Abstract
Free full text

A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling
Abstract
Type I IFNs are crucial components of the innate immune response to viral attack. They are rapidly synthesized and secreted after infection with human cytomegalovirus (CMV) and trigger a signal transduction pathway that involves successive activation and nuclear translocation of signal transducer and activator of transcription 1 (STAT1) and STAT2. The activated STATs, together with the IFN regulatory factor 9 protein, form a trimeric transcription complex (IFN-stimulated gene factor 3) that stimulates expression of numerous IFN-responsive genes, many of which exhibit antiviral activity. Here we demonstrate that the viral 72-kDa IE1 protein (IE1-72kDa) confers partial resistance to the antiviral activity of type I IFNs upon CMV. Accordingly, IFN-responsive transcripts accumulate to substantially increased levels after infection with an IE1-deficient mutant as compared with wild-type virus, and ectopic expression of the viral protein in stably transfected cells is sufficient to block their induction. We further show that IE1-72kDa forms a physical complex with STAT1 and STAT2 in nuclei of infected cells and in vitro and prevents association of STAT1, STAT2, and IFN regulatory factor 9 with promoters of IFN-responsive genes in vivo. Our results indicate that the viral protein blocks an intranuclear step after nuclear translocation and before DNA binding of IFN-stimulated gene factor 3, presumably by interfering with the integrity and/or correct subnuclear localization of the protein complex. This study identifies the CMV IE1-72kDa protein as a viral antagonist of the cellular innate immune response, inhibiting IFN-dependent STAT signaling by means of an unprecedented molecular mechanism.
Type I IFNs are multifunctional cytokines that represent key components of the innate immune response to viral infection (1–4). Synthesis of IFN-α and/or IFN-β is rapidly induced after the initial interaction of various viruses with their host cells. Secreted IFN-α/β binds to its cognate receptor on target cells, triggering a signaling cascade that involves two members each of the Janus kinase (Jak) and signal transducers and activators of transcription (STAT) families, Jak1/Tyk2 and STAT1/STAT2, respectively. Upon Jak1/Tyk2-mediated tyrosine phosphorylation, STAT1 and STAT2 heterodimerize and translocate to the nucleus, where they bind to IFN regulatory factor 9 (IRF9) to form the transcription complex IFN-stimulated gene factor 3 (ISGF3). ISGF3, in turn, sequence-specifically binds to an IFN-stimulated response element (ISRE) that is present in numerous type I IFN-simulated genes (ISGs), many of which exhibit antiviral activity (5–8).
Human cytomegalovirus (CMV) is a ubiquitous opportunistic pathogen causing morbidity and mortality in people with immature or compromised immune systems (9, 10). In immunocompetent hosts CMV infection is effectively controlled by both adaptive and innate immune functions (11, 12). Although CMV initially triggers the accumulation of many IFN-stimulated mRNAs, it is eventually able to disarm this antiviral response, at least in part (13–19). The UL83-coded virion constituent pp65 was the first CMV gene product shown to block the induction of some ISGs (20, 21). However, experimental evidence suggested that CMV encodes at least one additional gene product, synthesized at immediate-early (IE) times after infection, that suppresses the up-regulation of IFN-dependent gene expression (15).
The initial and most abundant viral IE transcript originates from the major IE transcription unit and gives rise to a nuclear phosphoprotein referred to as IE1-72kDa (72-kDa IE1 protein) (9, 22). This protein is required for fully productive CMV replication in cultured fibroblasts at low-viral-input multiplicities but is dispensable for viral growth under conditions where one cell becomes infected by multiple particles (23, 24). The mechanism through which IE1-72kDa facilitates CMV infection involves transcriptional activation of IE and early viral genes, a process that is mediated, at least in part, through changes in histone acetylation (23–26). Here we demonstrate that the viral IE1-72kDa protein efficiently suppresses activation of cellular ISGs by targeting STAT1 and STAT2, thereby counteracting the type I IFN-mediated antiviral host cell response against CMV.
Results
IE1 Confers Partial Resistance to Type I IFNs on CMV.
Recent work has suggested that CMV synthesizes one or more IE gene products able to suppress the type I IFN-dependent host cell response (15). To test the idea that the most abundant viral IE protein, IE1-72kDa, counteracts this response, we compared wild-type and IE1-deficient mutant CMVs for their susceptibility to exogenous IFN-α. For these experiments, we used high-viral-input multiplicities that permit wild-type-like viral replication in human fibroblasts in the absence of IE1 (23, 24). Compared with wild-type, the IE1-deficient virus displayed an ≈40- or >370-fold increased sensitivity to 250 or 1,000 units/ml IFN-α, respectively (Fig. 1A). In fact, mutant virus production was completely abolished in the presence of 1,000 units/ml IFN-α, whereas replication of wild-type virus was attenuated but not fully blocked under these conditions (Fig. 1A).
