Abstract
Free full text

Tristetraprolin and Its Family Members Can Promote the Cell-Free Deadenylation of AU-Rich Element-Containing mRNAs by Poly(A) Ribonuclease
Abstract
Eukaryotic mRNA stability can be influenced by AU-rich elements (AREs) within mRNA primary sequences. Tristetraprolin (TTP) is a CCCH tandem zinc finger protein that binds to ARE-containing transcripts and destabilizes them, apparently by first promoting the removal of their poly(A) tails. We developed a cell-free system in which TTP and its related proteins stimulated the deadenylation of ARE-containing, polyadenylated transcripts. Transcript deadenylation was not stimulated when a mutant TTP protein was used that was incapable of RNA binding, nor when a mutant ARE was present that did not bind TTP. The ability of TTP to promote transcript deadenylation required Mg2+, but not ATP or prior capping of the RNA substrate. Cotransfection and additivity studies with the poly(A) RNase (PARN) demonstrated that TTP promoted the ability of this enzyme to deadenylate ARE-containing, polyadenylated transcripts, while having no effect on transcripts lacking an ARE. There was no effect of TTP to act synergistically with enzymatically inactive PARN mutants. We conclude that TTP can promote the deadenylation of ARE-containing, polyadenylated substrates by PARN. This interaction may be responsible for the ability of TTP and its family members to promote the deadenylation of such transcripts in intact cells.
Steady-state levels of cellular mRNAs are determined by the balance between their biosynthesis and turnover. Different mRNAs can exhibit marked differences in turnover rates within the same cell, and the turnover rates of individual mRNAs can also vary significantly in response to changes in the cellular environment. In mammalian cells, the earliest step in mRNA turnover is thought to be removal of the poly(A) tail, or deadenylation, and this process in turn largely determines the overall decay rate of the mRNA (15, 16, 31, 36).
It has been appreciated for many years that cis-acting AU-rich elements (AREs), often within the 3′-untranslated regions (3′-UTR) of the mRNA, can confer decreased stability on the mRNAs that contain them (32). These AREs have been classified according to the grouping of A and U residues within the motif (10, 36), and a recently updated database of these motifs has been established (1, 2). One type of ARE (36, 37) contains tandem repeats of the pentameric motif AUUUA; examples are present in the mRNAs encoding the clinically significant cytokines tumor necrosis factor alpha (TNF-α), granulocyte-macrophage colony-stimulating factor (GM-CSF), and interleukin-3 (IL-3). In these three well-studied cases, removal of the ARE makes the mRNA more stable, whereas transplantation of the ARE to previously stable mRNAs makes those mRNAs less stable (19, 37).
Much attention has therefore been focused on cellular proteins that bind to these AREs as potential regulators of mRNA stability (15, 36). One such ARE-binding protein is tristetraprolin (TTP), the prototype of a small family of tandem CCCH zinc finger proteins (5). Studies from TTP knockout mice and their control littermates have shown that TTP can promote the destabilization of TNF-α and GM-CSF mRNAs in primary macrophages and bone marrow-derived stromal cells, respectively, suggesting that TTP is a normal, physiological regulator of steady-state levels of these mRNAs in specific cell types (7, 8). TTP itself can be regulated at several levels, including transcription, phosphorylation, and subcellular localization, providing for potentially complex regulation of TNF-α and GM-CSF mRNA levels in appropriate cells (24, 25, 28, 29, 33, 35).
One clue to the mechanism of TTP's ability to destabilize class ARE-containing mRNAs came from studies of GM-CSF mRNA stability in primary bone marrow-derived stromal cells from TTP knockout mice (7). A striking finding was that GM-CSF mRNA was not only much more stable in the cells from the TTP knockout mice than in the control cells but also that there was also a marked increase in the ratio of the fully polyadenylated mRNA to the deadenylated mRNA body in the TTP-deficient cells compared to the control cells. This implicated TTP in the deadenylation of GM-CSF mRNA, even though the ARE TTP-binding site is separated from the poly(A) tail in the mouse GM-CSF mRNA by ca. 54 bases (see GenBank accession no. X02333).
