mediated recognition of target cells. There was no adverse effect observed on the T lymphocytes subjected to RNA electroporation evaluated by cell growth rate, annexin-V staining of apoptotic cells, BrdU incorporation, tumor antigen-specific recognition or antigen-specific TCR affinity. The results of this study indicate that mRNA electroporation provides a powerful tool to introduce genes into both human and murine primary T lymphocytes.
INTRODUCTION
It has been very difficult to modify primary T lymphocytes genetically using non-viral vector-based gene transfer delivery systems. Therefore, investigators have generally utilized viral vector-based transduction, especially retroviral and lentiviral vectors [1,2], in experiments that required significant transgene expression in primary T cells. However, the use of viral vector-based gene transfer is limited by the time and cost required for vector production and restrictions on the size and number of genes that can be packaged by these vectors. In addition, potential safety issues could limit the use of viral vectors in certain clinical application [3]. Therefore the development of efficient nonviral gene transfer technologies for T lymphocytes is needed to facilitate both T lymphocyte-based research and potential clinical applications.
The use of plasmid vectors for the ex vivo genetic modification of nontransformed human tissue/cell types for therapeutic purposes has been limited by the low efficiency of currently available plasmid transfection systems and by the toxicity that many plasmid transfection reagents have for primary cell types, in particular lymphocytes [4–7]. Since primary T lymphocytes are notoriously refractory to most kinds of nonviral DNA delivery methods, electroporation of plasmid DNA encoding target genes is becoming the most commonly used strategy to transiently introduce a gene of interest into T lymphocytes. Bell et al. [4] demonstrated 57% transgene expression with 42% viability for electroporation of in vitro-activated DO11.10 murine T lymphocytes with a relatively high amount of plasmid DNA (100 μg DNA per 5–7.5 × 106 T cells). Van Tendeloo et al. [5] electroporated plasmid DNA into phytohemagglutinin (PHA)-stimulated human primary peripheral blood lymphocytes (PBLs) at optimized conditions and observed 16% gene expression with 70% mortality. Lai et al. [8] reported that 20–35% transgene expression with 75% viability was detected by transfecting resting and activated murine CD4 T cells using Nucleofection, a modified electroporation method. Nucleofection technology has a number of limitations. With this technology special buffers (supplied only by the manufacturer) are required to be used with a specific electroporation apparatus and preset programs limit the user’s ability to develop alternative electroporation conditions.
The concept of using RNA molecules as therapeutic agents has received increasing attention during the past decade [9]. The fact that successful electroporation of RNA into human and murine dendritic cells with high transgene expression could be easily achieved inspired us to investigate if RNA could be used in transfection of T lymphocytes [10,11].
In this study, we investigated the use of RNA electroporation in terms of transfection efficiency, cell viability, and impact of RNA electroporation on the function of T cells. Moreover, we extended the application of this technology to primary murine T lymphocytes and demonstrated that murine T lymphocytes from spleen, which are, in general, more refractory to gene transfer than human T lymphocytes, could also be transfected with very high efficiency using RNA electroporation.
RESULTS
RNA Electroporation of Human Primary T Cells with High Gene Transfer Efficiency
We determined optimized gene transfer conditions by altering different electroporation parameters and by evaluating transfection efficiency and cell viability as determined by flow-cytometric analysis for both unstimulated peripheral blood mononuclear cells (PBMCs) and stimulated human PBLs. Of the conditions tested, a voltage of 400 V combined with a time of 500 μs resulted in high green fluorescent protein (GFP) expression (93%) with minimal toxicity (82% viability) to stimulated T cells (Table 1). Using unstimulated PBMCs, a voltage of 300 V combined with a time of 10 ms resulted in 95% GFP expression with 57% viability relative to nonelectroporated PBMCs (Table 1).
TABLE 1
Optimization of electroporation conditions for resting and stimulated T cells
| Resting human PBLsa | Stimulated human PBLsb | Resting murine splenocytesc | ||||||
|---|---|---|---|---|---|---|---|---|
| Settings | % GFP | MFI | Viability (%) | % GFP | MFI | Viability (%) | % GFP | Viability (%) |
| 200 V/50 ms | 44 | 170 | 33 | 91 | 746 | 60 | 21 | 9 |
| 200 V/20 ms | 69 | 176 | 62 | 93 | 538 | 78 | 18 | 41 |
| 200 V/10 ms | 69 | 129 | 62 | 83 | 212 | 86 | 17 | 50 |
| 300 V/10 ms | 95 | 286 | 57 | nd | nd | nd | 52 | 28 |
| 300 V/5 ms | nd | nd | nd | nd | nd | nd | 49 | 41 |
| 360 V/5 ms | 88 | 214 | 47 | nd | nd | nd | 56 | 15 |
| 360 V/2 ms | nd | nd | nd | 78 | 437 | 70 | nd | nd |
| 360 V/1 ms | nd | nd | nd | 90 | 153 | 81 | 29 | 52 |
| 400 V/0.5 ms | 30 | 24 | 68 | 93 | 100 | 82 | 15 | 57 |
| 500 V/2 ms | nd | nd | nd | 58 | 236 | 54 | nd | nd |
| 500 V/1 ms | nd | nd | nd | 90 | 227 | 63 | nd | nd |
| 500 V/0.5 ms | nd | nd | nd | 94 | 123 | 70 | nd | nd |
| No EP | 0.5 | 8 | 73 | 0.6 | 4 | 82 | 0.5 | 72 |
GFP expression of CD3+ cells and cell viability (by PI exclusion) were detected 24 h postelectroporation. nd, not determined. Data are representative of two independent experiments.