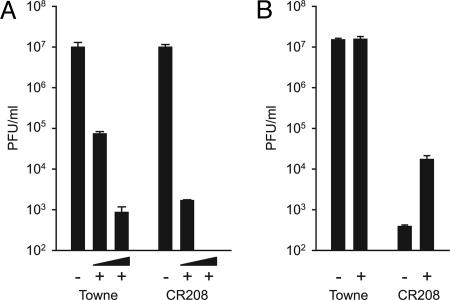
Differential effects of exogenous IFN-α or neutralization of endogenous IFN-β on wild-type (Towne) and IE1-deficient (CR208) mutant virus yields. Experiments were performed in triplicate, and bars represent mean values with standard errors. (A) MRC-5 cells treated with 250 or 1,000 units/ml recombinant IFN-α (+) or untreated cells (−) were infected at a multiplicity of 5 plaque-forming units (PFU) per cell. Virus yields were determined 72 h after infection. (B) MRC-5 cells treated with 500 neutralizing units/ml of an IFN-β-specific antiserum (+) or untreated cells (−) were infected at a multiplicity of 0.1 PFU per cell. Virus yields were determined 7 days after infection.
IFN-β is known to be the primary IFN produced upon virus infection in fibroblasts. Therefore, we also monitored CMV virion production in the presence of an IFN-β-neutralizing antiserum in the medium of infected fibroblast cultures. In contrast to the IFN-α assays, low-viral-input multiplicities were used in these experiments, resulting in attenuated growth of the mutant virus compared with wild-type (23, 24, 26). Interestingly, wild-type virus replication was not affected by the presence of the antiserum, indicating that CMV infection in fibroblasts is normally not limited by autocrine or paracrine IFN-β activity (Fig. 1B). However, mutant virus yields were increased by >50-fold upon antibody-mediated neutralization of IFN-β (Fig. 1B), suggesting that the IE1-specific growth defect at low-viral-input multiplicities is at least partly due to IFN-dependent antiviral mechanisms. Taken together, these results strongly indicate that IE1 is a major determinant of the type I IFN resistance of CMV.
IE1 Antagonizes Type I IFN-Stimulated Cellular Gene Expression.
To examine whether the viral IE1-72kDa protein antagonizes the antiviral activity of type I IFNs by interfering with activation of ISGs, we quantified mRNAs from two ISRE-controlled cellular genes, ISG54 and MxA, in cells infected with wild-type or IE1-deficient mutant viruses. ISG54 and MxA were chosen because these ISGs have been repeatedly reported to be up-regulated by CMV infection of fibroblasts (14–16,18,20). Addition of IFN-α resulted in a strong (>40-fold) induction of ISG54 in noninfected cells (Fig. 2A). We could also observe significant ISG54 activation 24 h after infection with the wild-type virus, and this effect was clearly enhanced by simultaneous treatment with IFN-α (Fig. 2A). However, activation of ISG54 transcription was substantially stronger in cells infected with the mutant as compared with the wild-type virus in both untreated and IFN-α-stimulated cells (Fig. 2A). Similar effects were observed for the MxA gene, although the corresponding transcript was less efficiently induced by IFN-α or CMV infection, in agreement with previous observations (Fig. 2A) (15, 16). Interestingly, there were no major differences in activation of the IFN-β gene by wild-type and mutant viruses (Fig. 2A). Moreover, in a time-course experiment, significant differences in ISG expression between wild-type and mutant viruses were detected at 48 and 24 h but not at 6 h after infection, where the ISG54 transcript was strongly induced in both the presence and the absence of IE1 (Fig. 2B). Likewise, when de novo protein synthesis was blocked with cycloheximide, the extent of ISG54 induction was very similar in infections with the two viruses (data not shown), ruling out the possibility that constituents of the virus stocks make the IE1-null mutant a more potent ISG inducer than wild-type.