The phenomenon of TTP-induced deadenylation was studied in more detail in cotransfection studies, which revealed that (i) native TTP but not a variety of tandem zinc finger (TZF) domain mutants promoted the deadenylation of mRNAs containing normal AREs (19, 22); (ii) certain ARE mutants that prevented TTP binding also were resistant to TTP-promoted deadenylation (22); (iii) the two other known members of the TTP protein family in mammals, ZFP36L1 and ZFP36L2, could also promote the deadenylation of class II ARE-containing mRNAs (19, 21); (iv) the TTP-induced deadenylation was accompanied by complete turnover of the mRNAs at low concentrations of expressed TTP, whereas high concentrations caused accumulation of the deadenylated mRNA body (19, 20); and (v) dephosphorylation of TTP could increase its apparent binding affinity for its ARE target (6).
In order to unravel the molecular steps involved in TTP-stimulated mRNA deadenylation and breakdown, it was necessary to develop a cell-free, TTP-dependent deadenylation assay for ARE-containing mRNAs. To do this, we took advantage of the fact that human 293 cells do not express detectable levels of TTP or its two known related human proteins, ZFP36L1 or ZFP36L2. In this cell type, coexpression of TTP or its related proteins with ARE-containing mRNAs leads to their deadenylation and subsequent destruction (19), indicating that the TTP-dependent deadenylation machinery is present and “activatable” in these cells. In the present study, we describe the development of such a cell-free deadenylation system in 293 cell extracts, and show that the presence of TTP or its related proteins can effectively activate the poly(A) RNase (PARN) in a manner that is specific for mRNA substrates that contain AREs composed of tandem AUUUA pentamers. These data indicate that TTP and its related proteins can bind selectively to ARE-containing substrate mRNAs and then specifically promote their deadenylation by the ubiquitous PARN enzyme. If this mechanism obtains in intact cells, it provides a means for the selective degradation of ARE-containing mRNAs by PARN or a related enzyme, while leaving the majority of polyadenylated transcripts unaffected.
MATERIALS AND METHODS
Plasmid constructs.
The expression plasmid CMV.hTTP.tag, its zinc finger mutant C124R, and its TZF domain alone [CMV.hTTP(97-173).tag] were made as described previously (21). The numbering system for the TTP mutants used the GenBank RefSeq for TTP, NP_003398. CMV.cMG1.tag and CMV.xC3H-3.tag were as described previously (21). CMV.hPARN.flag, which expressed the human poly(A)-dependent 3′ exoribonuclease (i.e., PARN), was constructed as follows. A cDNA coding for the open reading frame of PARN was made by reverse transcription-PCR from HeLa cell total RNA. The 5′ primer for the PCR amplification was 5′-ACGTggtaccGCCGAGATGAACCCCAGTG-3′, and the 3′ primer was 5′-GTGCCACCGGTGTTCCAACTGTTGATTACAAGGACGACGATGACAAGTAAGctcgagCAT; the lowercase letters indicate the restriction sites for Asp718 and XhoI, respectively. The underlined letters in the 3′ primer represent the coding sequence for the FLAG epitope. The resulting PCR product was digested with the enzymes and ligated into the Asp718 and XhoI sites of the expression vector CMV.BHG3′/BS+ (20). The correct sequence of the hPARN.flag DNA insert was confirmed by dRhodamine terminator cycle sequencing (Perkin-Elmer, Foster City, Calif.), and the sequence of hPARN was identical to bp 58 to 1974 (coding for the first to the last amino acids) of GenBank accession no. NM_002582.1; the protein sequence is listed as NP_002573. Mutations were introduced into the PARN cDNA sequence as described previously (25); correct sequences of all mutations were verified by dRhodamine terminator cycle sequencing.
Transfection of HEK 293 cells and preparation of cell extracts.