We next increased the amount of GFP RNA electroporated into stimulated PBLs at 400 V/500 μs to determine the effect of RNA concentration on gene transfer efficiency and cell viability. The mean fluorescence intensity was increased with the amount of input RNA used, up to an RNA dose of 8 μg/1 × 106 T cells (Fig. 1A). There was no change in cell viability at any RNA amount. PBLs can respond to minimal amounts of input RNA. At the lowest amount tested, 60 ng of RNA electroporated into 1 × 106 PBLs produced discernable GFP expression detectable at 24 h postelectroporation (Fig. 1B). We also determined the kinetics of GFP expression following electroporation. GFP expression in electroporated T cells peaked at 24–72 h and declined to background levels after 14 days (Fig. 1A). The onset of gene expression occurred shortly after electroporation. As shown in Fig. 1C, as early as 30 min after electroporation, GFP expression could be detected.
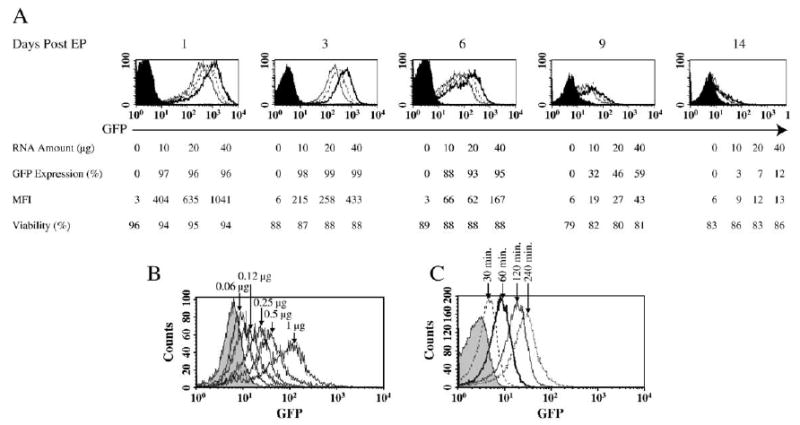
GFP expression in electroporated human PBLs. (A) 5 × 106 stimulated human PBLs were electroporated with increasing amounts of in vitro-transcribed (IVT) GFP RNA at 0 (electroporation without RNA, filled histogram), 10 (open histogram with solid line), 20 (open histogram with dotted line), and 40 μg (open histogram with dark solid line). Viability is the percentage of cells that excluded PI in the total population. GFP expression and mean fluorescence intensity (MFI) of the viable cells were measured on the days indicated. (B) Two times dilution of GFP RNA (starting from 1 μg) as indicated was electroporated into 1 × 106 (in 50-μl volume) stimulated PBLs. 24 h postelectroporation, GFP expression was detected by flow cytometry. (C) Stimulated PBLs were electroporated and immediately cultured in prewarmed culture medium (CM). At various time points as indicated, the samples were collected and fixed in 2% paraformaldehyde at 4°C and subjected to flow cytometry analysis to detect the GFP expression.
Effects of RNA Electroporation on T Cell Growth, Proliferation, Apoptosis, and Function
Although we observed no effect on cell viability with RNA electroporation under optimized conditions, we evaluated potential interference of electroporation on T cell function further. To determine whether RNA electroporation of T cells under optimized condition affects T cell growth and proliferation, we counted T cells at different time points after electroporation with increasing amounts of RNA as indicated (Fig. 2A). Compared to T cells that were electroporated without RNA or were not electroporated, cell growth rate postelectroporation was not affected (Fig. 2A). There was no difference in cell number between groups at any time point observed (up to 14 days postelectroporation), indicating that there was no death or irreversible damage of the T cells caused by RNA electroporation. To test further the effect of RNA electroporation on T cell proliferation, we monitored BrdU incorporation with and without TCR stimulation using OKT3 antibody. Results demonstrated that, from day 1 to day 4 after electroporation, there was no significant difference observed for BrdU incorporation between all the groups tested (Table 2).