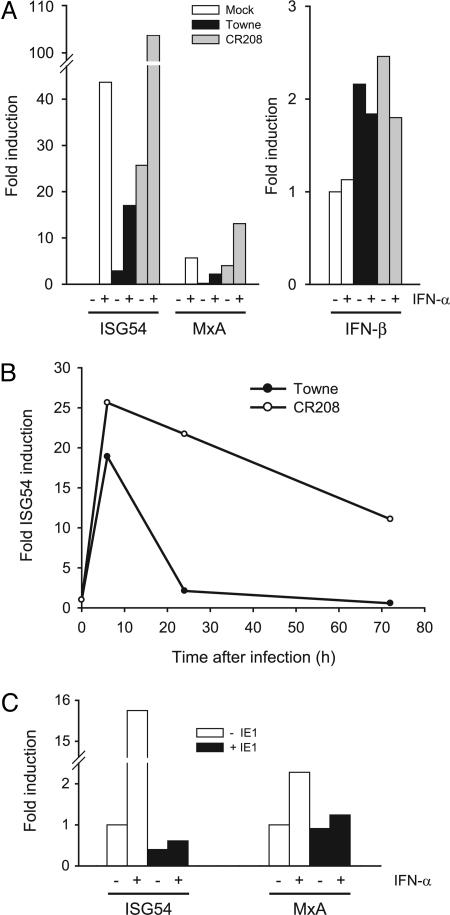
IE1 expression antagonizes induction of ISGs. (A) MRC-5 cells treated with 1,000 units/ml recombinant IFN-α (+) or untreated cells (−) were infected with wild-type (Towne) or mutant (CR208) CMV at a multiplicity of 1 PFU per cell or mock-infected. ISG54/MxA (Left) and IFN-β (Right) mRNAs were quantified 24 h or 6 h after infection, respectively, by kinetic RT-PCR. (B) MRC-5 cells were infected with wild-type (Towne) or mutant (CR208) CMV at a multiplicity of 1 PFU per cell, and ISG54 mRNA was quantified by kinetic RT-PCR at 6, 24, and 72 h after infection. (C) The cell lines ihfie1 (+IE1) and ihf (−IE1) were treated with 1,000 units/ml recombinant IFN-α (+) or were left untreated (−), and ISG54 or MxA mRNAs were quantified by kinetic RT-PCR.
To examine whether the observed effects were direct rather than indirect downstream consequences of IE1 expression, we also analyzed ISG54 and MxA mRNA accumulation in IFN-α-stimulated human fibroblasts stably expressing the IE1-72kDa protein outside the viral context (Fig. 2C). Strikingly, induction of ISG54 and MxA by IFN-α was almost completely blocked by IE1 expression, indicating that the viral protein alone is sufficient for effective inhibition of IFN-dependent ISG activation. Because the fibroblasts used in this experiment (ihf and ihfie1 cells) express human papillomavirus E6 and E7 proteins (23, 24) we also performed these assays in MRC-5 and H1299 cells stably transfected with IE1 and obtained similar, although less pronounced, inhibitory effects on ISG induction (data not shown).
Although we cannot exclude that synergism between the CMV protein and E6/E7 gene products enhances the magnitude by which ISG activation is suppressed, taken together these data clearly suggest that IE1-72kDa is required and sufficient to inhibit type I IFN-dependent induction of at least a subset of ISGs. However, the viral protein appears to have little effect on the initial (IFN-independent) IFN-β and ISG activation triggered by the virus. These observations strongly suggest that IE1-72kDa counteracts ISG induction by antagonizing IFN signaling.
IE1 Does Not Interfere with Expression, Phosphorylation, or Nuclear Targeting of ISGF3 Components.
Negative regulation of IFN signaling could provide one mechanistic explanation for the observed defect in ISG activation in the presence of IE1-72kDa. The effects of IE1 expression on IFN-α signal transduction were tested by examining expression, phosphorylation, and nuclear accumulation of all three protein constituents that form the ISGF3 complex in IE1-expressing fibroblasts versus control cells. Western blot analyses revealed that steady-state protein levels of STAT1, STAT2, and IRF9 were not affected by IE1-72kDa before or after induction with IFN-α (Fig. 3A). Moreover, immunoblotting with phosphotyrosine-specific antibodies indicated that IFN-α-dependent posttranslational activation of STAT1 and STAT2 is not inhibited by IE1 (Fig. 3A). Indirect immunofluorescence microscopy was used to examine the subcellular localization of ISGF3 components with and without IFN-α treatment. Irrespective of IE1 expression, IFN-α triggered nuclear translocation of STAT2 and nuclear accumulation of tyrosine-phosphorylated STAT1 (Fig. 3B). The subcellular distribution of total STAT1 protein was also examined and showed no IE1-dependent differences (data not shown). Moreover, no changes in IRF9 localization were observed between IE1-expressing and control cells (Fig. 3B). These results indicate that IFN-α-responsive signal transduction to the nucleus remains intact in the presence of IE1, suggesting that the viral protein interferes with an intranuclear step in ISG induction. This idea is in agreement with the well documented nuclear localization of the IE1-72kDa protein (9, 22, 27).