HEK 293 cells (referred to as 293 cells) were maintained, and transient transfection of 1.2 × 106 cells (in 100-mm plates) with plasmid constructs in calcium phosphate precipitates was performed as described previously (20). Unless otherwise indicated, 0.2 μg of hTTP, 2 μg of TZF, or 0.5 μg of hPARN expression plasmid was added to each plate of cells. Vector DNA (BS+) was added to each to make the total amount of cotransfected DNA 5 μg per plate. At 24 h after the removal of the transfection mixture, the cell monolayers were rinsed with ice-cold Ca2+- and Mg2+-free phosphate-buffered saline and then scraped into phosphate-buffered saline. After centrifugation at 600 × g for 3 min at 4°C, the cell pellet was gently rinsed in ice-cold diethyl pyrocarbonate-treated water containing 8 μg of leupeptin/ml, 0.5 mM phenylmethylsulfonyl fluoride, and 2 mM dithiothreitol. The cells were resuspended in the same solution (0.5 ml per 100-mm plate of cells) on ice for 3 min and then passed three times through a 27G needle. Buffer was then added to the cell lysates to achieve a final concentration of 10 mM HEPES (pH 7.6), 40 mM KCl, and 5% glycerol. The lysates were centrifuged at 100,000 × g for 45 min at 4°C, and then glycerol was added to the supernatant to achieve a final concentration of 15%. The cell extracts were stored at −70°C.
For some experiments, the fusion proteins hTTP.flag and hPARN.flag were isolated from the 100,000 × g supernatants (before the addition of glycerol) by using the FLAG immunoprecipitation kit (Sigma) and following the manufacturer's protocol. The resulting eluates were adjusted to a final concentration of 10 mM HEPES (pH 7.6), 40 mM KCl, and 15% glycerol and then stored at −70°C.
Cell-free deadenylation assays. (i) Preparation of RNA probes.
Plasmid pTNFα 1309-1332 (bp 1309 to 1332 of GenBank accession no. X02611) was constructed as described previously (20). Plasmid TNF-α 1309-1332 (A)50/SK− was made by inserting 50-bp (50 T's in the strand to be transcribed from) double-stranded oligonucleotides into the XbaI-EagI cloning sites of pTNFα 1309-1332 (Fig. (Fig.1A).1A). A template for RNA probe ARE-A50 was PCR amplified from this plasmid with primers M13 Forward and T50Xba. The resulting double-stranded template was sequenced by dRhodamine terminator cycle sequencing to make sure that the ARE was followed by 50 A's so that the transcribed probe would end with a string of A's with no other nucleotides 3′ of the poly(A) tail. Mutant A/C TNF-α 1309-1332 (A)50/SK− was made by substituting the flanking A's in the AUUUA motif with C's, and probe A/C ARE-A50 was made by the method described above. The templates for probe ARE and V were prepared by linearizing pTNFα 1309-1332 (A)50/SK− with XbaI and EcoRV, respectively.

Plasmid constructs and RNA probes. Constructs were made as described in Materials and Methods. Dashed lines in the schematic plasmids pictured at the top of each panel represent vector SK− sequence, whereas the solid lines represent the inserted ARE or poly(A) sequences, as indicated. The RNA probes are represented beneath each plasmid; in these diagrams, the dashed line represents sequence transcribed from SK−, the open box is an ARE, the solid box is a mutated ARE, the solid line is the normal 3′-UTR sequence 3′ of the ARE, and the box containing A's represents the poly(A) tail. (A) pTNFα 1309-1332 (A)50/SK− contains the TNF-α ARE sequence (bp 1309 to 1332 of GenBank accession no. X02611) between the EcoRV and XbaI sites of SK− as indicated. A double-stranded oligonucleotide encoding 50 A's was inserted between the XbaI and EagI sites. pTNFα A/C 1309-1332 (A)50/SK− was identical to pTNFα 1309-1332 (A)50/SK− except the A's in the ARE were mutated to C's. (B) (A)50/SK− contains a double-stranded oligonucleotide encoding 50 A's inserted between the XbaI and EagI sites. (C) pGM-CSF 668-775 (A)50/SK− contains the 3′ portions of the 3′-UTR of the mouse GM-CSF cDNA. The ARE is located from bp 668 to 722 of the cDNA, whereas the number 775 corresponds to the 3′ end of the cDNA. All probes were transcribed from templates prepared by PCR amplification or enzyme linearization of these plasmids by using RNA polymerase T7, as described in Materials and Methods.