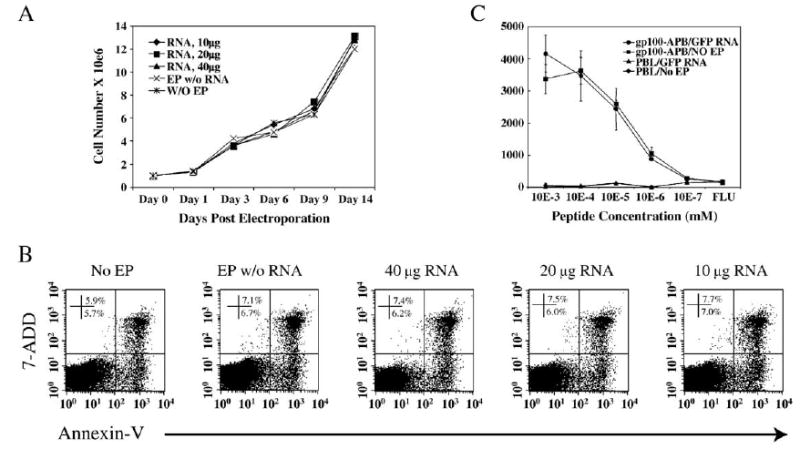
Effects of electroporation on T cell growth and function. (A) 5 × 106 stimulated PBLs were electroporated with increasing amounts of IVT GFP RNA as indicated. Samples were collected at different times postelectroporation and the cell numbers were counted and growth curves plotted. (B) PBLs were electroporated as in A. Three days after electroporation, the numbers of annexin-V- and 7-AAD-positive cells (% of total cells) were determined. The results shown were plotted without gating (representative of two experiments). (C) The melanoma tumor antigen gp100-specific TCR-transduced PBL line (gp100-APB) or nontransduced PBLs as control (PBL) were electroporated with GFP (gp100-APB/GFP RNA) and compared to PBLs electroporated without RNA (gp100-APB/NO EP). Serially diluted gp100 peptide-pulsed T2 cells were cocultured with the electroporated PBLs. IFN-γ secretion was detected by ELISA (representative of two experiments).
TABLE 2
BrdU incorporation of RNA-electroporated PBLs
| RNA EP | OKT3+IL-2EP w/o RNA | No EP | RNA EP | IL-2 only EP w/o RNA | No EP | |
|---|---|---|---|---|---|---|
| Day 1 | 21 | 18 | 21 | 10 | 10 | 10 |
| Day 2 | 58 | 54 | 58 | 27 | 23 | 24 |
| Day 3 | 51 | 56 | 57 | 23 | 22 | 21 |
| Day 4 | 39 | 43 | 40 | 22 | 19 | 17 |
PBLs were stimulated with OKT3 for 3 days and cultured in the presence of IL-2 for an additional 7 days. 10 μg IVT mCD62L RNA was electroporated into 5 × 106 PBLs. PBLs without electroporation and PBLs electroporated without RNA were used as controls. Immediately after electroporation, the cells were separated into two groups and cultured with or without OKT3. The values presented represent the percentage of the cells that incorporated BrdU.
It was reported that transfection of T cells with plasmid DNA caused apoptosis of transfected cells [7]. We examined annexin-V and 7-AAD staining after the T cells were electroporated with RNA. Three days postelectroporation, we detected the annexin-V- and 7-AAD-positive cells by flow cytometry. We did not observe any increase in apoptotic cells (annexin-V+/7-AAD−) or dead cells (annexin-V+/7-AAD+) for electroporated T cells compared to the T cells that were not electroporated, even when RNA was used at the higher dose (40 μg RNA/5 × 106 cells) (Fig. 2B). This result was consistent with our observations of cell growth shown in Fig. 2A, indicating that there was no direct cell death or apoptosis induced when the T cells were electroporated with RNA under optimized conditions (400 V/500 μs).
To test if RNA electroporation impairs the function of the T cells, we produced a melanoma tumor antigen-specific T cell line by engineering PBLs with an anti-gp100 TCR [13] and subjecting them to electroporation. We determined the gp100 antigen-specific T cell recognition of tumor cell lines or peptide pulsed T2 cells postelectroporation (Table 3). The RNA-electroporated T cell line recognized both tumor (526mel, HLA-A2+, gp100+) and gp100 peptide-pulsed T2 cells as efficiently as the nonelectroporated T cell line. The peptide affinity of the T cell line after electroporation was tested by a peptide titration assay. Both RNA-electroporated and nonelectroporated T cell lines recognized peptide at the same level, between 0.1 and 1 nM peptide (Fig. 2C).
TABLE 3
Electroporation of tumor antigen-specific T cells (IFN-γ, pg/ml)
| T2-ESO | T2-gp100 | T2 | 526 | 888 | |
|---|---|---|---|---|---|
| gp100-APB/GFP RNA | 18 (±5) | 3015 (±136) | 0 | 384 (±70) | 15 (±2) |
| gp100-APB/NO EP | 95 (±35) | 3970 (±42) | 65 (±68) | 415 (±51) | 18 (±5) |
gp100-specific TCR-transduced PBLs were electroporated with IVT GFP RNA (gp100-APB/GFP RNA). Nonelectroporated gp100 TCR-transduced PBLs were used as controls (gp100-APB/NO EP). Twenty-four hours postelectroporation, the cells were cocultured with a gp100 peptide-pulsed T2 cell line (T2-gp100), using NY-ESO-1 p157-165v-pulsed T2 (T2-ESO) as control, and melanoma cell lines (526, HLA-A2+; 888, HLA-A2−). After overnight coculture, the supernatants were collected and subjected to ELISA to determine IFN-γ secretion. Values are means of triplicate samples (SD). Data are representative of two experiments.