IE1-72kDa fails to affect accumulation, phosphorylation, and subcellular localization of ISGF3 components. (A) Western blots showing steady-state levels of the indicated proteins in ihf or ihfie1 cells at various times before and after treatment with 1,000 units/ml recombinant IFN-α. (B) Indirect immunofluorescence images showing typical subcellular localization of the indicated proteins in ihf or ihfie1 cells before and after treatment for 1 h with 1,000 units/ml recombinant IFN-α. A rabbit-specific Alexa Fluor 488 conjugate (Molecular Probes) was used as secondary antibody. Before analysis, IE1 expression was determined by immunofluorescence to be present in ≈99% of ihfie1 cells. (Magnification: ×500.)
IE1 Blocks IFN-Induced Association of ISGF3 with Chromatin.
Next, we asked whether the nuclear presence of IE1-72kDa affects IFN-induced sequence-specific association of ISGF3 components with the ISRE promoter element. To test this possibility in an in vivo setting with native chromatin, we performed chromatin immunoprecipitation (ChIP) assays using STAT1-, STAT2-, and IRF9-specific antibodies in IE1-expressing fibroblasts versus IE1-negative control cells. Relative amounts of DNA corresponding to an ISRE-spanning region of the ISG54 promoter were first analyzed by standard PCR (data not shown) and then exactly quantified by real-time PCR assay (Fig. 4A). As soon as 15 min after addition of IFN-α, an ≈30-fold increase in STAT2 association with the ISG54 promoter was detected in ihf cells relative to nontreated cells (Fig. 4A). This effect was further enhanced at 30 and 60 min after IFN-α treatment (≈70-fold increase at 60 min; Fig. 4 A and B). In contrast, in the presence of IE1-72kDa, STAT2 interaction with chromatin was only modestly induced by a factor of ≈2.5 to ≈6.5 at 15 or 60 min after IFN-α treatment, respectively (Fig. 4 A and B). Likewise, the viral protein also proved to substantially antagonize the IFN-activated association of STAT1 and IRF9 with DNA (Fig. 4B). Similar ChIP data with respect to IE1-dependent inhibition of STAT2 DNA interaction were obtained in wild-type versus mutant virus-infected MRC-5 cells (Fig. 4C). Together these results imply that the IE1-72kDa protein counteracts type I IFN-induced signaling at a step after nuclear translocation but before sequence-specific DNA interaction of ISGF3.

IE1-72kDa interferes with DNA association of ISGF3 components. (A) ChIP assays using STAT2-specific antibodies, quantified by real-time PCR. Amounts of DNA coprecipitated from ihf or ihfie1 cells at 0, 15, 30, or 60 min after IFN-α treatment (1,000 units/ml) were normalized to input DNA and plotted as mean values relative to nontreated samples (0 min, set to 1). (B) ChIP assays using STAT1-, STAT2-, or IRF9-specific antibodies. Amounts of DNA coprecipitated with polyclonal antibodies directed against the indicated proteins from ihf or ihfie1 cells at 60 min after IFN-α treatment (1,000 units/ml) were quantified by real-time PCR, normalized to input DNA, and plotted as mean values relative to nontreated samples (set to 1). (C) STAT2-specific ChIP assays from MRC-5 cells that were mock-treated or infected with wild-type Towne or CR208 viruses at a multiplicity of 1 PFU per cell for 12 h. Amounts of coprecipitated DNA were quantified by real-time PCR, normalized to input, and plotted as mean values relative to mock-infected samples (set to 1).
Colocalization and Physical Interaction Between IE1 and STAT1/STAT2.
When we compared the immunofluorescence staining patterns of IE1-72kDa and STAT2 in IFN-treated ihfie1 cells we noticed a striking nuclear colocalization of these two proteins. In most cells (>90%) the IE1 protein displayed a diffuse nucleoplasmic localization, and in these cells STAT2 was diffusely distributed throughout the nucleus as well (Fig. 5A). However, in cells undergoing mitosis, a predominant targeting of IE1-72kDa to condensed chromatin was evident. This observation is in agreement with previous findings (27–29). Strikingly, STAT2 was observed to colocalize with the viral protein at chromosomes in all mitotic cells examined (Fig. 5A). Moreover, in a minority (≈5%) of interphase nuclei, the IE1 protein displayed a dot-like staining pattern. At these dots, IE1-72kDa was found to colocalize extensively with the promyelocytic leukemia (PML) protein (data not shown), indicating that they represent subnuclear multiprotein complexes known as PML bodies or nuclear domain 10 (ND10). IE1-72kDa has long been known to associate with ND10, triggering their disruption, but in a small subset of cells the viral protein stably associates with these nuclear domains (27, 30, 31). Importantly, the IE1 and STAT2 immunostainings overlapped exactly at the ND10 structures in >98% of cells showing this pattern (Fig. 5A). In contrast, STAT2 was rarely or never found to localize to mitotic chromatin or nuclear dots, respectively, in IFN-stimulated cells not expressing IE1-72kDa (ihf cells). Instead, STAT2 nuclear staining was mostly diffuse in these cells (data not shown). These observations indicate that IE1-72kDa might delocalize STAT2 within the nucleus, thereby inactivating this important mediator of IFN signaling. Likewise, we found an immunofluorescent costaining of STAT1 and IE1-72kDa at nuclear dots in most cells (>90%) with this staining pattern, whereas colocalization with condensed chromatin during mitosis was never detected between these two proteins (Fig. 5B). Again, STAT1 was found relatively evenly distributed throughout the nucleoplasm in all ihfie1 cells with diffuse nuclear localization of the IE1 protein and in all ihf cells tested (Fig. 5B and data not shown). With respect to IRF9, a weaker association between the subnuclear localizations of the cellular and viral protein was observed as compared with STAT1 and STAT2. IRF9 only partially colocalized with IE1-72kDa at ND10 structures in a subset (≈67%) of nuclei (Fig. 5C). In mitotic cells, IRF9 was frequently found to associate with the cellular spindle apparatus but never localized to chromosomes (Fig. 5C). Finally, as was the case with STAT1 and STAT2, in cells that displayed even nuclear distribution of the IE1 protein or that were IE1-negative, IRF9 was localized in a nuclear diffuse fashion (Fig. 5C and data not shown).