Plasmid pA50/SK− was made by inserting 50 bp (50 T's in the strand to be transcribed from) of double-stranded oligonucleotides into the XbaI-EagI cloning sites of vector SK− (Fig. (Fig.1B).1B). The template for in vitro transcription of probe A50 was PCR amplified from pA50/SK− with primers M13 Forward and T50Xba and verified by sequencing.
Plasmid mGM-CSF 668-775(A)50/SK− (Fig. (Fig.1C)1C) contained 3′-UTR sequences from mouse GM-CSF mRNA b668-775 (bp 3399 to 3506 of GenBank accession no. X03020) that was inserted into the XbaI and EcoRV cloning sites of SK−, followed by 50 A's inserted between the XbaI and EcoRV sites. The template for probe g668-775A50 was PCR amplified (primers M13 Forward and T50Xba) from the plasmid and was sequenced to verify the 3′ end as described above. The template for probe g668-775 was prepared by linearizing the plasmid with XbaI.
The capped RNA probes were transcribed in the presence of [α-32P]UTP (800 Ci/mmol) and Ribo m7G cap analog (Promega). Linearized plasmids or PCR amplification products were used as templates, and the Promega riboprobe in vitro transcription systems protocol was employed. The resulting products were separated from the free nucleotides by using G50 columns.
(ii) In vitro deadenylation assay.
The reaction mixtures were assembled on ice. The reaction was initiated by adding 50 μl of probe (4 × 104 cpm in an assay buffer consisting of 10 mM HEPES [pH 7.6], 40 mM KCl, and 5% glycerol) into a tube containing 5 μg of protein from the 100,000 × g extract in 50 μl of assay buffer. MgCl2 (3 mM) was present in the assay unless otherwise indicated. The mixtures were incubated on ice or at 37°C for the times indicated. EDTA was added to achieve a final concentration of 20 mM to terminate the reaction. The mixture was then extracted once with phenol-chloroform. An aliquot of 60 μl of the aqueous phase was mixed with 60 μl of 2× formamide stop solution (95% formamide, 0.05% bromophenol blue, 0.1% xylene cyanol) and then heated at 70°C for 5 min. Aliquots of reaction products were analyzed on a 6 or 8% acrylamide gel containing 7 M urea.
Analysis of RNA-protein complexes by electrophoretic mobility shift assays and Western blotting. (i) RNA electrophoretic mobility shift assay.
Extracts (100,000 × g) prepared from 293 cells transfected with either vector alone or expression constructs driven by the CMV promoter (5 μg of protein) were incubated with 2 × 105 cpm of RNA probe as described previously (20, 21).
(ii) Western blotting.
Cell extracts were mixed with 1/5 volume of 5× sodium dodecyl sulfate (SDS) sample buffer (4), boiled for 5 min and then loaded onto SDS-8% polyacrylamide gels. Western blotting was performed by standard techniques. Membranes were incubated in Tris-buffered saline-0.3% Tween 20 with a polyclonal antiserum HA.11 (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) (1:2,000), a monoclonal anti-FLAG M2 antibody (Sigma; 1:2,000), or anti-PARN antiserum (a generous gift from Mike Wormington, University of Virginia) (11, 18).
(iii) Protein cross-linking.