Induction of Tumor Antigen Recognition by TCR RNA Electroporation
Since primary human T cells could be transfected with high efficiency by RNA electroporation without any toxicity, RNA electroporation may provide a powerful tool for the analysis of functional genes in T cells. We electroporated mRNAs encoding functional α and β chains of HLA-A2-restricted TCRs that recognize NY-ESO-1- or MART-1-specific tumor antigen epitopes into OKT3-stimulated PBLs. We examined Vβ usage for both NY-ESO-1 (Vβ8) and MART-1 (Vβ12) by flow cytometry, and Vβ expression was 65 and 45%, respectively, and transferred Vβ genes were expressed in both CD8+ and CD8− (mainly CD4+) populations (Fig. 3A).
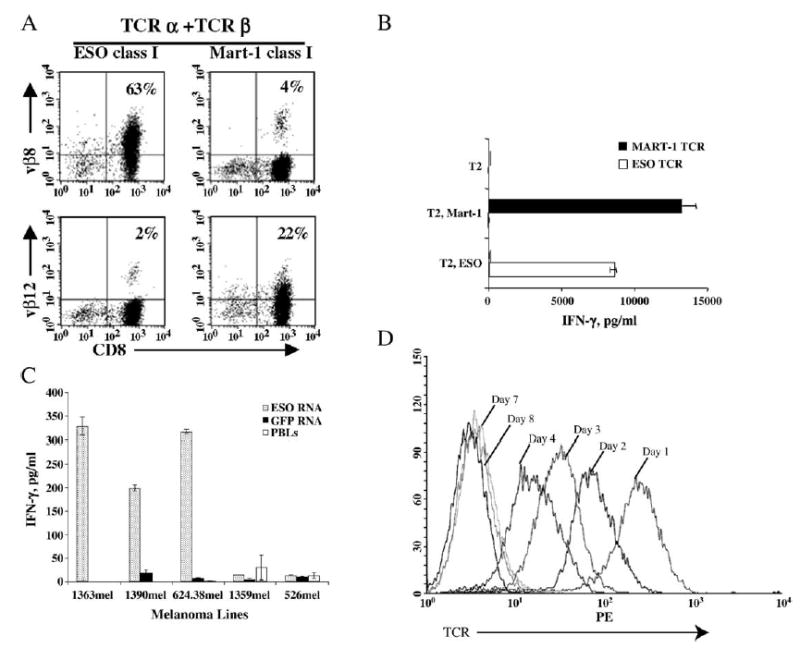
Tumor antigen-specific recognition by TCR RNA-electroporated T cells. (A) PBLs were stimulated with OKT3 for 3 days. CD4 T cells were depleted and continued in culture for an additional 3 days; the cells were harvested and electroporated with mRNA of both α and β TCR for NY-ESO-1 (ESO class I) or MART-1 (MART-1 class I). After overnight culture, the cells were stained with CD8 Ab and Ab against Vβ8 (NY-ESO-1) or Vβ12 (MART-1). (B) PBLs were electroporated as described above with MART-1 TCR RNA (MART-1 TCR), or NY-ESO-1 TCR RNA (ESO TCR). After 2 h culture in CM, the cells were cocultured with peptide-pulsed T2 cells. IFN-γ secretion was detected by ELISA. (C) Tumor recognition of TCR RNA-electroporated T cells. PBLs were stimulated with OKT3 for 2 days and cultured in CM with IL-2 for an additional 18 days before electroporation of NY-ESO-1 TCR RNA or GFP RNA. Four hours postelectroporation, the electroporated PBLs or nonelectroporated PBLs were cocultured with panels of melanoma cell lines described under Materials and Methods. IFN-γ secretion was measured from the culture supernatant by ELISA after overnight coculture. (D) PBLs were stimulated with OKT3 for 3 days and electroporated with p53-specific TCR (α and β chains) or GFP RNA. TCR expression was detected by flow cytometry at different days after electroporation as indicated (representative of three experiments).
To determine if the transfected α and β chains associated into a functional TCR, we incubated electroporated cells with peptide-pulsed target cells. IFN-γ secretion detected by ELISA in cocultures of TCR-transfected PBLs with peptide-pulsed T2 cells or peptide-pulsed autologous dendritic cells demonstrated peptide-specific recognition when the appropriate mRNAs encoding α, β chains of NY-ESO-1 and MART-1 TCR were electroporated (Fig. 3B). Moreover, we tested panels of melanoma cell lines against NY-ESO-1 TCR RNA- or GFP-electroporated PBLs (Fig. 3C). GFP RNA-electroporated PBLs (or nonelectroporated PBLs) recognized none of the melanoma cell lines tested. Anti-NY-ESO-1 TCR-electroporated PBLs recognized the melanoma cell lines that express both NY-ESO-1 and HLA-A2 (1363mel, 1390mel, and 624.38mel), but not NY-ESO-1-negative (526mel) or HLA-A2-negative (1359mel) lines.