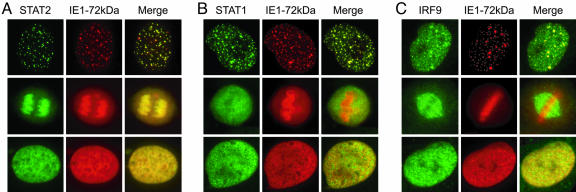
Nuclear colocalization of IE1-72kDa with ISGF3 proteins. Subconfluent ihfie1 cells were treated with 1,000 units/ml IFN-α for 1 h and subjected to double-labeling indirect immunofluorescence analysis using an IE1-specific mouse antibody in combination with rabbit sera directed against STAT2, STAT1, or IRF9, as indicated, followed by incubation with anti-mouse Alexa Fluor 546 and anti-rabbit Alexa Fluor 488 conjugates. Images were acquired on a Leica DMXR microscope. Typical nuclei showing dot-like, mitotic chromatin-associated or nuclear diffuse localization of IE1-72kDa and corresponding STAT2 (A), STAT1 (B), or IRF9 (C) stainings are shown. (Magnification: ×500.)
The immunolocalization results indicated that IE1-72kDa might be present in a physical complex with one or more components of ISGF3. To test this possibility, we performed coimmunoprecipitation experiments. As shown in Fig. 6A, complex formation between the IE1 protein and STAT2 was readily detected in extracts from wild-type CMV-infected cells. Importantly, a set of control experiments ruled out the possibility of nonspecific STAT2 precipitation in the absence of IE1 antigen or antibodies (Fig. 6A). Besides STAT2, the p84 and p91 isoforms of STAT1 could also be demonstrated to specifically coprecipitate with the IE1 protein from infected cells (Fig. 6A). However, compared with STAT2, it was more difficult to detect STAT1 above background in immunocomplexes with IE1-72kDa. In contrast, several attempts to demonstrate specific binding between the viral protein and IRF9 by coimmunoprecipitation failed (Fig. 6A).
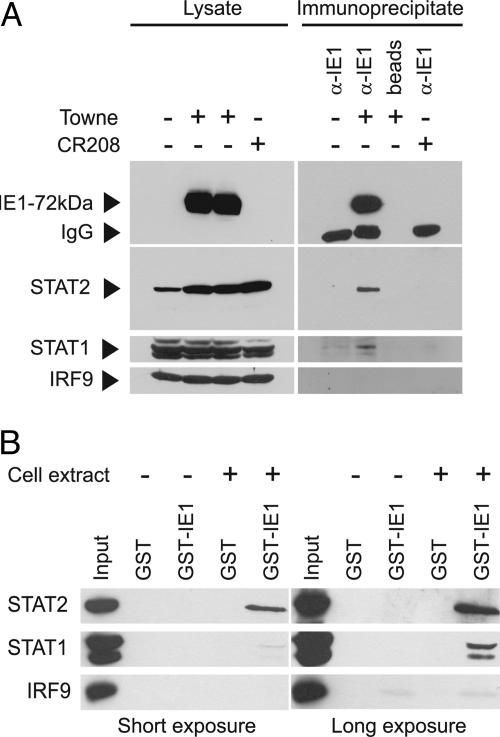
Physical interaction of IE1-72kDa with ISGF3 components. (A) In vivo binding analyzed by coimmunoprecipitation assay. MRC-5 cells were mock-infected or infected at a multiplicity of 3 PFU per cell with CMV Towne or CR208 for 24 h. Immunoprecipitations were performed by using an IE1-specific antibody or empty Sepharose beads. Proteins from whole-cell lysates (≈5% of material used as input for immunoprecipitations; Left) or immunoprecipitates (Right) were separated in SDS/10% polyacrylamide gels and detected by Western blotting with antibodies as indicated. IgG, Ig heavy chains. (B) In vitro binding analyzed by GST capture assay. Equal amounts of GST or GST-IE1 proteins were reacted with whole-cell extracts prepared from MRC-5 cells (+) or GST pull-down lysis buffer (−), and complexes or whole-cell lysates (Input) were separated in SDS/10% polyacrylamide gels. STAT1, STAT2, or IRF9 proteins were detected by Western blotting, and two different film exposures are shown.
In addition to these in vivo binding studies in CMV-infected cells, we further examined the interaction between IE1-72kDa and ISGF3 outside the virus context employing in vitro capture assays. To this end, we mixed cell extracts with a GST-IE1 fusion protein expressed in Escherichia coli. Consistent with the results from our coimmunoprecipitations, the pull-down assays demonstrated specific binding of IE1-72kDa with STAT1 and STAT2 but not IRF9 (Fig. 6B). These experiments also confirmed a higher-affinity interaction of IE1-STAT2 compared with IE1-STAT1 (Fig. 6B; compare input to output signals at short and long exposures). In sum, these results show that the CMV IE1-72kDa protein can physically interact with STAT1 and STAT2 without the need for additional viral factors.
Discussion
The infectious cycle of herpesviruses including CMV starts with virion binding to the cell surface. Postattachment events, such as membrane fusion, trigger antiviral molecular processes resembling a type I IFN response. As part of this response, transcription of IFN genes and several ISGs is up-regulated through processes that involve IRF1 and IRF3 in combination with other transcriptional activators. After gene induction, IFN-α and IFN-β proteins are secreted and signal through the IFN-α/β receptor via the Jak–STAT pathway to ISRE-regulated genes. The IFN response provides a first line of defense against infection by generating an intracellular environment that restricts viral replication and signals the presence of a viral pathogen to the adaptive arm of the immune response. Because these innate antiviral mechanisms are exceedingly potent and rapid, it is not surprising that many if not all viruses have evolved ways to either preclude the synthesis of IFNs or evade downstream antiviral events (1–4).
Recent studies have shown that herpesviruses encode several proteins that counteract the host IFN response. In CMV, the UL83-coded pp65 protein was reported to inhibit expression of ISGs and other antiviral genes via pathways that affect IRF1/NFκB (20) or IRF3 (21). Besides, CMV IE2-86kDa antagonizes virus-induced IFN-β production (32), and TRS1/IRS1 proteins block IFN-induced dsRNA-activated antiviral pathways (33). As opposed to all of these CMV gene products, several lines of evidence strongly support the view that the IE1-72kDa protein counteracts STAT-mediated IFN signaling, rather than interfering with the initial virus-induced activation of IFN and ISG transcription, which is independent of IFN secretion. (i) Initial activation of ISG transcription upon virus infection occurs irrespective of the presence or absence of IE1. Instead, IE1-mediated suppression of CMV-dependent ISG induction follows a delayed temporal pattern (Fig. 2B). (ii) IE1-72kDa efficiently inhibits ISGF3 DNA binding and ISG activation triggered by exogenous IFN-α (Figs. 4 and and22C) and confers partial type I IFN resistance upon CMV (Fig. 1). (iii) Confirming previous results (32), IE1 expression does not negatively affect CMV-induced IFN-β transcription (Fig. 2A). (iv) IE1 interacts with STAT1 and STAT2 (Figs. 5 and and6).6). In the latter respect, IE1-72kDa may exhibit functional similarities to a recently described STAT2-interacting protein of murine cytomegalovirus, M27 (34). However, this protein was shown to bind exclusively to STAT2 (and not STAT1), targeting it for degradation, whereas steady-state levels of all ISGF3 components are unaffected by IE1 expression (Fig. 3). Based on the fact that we could detect specific binding of IE1-72kDa with STAT1 and STAT2 but not IRF9 (Fig. 6), the viral protein may hinder or disrupt the interaction of STAT1/STAT2 with IRF9 inside the nucleus, thereby precluding association of trimeric ISGF3 complexes with ISG promoters (Fig. 4). Currently, we do not know whether the viral protein interacts with both STAT proteins directly. The fact that we could reproducibly detect more efficient binding to STAT2 as compared with STAT1 (Fig. 6) might indicate that the interaction of IE1-72kDa with STAT2 is direct whereas STAT1 binding occurs collaterally by means of STAT1/STAT2 dimerization. In this context, it is also conceivable that an indirect, STAT-mediated association of IE1-72kDa with IRF9 is below the detection limit of our binding assays. In support of this view, immunocytochemistry data indicate that the IE1 protein colocalizes not only with STAT1 and STAT2 but also with IRF9, although less extensively (Fig. 5). Moreover, at least in a subset of cells, the viral protein appears to sequestrate the cellular regulators at subnuclear dot structures and/or condensed chromatin. Thus, alternatively to interfering with ISGF3 integrity, IE1-72kDa might prevent association of intact trimeric complexes with ISRE DNA by redirecting them within the nucleus.