Since GFP is known to be an extremely stable protein, it may not be representative of other proteins. Therefore, we tested the kinetics of TCR gene expression using an anti-p53 TCR gene. We monitored the duration of expression in T cells following RNA electroporation by assaying for TCR expression by FACS and by determining functional activity by recognition of peptide-pulsed T2 cells. Data in Fig. 3D demonstrate that TCR expression was easily detected at 4 days postelectroporation and continued to be above antibody background staining out to 8 days postelectroporation. Table 4 shows the IFN-γ secretion by PBLs electroporated with anti-p53 TCR mRNA, in comparison to GFP RNA as a control. A high level of IFN-γ production (66,600 pg/ml) was induced when electroporated PBLs were cocultured with the peptide-pulsed T2 cell line 1 day postelectroporation. At day 7 postelectroporation, specific IFN-γ was still detected at a level fivefold above background and returned to background levels at the next time point assayed (8 days).
TABLE 4
Duration of antigen-specific recognition by p53 TCR-electroporated PBLs
| Days post-EP (IFN-γ, pg/ml) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Peptide/T2 | EP/PBL | 1 | 2 | 3 | 4 | 5 | 7 | 8 | |
| p53 (264–272) | GFP | 440 (±200) | 510 (±261) | 531 (±115) | 1035 (±1090) | 0 | 85 (±15) | 60 (±14) | |
| p53 | 66,600 (±4640) | 45,141 (±4350) | 33,665 (±2901) | 19,345 (±1985) | 2351 (±275) | 530 (±190) | 152 (±17) | ||
| NY-ESO-1 | GFP | 358 (±86) | 498 (±24) | 551 (±51) | 492 (±140) | 27 (±0) | 55 (±5) | 64 (±28) | |
| p53 | 632 (±124) | 1347 (±129) | 499 (±107) | 421 (±250) | 0 | 45 (±30) | 107 (±10) | ||
| No peptide | GFP | 400 (±294) | 666 (±435) | 1010 (±890) | 505 (±230) | 35 (±0) | 75 (±15) | 57 (±19) | |
| p53 | 440 (±94) | 1218 (±159) | 480 (±56) | 335 (±210) | 60 (±0) | 60 (±10) | 107 (±5) | ||
PBLs were stimulated with OKT3 for 3 days and electroporated with p53-specific TCR or GFP RNA. Electroporated PBLs were cocultured with peptide-pulsed T2 cell line at the days indicated and IFN-γ secretion was detected by ELISA. Values are means of triplicate samples (SD). Data are representative of two experiments.
RNA Electroporation of Murine Splenocyte T Cells
Murine T lymphocytes are more resistant to most gene delivery methods than human T cells. To test if murine T cells are amenable to RNA electroporation, we harvested splenocytes from two different strains of mice (C57BL/6 or C3H) and subjected them to electroporation. The results from optimization tests demonstrated that 15–56% of murine CD3 T cells expressed GFP following electroporation of unstimulated splenocytes (Table 1). When unstimulated splenocytes were electroporated, both CD3+ T cells and CD3− non-T cells (mostly B cells, NK cells, and monocytes) could be efficiently transfected (Fig. 4A). To activate T cells, we stimulated murine splenocytes with 2 μg/ml ConA in the presence of 2 ng/ml IL-7 for 3 days and electroporated them with GFP RNA. As shown in Fig. 4B, both CD8 and CD4 T cells from ConA-stimulated splenocytes from C57BL/6 mice were efficiently transfected; the transfection efficiency was 78 and 60% for CD8 and CD4, respectively. Similar results were observed with GFP RNA-electroporated splenocytes from C3H mice (data not shown). To test if antigen-specific murine T cells also could be efficiently transfected via RNA electroporation, we stimulated splenocytes from the anti-gp100 transgenic mouse strain Pmel, or a cross of Pmel with a CD62L knockout mice, in vitro with 1 μg/ml gp100 peptide for 7 days and then electroporated them with GFP or CD62L RNA. Over 92% of transgene expression was detected for both GFP and CD62L 24 h after electroporation (Fig. 4C). To test the duration of expression in murine T cells, we electroporated peptide-stimulated T cells from Pmel TCR/CD62L KO transgenic mice with murine CD62L mRNA. At 9 days postelectroporation, mouse CD62L expression continued to be detected above background staining (data not shown).