To our knowledge, no other viral protein besides CMV IE1-72kDa is currently known to antagonize an intranuclear step in STAT signaling. IE1 may therefore represent a novel viral tool to probe Jak–STAT signal transduction pathways. Our data also indicate that the ability of IE1-72kDa to counteract the type I IFN response is crucial for efficient productive CMV replication (Fig. 1). Thus, our work may pave the way for new antiviral intervention strategies that target the viral IE1 protein and/or cellular IFN signaling pathways.
Materials and Methods
Cell Culture and Virus Infections.
Human MRC-5 embryonic lung fibroblasts (European Collection of Cell Cultures) were cultured in Dulbecco’s modified Eagle’s medium containing 10% FCS. MRC-IE1 cells were described in ref. 26. The IE1-deficient CMV (CR208) and its parental wild-type strain (Towne) were provided by Ed Mocarski (Stanford University, Stanford, CA) and Richard Greaves (Imperial College, London). Wild-type and mutant CMVs were grown and titered on MRC-5 or MRC-IE1 cells, respectively, by standard plaque assay. The life-extended IE1-72kDa-expressing fibroblast cell line ihfie1.3 (herein referred to as ihfie1) and the corresponding control cells (ihf-2, herein referred to as ihf) have also been described (23, 24). Human recombinant IFN-α and a neutralizing rabbit polyclonal antibody directed against human IFN-β (AB1431) were obtained from R & D Systems and Chemicon, respectively. For viral growth and cellular mRNA analyses, cells were pretreated with these reagents for 24 h, and treatment was continued over the whole course of the experiment.
Protein–Protein Interaction Assays, Western Blotting, and Immunofluorescence.
Immunoprecipitations were performed as described in ref. 26. For capture assays, the IE1 cDNA was cloned into vector pGEX-KG (35) via SmaI and EcoRI sites, and the viral protein was expressed as a fusion with GST in E. coli strain M15[pRep4] (Qiagen). Affinity purification using glutathione Sepharose (Amersham Pharmacia) was performed according to the manufacturer’s instructions. Subsequently, 5 μg of glutathione Sepharose-bound GST or GST-IE1 protein were incubated with total cell extract from a 15-cm dish of MRC-5 fibroblasts in GST pull-down lysis buffer [50 mM Tris-HCl, pH 7.4/100 mM NaCl/10% glycerol/0.5% Triton X-100/protease inhibitor mixture (Complete Mini; Roche)] for 2 h at 4°C. Sepharose beads were washed four times with wash buffer (50 mM Tris, pH 7.4/100 mM NaCl/0.1% Triton X-100) and subjected to SDS/PAGE.
For Western blotting, Sepharose beads from immunoprecipitations or GST capture assays or whole-cell protein extracts prepared in lysis buffer (50 mM Tris-HCl, pH 8.0/150 mM NaCl/0.1% SDS/1% Nonidet P-40/0.5% sodium deoxycholate/protease inhibitor mixture) were mixed with 2× sample buffer (100 mM Tris-HCl, pH 6.8/4% SDS/0.2% bromophenol blue/20% glycerol/0.2 M 2-mercaptoethanol) followed by heating at 95°C for 5 min. Proteins were then assayed as described (27). Indirect immunofluorescence assays were also performed as described (27). The following primary antibodies were used in this study: mouse anti-CMV IE1 (1B12) (36), mouse or rabbit anti-IRF9 (ISGF3γ, BD Transduction Laboratories, and H-143, Santa Cruz Biotechnology, respectively), mouse anti-STAT1α p91 (C-111; Santa Cruz Biotechnology), rabbit anti-STAT1 p84/p91 (E-23; Santa Cruz Biotechnology), rabbit anti-p-STAT1 (9171; Cell Signaling Technology), rabbit anti-STAT2 (H-190 and/or N-17; Santa Cruz Biotechnology), rabbit anti-p-STAT2 (07-224; Upstate Biotechnology), and rabbit anti-α-tubulin (H-300; Santa Cruz Biotechnology).
ChIP and Real-Time Quantitative PCR.
ChIP assays were performed according to a previously published protocol (26). Protein from 5 × 106 ihf or ihfie1 cells was immunoprecipitated with a mixture of two polyclonal rabbit antisera directed against STAT2 (H-190/N-17) or antibodies against STAT1 (E-23) or IRF9 (H-143). Quantitative real-time PCR amplifications were carried out in triplicate as described (26). Relative changes in coprecipitated DNA were normalized to input DNA and calculated by using the relative quantification strategy described in Roche Applied Science Technical Note No. LC 13/2001. The PCR-amplified region comprised a 200-bp sequence covering the ISRE element and transcription initiation site of the human ISG54 promoter (nucleotides −169 to +30 relative to the transcription start site). The primer sequences were 5′-GGAGGAAAAAGAGTCCTCTA-3′ (ISG54P forward) and 5′-AGCTGCACTCTTCAGAAA-3′ (ISG54P reverse).
For quantification of viral transcripts, total RNA was isolated from 2.5 × 106 cells with TRIzol (Invitrogen). RNA was further purified by using the RNeasy Mini kit, including a DNase digestion step (Qiagen). Five micrograms of purified RNA was converted into cDNA by using an oligo(dT) primer and SuperScript III enzyme (Invitrogen), and quantitative real-time PCR was performed as described above employing 2 μl of 1:10 diluted first-strand cDNA and the following primer pairs: 5′-GACATCCCTGAGGAGATTAAG-3′ (IFN-β forward), 5′-ATGTTCTGGAGCATCTCATAG-3′ (IFN-β reverse), 5′-ACGGTATGCTTGGAACGATTG-3′ (ISG54 forward), 5′-AACCCAGAGTGTGGCTGATG-3′ (ISG54 reverse), 5′-TCTTCATGCTCCAGACGTAC-3′ (MxA forward), and 5′-CCAGCTGTAGGTGTCCTTG-3′ (MxA reverse).
Acknowledgments
We thank Richard Greaves and Ed Mocarski for generous gifts of reagents, Ines Tschertner (Universität Regensburg) for technical assistance, Alexandra Nitzsche and Dietmar Gross (Universität Regensburg) for experimental help, and Hans Wolf (Universität Regensburg) for invaluable support.
Abbreviations
| ChIP | chromatin immunoprecipitation |
| CMV | human cytomegalovirus |
| IE | immediate-early |
| IE1-72kDa | 72-kDa IE1 protein |
| IRF | IFN regulatory factor |
| ISG | IFN-stimulated gene |
| ISGF3 | ISG factor 3 |
| ISRE | IFN-stimulated response element |
| Jak | Janus kinase |
| PFU | plaque-forming unit |
| STAT | signal transducer and activator of transcription. |
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0600007103
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/103/10/3840.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
cGAS-STING-TBK1 Signaling Promotes Valproic Acid-Responsive Human Cytomegalovirus Immediate-Early Transcription during Infection of Incompletely Differentiated Myeloid Cells.
Viruses, 16(6):877, 30 May 2024
Cited by: 1 article | PMID: 38932169 | PMCID: PMC11209474
ScRNA-seq reveals novel immune-suppressive T cells and investigates CMV-TCR-T cells cytotoxicity against GBM.
J Immunother Cancer, 12(4):e008967, 30 Apr 2024
Cited by: 0 articles | PMID: 38688579 | PMCID: PMC11086384
IE1 of Human Cytomegalovirus Inhibits Necroptotic Cell Death via Direct and Indirect Modulation of the Necrosome Complex.
Viruses, 16(2):290, 13 Feb 2024
Cited by: 2 articles | PMID: 38400065 | PMCID: PMC10893529
Insights into the Transcriptome of Human Cytomegalovirus: A Comprehensive Review.
Viruses, 15(8):1703, 08 Aug 2023
Cited by: 2 articles | PMID: 37632045 | PMCID: PMC10458407
Review Free full text in Europe PMC
Chromatin control of human cytomegalovirus infection.
mBio, 14(4):e0032623, 13 Jul 2023
Cited by: 6 articles | PMID: 37439556 | PMCID: PMC10470543
Review Free full text in Europe PMC
Go to all (121) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2.
Virology, 283(2):230-239, 01 May 2001
Cited by: 162 articles | PMID: 11336548
IFN-type-I-mediated signaling is regulated by modulation of STAT2 nuclear export.
J Cell Sci, 119(pt 6):1092-1104, 28 Feb 2006
Cited by: 19 articles | PMID: 16507591
IRF-9/STAT2 [corrected] functional interaction drives retinoic acid-induced gene G expression independently of STAT1.
Cancer Res, 69(8):3673-3680, 07 Apr 2009
Cited by: 50 articles | PMID: 19351818
The interferon receptors.
Semin Oncol, 24(3 suppl 9):S9-18-S9-40, 01 Jun 1997
Cited by: 15 articles | PMID: 9208871
Review