RNA electroporation of murine splenocyte T cells. (A) 2.5 × 106 freshly prepared C57BL/6 splenocytes were electroporated with 5 μg GFP RNA. GFP expression in T lymphocytes was detected by flow cytometry after 24 h culture. The numbers indicate the percentage of cells within each quadrant. The numbers in parentheses represent the percentage of GFP expression cells in CD3+ and CD3− lymphocytes. (B) ConA-stimulated splenocytes from C57BL/6 mice were electroporated with GFP RNA, and GFP expression was detected 24 h postelectroporation. The numbers in parentheses represent the percentage of GFP expression cells in CD4 (60%) or CD8 (78%) T cells. (C) GFP or mCD62L was electroporated into gp100 peptide-stimulated splenocytes from Pmel TCR transgenic mice (Pmel) or Pmel TCR transgenic/CD62L knockout mice (Pmel/CD62L KO). Twenty-four hours after electroporation, the cells were stained for antibody against murine CD62L and the expression of CD62L and GFP was detected by FACS (representative of three experiments).
DISCUSSION
In this study, we used an RNA electroporation system to improve transfection efficiency and reduce or eliminate transfection-related toxicity. A transfection efficiency of over 90% could be easily achieved for human PBLs. Similar results were observed in murine T cells that were stimulated with ConA or antigen-specific peptide. The results from optimization experiments altering electroporation parameters for stimulated human PBLs demonstrated that, except at high voltage and long pulse lengths (500 V/2 ms and 500 V/5 ms) the viability of transfected T cells was 63–86% at 24 h postelectroporation (Table 1) and transgene expression of 99% was achieved at 3 days postelectroporation (Fig. 1A). The length of time poststimulation does not significantly influence RNA electroporation as similar efficiencies can be achieved with cells stimulated from 2 to 18 days. Recently, Smits et al. electroporated PHA-stimulated human PBLs with GFP RNA and 50% of transgene expression was detected using flow cytometry [17]. Differences in PBL stimulation, model of electroporator, and/or the parameters for electroporation may account for the higher transfection efficiency achieved herein.
RNA electroporation may have distinct advantages over plasmid DNA-based electroporation. Following electroporation of plasmid DNA, the plasmid must enter the nucleus before the DNA can be transcribed into mRNA. Transcription efficiency is largely dependent on the promoter used in the plasmid vector and can be greatly influenced by target cell type-specific transcription factors, which ultimately determine transgene expression efficiency. By introducing mRNA directly into the cytoplasm of the cells, transcription is bypassed and the target gene can be immediately translated into protein. Indeed, we observed easily detectable GFP expression within 30 min postelectroporation (Fig. 1C). RNA electroporation also introduces a molecule (although completely synthetic) that is almost identical to native mRNAs (in vitro-transcribed RNAs have both poly(A) tails and 5′-methlylated cap analogues). This may be a critical distinction between RNA and plasmid DNA, as bacterial-based plasmids are known to contain “danger signals” and signal through Toll-like receptors leading to alterations in T cell biology [18]. These difference in how the T cells recognize RNA versus plasmid DNA may in part account for the difference in cell viability following electroporation of these two different nucleic acid species.
Any transfection method, including electroporation, may induce unforeseen adverse affects on transfected T cells in terms of viability, proliferation, or functionality. The optimized electroporation conditions used in this study did not significantly alter T cell viability or cell proliferation and did not induce apoptosis (Fig. 2 and Tables 2 and and3).3). Functionality of electroporated T cells was verified by testing electroporation of antigen-specific T cell recognition of limiting dilutions of gp100 peptide (Fig. 2). The most important potential application of this technology would be to engineer T cells with new biological functions. When T cells without tumor recognition were electroporated with in vitro-transcribed α and β TCR mRNAs, electroporated T cells were redirected to recognize not only tumor antigen peptide-pulsed APC (T2 cell line and dendritic cells), but also tumor antigen-expressing melanoma lines (Fig. 4). This simplified method of redirecting tumor antigen recognition of T lymphocytes by RNA electroporation makes it possible to screen TCR α and β cDNA libraries rapidly for functional TCR α/β pairs.
Promising results have recently been reported in adoptive immunotherapy trials for the treatment of patients with metastatic cancer [19–21]. Engineering T cells with new functions using RNA electroporation may provide a powerful tool for altering T cell biology in circumstances in which long-term transgene expression is not necessary or may be undesirable. Molecules such as some cytokine genes and their receptors (IL-2, IL-15, and IL-7) [22,23] or costimulatory molecules (CD28, CD27, 4-1BB, and OX40) [24–26] can influence T cell functionality and homeostasis but do not need to be constitutively expressed. Furthermore, some molecules, such as CD62L and CCR7, are important for T cell homing and migration [15], but long-term expression of such molecules on transferred T cells may be undesirable. CD62L expression is associated with T cell migration to lymph nodes, but continuous expression of this molecule on the T cell surface could prevent T cells from migration out of the lymph node to the effector site [27]. The molecules Hlx, which promotes CD4 T lymphocyte polarization from Th2 to Th1 [28], and telomerase reverse transcriptase, which extends the replicative potential of T cells [29], are also possible candidates to be transferred into T cells for the adoptive immunotherapy of cancer or infectious diseases. Finally, the down-regulation of genes that may have a negative effect on T cell function (e.g., CTLA-4 or Foxp3) could be achieved by electroporation of siRNAs.
This highly reproducible and cost-effective technology is an attractive methodology for clinical development because it permits the use of naked RNA and does not involve additional reagents that must pass rigorous production standards and quality control testing. mRNA electroporation also provides a valuable alternative to viral transduction of T lymphocytes, which implies a more complex and laborious manipulation. Moreover, there may be lower safety concerns compared to viral vector transduction or plasmid DNA transfection that can result in the inactivation or activation of genes as a result of random integration of the introduced DNA.
In summary, mRNA electroporation provides a powerful tool to introduce genes easily into both human and murine primary T lymphocytes with high efficiency and low toxicity. The availability of 96-well electroporation cuvettes and the MaxCyte system [30] make electroporation amenable to high-throughput screening protocols and potentially useful for large-scale genetic modification of T lymphocytes required for clinical applications.
MATERIALS AND METHODS
Patient PBMCs, cell lines, and murine splenocytes
All of the PBLs used in this study were cryopreserved PBMCs obtained by leukophoresis of metastatic melanoma patients treated at the Surgery Branch, National Cancer Institute (NIH, Bethesda, MD, USA). T2 is a lymphoblastoid cell line deficient in TAP function, whose HLA class I proteins can be easily loaded with exogenous peptides [13]. Melanoma lines 1363mel, 1390mel, 624.38mel, and 526mel were generated at the Surgery Branch from resected tumor lesions, as previously described [13]. All the cell lines described above were maintained in R/10 (RPMI 1640 (Invitrogen, Inc., Carlsbad, CA, USA) supplemented with 10% fetal calf serum (Biofluid, Inc., Gaithersburg, MD, USA)). Culture medium (CM) for human T lymphocytes was RPMI with 0.05 mM mercaptoethanol, 300 IU/ml IL-2 (Chiron Corp., Emeryville, CA, USA), plus 10% human AB serum (Valley Biomedical, Winchester, VA, USA). Murine splenocytes, collected from 6- to 8-week-old C57BL/6 (The Jackson Laboratory, Bar Harbor, ME, USA) or Pmel-1 TCR transgenic (Tg) mice [14] or Pmel-1 TCR Tg mice crossed with C57BL/6-Sell−/− (Pmel-1 CD62L−/−) [15] were stimulated before electroporation as described below.
Electroporation of in vitro-transcribed mRNA
mRNA encoding the GFP was prepared from pGEM4Z/EGFP/A64 (kindly provided by Dr. Eli Gilboa, Duke University Medical Center, Durham, NC, USA) as previously described [11]. Murine CD62L mRNA was generated from pGEM4Z/mCD62L/A64 in which the EGFP of pGEM4Z/EGFP/A64 was replaced by murine CD62L cDNA. mRNA encoding TCR α and β chains of NY-ESO-1 and MART-1 were prepared from PCR products made by primer pairs that introduce the T7 promoter at the 5′ end to facilitate in vitro-transcription (IVT) of RNA. Templates for these amplifications were retroviral vectors MSGV-E1-APB (for NY-ESO-1) [12] and pMSGV1-AIB (for MART-1) [15], which harbor the cDNAs encoding these genes. cDNA encoding p53 TCR α and β chains were cloned from CTL clone p53/264.15.1, which was generated by immunization of HLA-A2 transgenic mice with p53-specific peptide [16] by the rapid amplification of cDNA ends method (GeneRacer Kit; Invitrogen) and subcloned into pGEM4Z/A64 vector for IVT of mRNA. The mMESSAGE mMA-CHINE High Yield Capped RNA Transcription Kit (Ambion, Inc., Austin, TX, USA) was utilized to generate IVT RNA. The IVT RNA was purified using an RNeasy Mini Kit (Qiagen, Inc., Valencia, CA, USA) and purified RNA was eluted in RNase-free water at 1–0.5 mg/ml. For the stimulation of T cells, human PBLs were stimulated with 50 ng/ml OKT3 for 3 days, murine splenocytes were stimulated either with 2 μg/ml ConA for 72 h (for wild-typeC57BL/6) or with1 mM hgp10025–33 and 30 IU/ml IL-2 for 7 days (for Pmel-1 Tg) before electroporation. For the electroporation of unstimulated PBMCs, cryopreserved cells were thawed and washed twice with OPTI-MEM (Invitrogen) and resuspended in OPTI-MEM at 2.5 × 107/ml. The cells (0.1 ml) were electroporated with 5 μg GFP RNA at the settings indicated in Table 1. The stimulated PBLs subjected to electroporation were also washed twice with OPTI-MEM and resuspended in OPTI-MEM at the final concentration of 2.5 × 107/ml. Cells and cuvettes were prechilled by putting them on ice for >5 min before electroporation. Subsequently, 0.05 to 0.2 ml of the cells was mixed with 2 μg/1 × 106 T cells of IVT RNA (or as indicated) and electroporated in a 2-mm cuvette (Harvard Apparatus BTX, Holliston, MA, USA), using an ECM830 Electro Square Wave Porator (Harvard Apparatus BTX). Immediately after electroporation, the cells were transferred to fresh CM with 300 IU/ml IL-2 and incubated at 37°C.
Peptide synthesis
Synthetic peptides used in this study were made using a solid-phase method on a peptide synthesizer (Gilson Co., Inc., Worthington, OH, USA) at the Surgery Branch of the NCI. The quality of each peptide was evaluated by mass spectrometry (Biosynthesis, Inc., Lewisville, TX, USA). The sequences of the peptide used in this study are as follows: NY-ESO-1p157–165V, SLLMWTTQV; NY-ESO-1p161–180, WITQCFLPV-FLAQPPSGQRR; modified hgp100p209–217, IMDQVPFSV; p53p264–272, LLGRNSFEV; and modified MART-1p27–35, ELAGIGILTV.
Flow-cytometric analysis
GFP expression and cell-surface expression of Vβ8, Vβ12, CD8, CD4, mouse TCR, and mCD62L were measured by flow cytometry using FITC-, PE-, or APC-conjugated Abs as directed by the suppliers of anti-TCR Vβ8 or Vβ12 (Immunotech, Westbrook, ME, USA) and all others (BD Biosciences, San Jose, CA, USA). 7-AAD and annexin-V (BD Biosciences) were used to detect dead or apoptotic cells and BrdU-incorporation assay was carried out using a FITC BrdU Flow Kit (BD Biosciences) following the manufacturer’s instruction.
Cytokine release assay
Human T cell cultures were tested for reactivity in a cytokine release assay using a commercially available IFN-γ ELISA kit (Endogen, Cambridge, MA, USA). IFN-γ release was measured with coculture of T cells with melanoma tumor lines or T2 pulsed with peptide (1 μg/ml or as indicated in the figure legends). Peptides were incubated with T2 cells for 3 h at 37°C, followed by washing (three times) before initiation of cocultures. For these assays, 1 × 105 responder cells (T cells) and 1 × 105 T2 or 5 × 104 tumor cells were incubated in a 0.2-ml culture volume in individual wells of 96-well plates. After overnight coculture, IFN-γ secretion was measured in culture supernatants diluted so as to be in the linear range of the assay.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.ymthe.2005.07.688
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1525001605014267/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.ymthe.2005.07.688
Article citations
Transcriptional rewiring in CD8<sup>+</sup> T cells: implications for CAR-T cell therapy against solid tumours.
Front Immunol, 15:1412731, 27 Sep 2024
Cited by: 0 articles | PMID: 39399500 | PMCID: PMC11466849
Review Free full text in Europe PMC
mRNA vaccine development and applications: A special focus on tumors (Review).
Int J Oncol, 65(2):81, 12 Jul 2024
Cited by: 0 articles | PMID: 38994758 | PMCID: PMC11251742
Review Free full text in Europe PMC
CAR-T-Cell-Based Cancer Immunotherapies: Potentials, Limitations, and Future Prospects.
J Clin Med, 13(11):3202, 29 May 2024
Cited by: 1 article | PMID: 38892913 | PMCID: PMC11172642
Review Free full text in Europe PMC
Engineering strategies to safely drive CAR T-cells into the future.
Front Immunol, 15:1411393, 19 Jun 2024
Cited by: 2 articles | PMID: 38962002 | PMCID: PMC11219585
Review Free full text in Europe PMC
Engineered T Cell Receptor for Cancer Immunotherapy.
Biomol Ther (Seoul), 32(4):424-431, 07 Jun 2024
Cited by: 1 article | PMID: 38844787 | PMCID: PMC11214961
Review Free full text in Europe PMC
Go to all (183) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines.
J Immunol, 174(7):4415-4423, 01 Apr 2005
Cited by: 131 articles | PMID: 15778407 | PMCID: PMC2174604
RNA-transfection of γ/δ T cells with a chimeric antigen receptor or an α/β T-cell receptor: a safer alternative to genetically engineered α/β T cells for the immunotherapy of melanoma.
BMC Cancer, 17(1):551, 17 Aug 2017
Cited by: 70 articles | PMID: 28818060 | PMCID: PMC5561563
High transfection efficiency of porcine peripheral blood T cells via nucleofection.
Vet Immunol Immunopathol, 144(3-4):179-186, 15 Oct 2011
Cited by: 4 articles | PMID: 22055481
Activation of tumor-specific T cells by dendritic cells expressing the NY-ESO-1 antigen after transfection with the cationic lipophosphoramide KLN5.
J Gene Med, 10(6):628-636, 01 Jun 2008
Cited by: 0 articles | PMID: 18338820
Funding
Funders who supported this work.
Intramural NIH HHS (2)
Grant ID: Z01 BC010763-01
Grant ID: Z99 CA999999