The 100,000 × g extracts (40 μg of protein), prepared from 293 cells transfected with vector alone or the expression constructs CMV.hTTP.tag, CMV.hPARN.flag, or both were incubated with or without 3 mM MgCl2 at 25°C for 20 min. Disuccinimidyl suberate (DSS; Pierce) was then added to achieve a final concentration of 0.1 mM, and the mixtures were rotated at 4°C for 45 min. The incubations were stopped by adding Tris (pH 8.0) to a final concentration of 0.1 M. Aliquots of the reaction products were loaded onto SDS-8% polyacrylamide gels. Western blotting was performed with antisera as described above.
RESULTS
RNA probes.
Figure Figure11 is a schematic depiction of the RNA probes used in the present study; details of their construction are in Materials and Methods. All of the probes contained the same 5′ sequence of 58 (or in one case 61) ribonucleotides transcribed from the multiple cloning sites (MCS) of vector SK− (Stratagene), which permitted the comparison of the degradation profiles of related probes. All probes contained a 5′-cap. Probe ARE-A50 (Fig. (Fig.1A)1A) consisted of 58 bases transcribed from the MCS of SK− and 24 bases of the core ARE from the mouse TNF-α mRNA (bp 1309 to 1332 of GenBank accession no. X02611), followed by 50 A's; the 3′ end of the RNA did not contain any non-A ribonucleotides (see Materials and Methods). Probe A/C ARE-A50 consisted of the same components as described for ARE-A50 except that the flanking A's of the AUUUA motif were replaced by C's. Probe ARE was identical to ARE-A50 except that it did not contain a poly(A) tail. In probe V, the RNA transcript only consisted of the 58 bases transcribed from the MCS of SK−. Probe A50 (Fig. (Fig.1B)1B) contained 61 bases of the SK− MCS 5′ of 50 A's. Probe g668-775A50 (Fig. (Fig.1C)1C) contained 58 bases of the SK− MCS, followed by the 3′-most 108 bases of the mouse GM-CSF mRNA (bp 3399 to 3506 of GenBank accession no. X03020), followed again by 50 A's. This region of the GM-CSF mRNA contains the ARE, as indicated in Fig. Fig.1.1. Probe g668-775 was identical except that it lacked the poly(A) tail.
Cell-free deadenylation of polyadenylated, ARE-containing probes.
These probes were then used to characterize the ability of 293 cell extracts to promote RNA deadenylation in a TTP-dependent manner. These cells do not express endogenous TTP; the extracts used for these experiments were from cells transiently transfected with CMV.hTTP.tag, its derivatives, or vector alone (20, 22). Similar results were obtained with extracts derived from a 20-min, 12,000 × g centrifugation after cell lysis with 0.5% NP-40, as with extracts homogenized in the absence of detergent and centrifuged for 100,000 × g for 45 min at 4°C. Therefore, all of the results shown here were obtained with the detergent-free, high-speed extracts. Similar results were also obtained with capped and uncapped RNA substrates; all data shown in the present study were obtained with capped substrates.
When probes A50, ARE, ARE-A50, and V were incubated with extracts from cells transfected with vector alone, all three probes showed similar patterns of slight degradation after 1 h at 37°C (Fig. (Fig.2A,2A, compare lanes 1 and 2, lanes 5 and 6, lanes 9 and 10, and lanes 13 and 14). However, when these probes were incubated in parallel with the same amount of extract protein prepared from cells transfected with the human TTP expression plasmid CMV.hTTP.tag, only probe ARE-A50 was markedly more degraded (Fig. (Fig.2A,2A, compare lanes 3 and 4, lanes 7 and 8, and lanes 11 and 12). The disappearance of probe ARE-A50 in the presence of TTP (lane 12) was accompanied by the appearance of a new band of smaller size (arrow) that migrated to the same position as probe ARE, indicating that the new band represented the accumulation of a deadenylated form of the ARE-A50 probe. Thus, under these experimental conditions, TTP appeared to promote the degradation of only the ARE-A50 probe and not the degradation of a non-ARE-containing poly(A) probe (A50) or an ARE-containing nonpolyadenylated probe (ARE). The migration position of the V probe (MCS sequences alone) is shown in lanes 13 and 14.
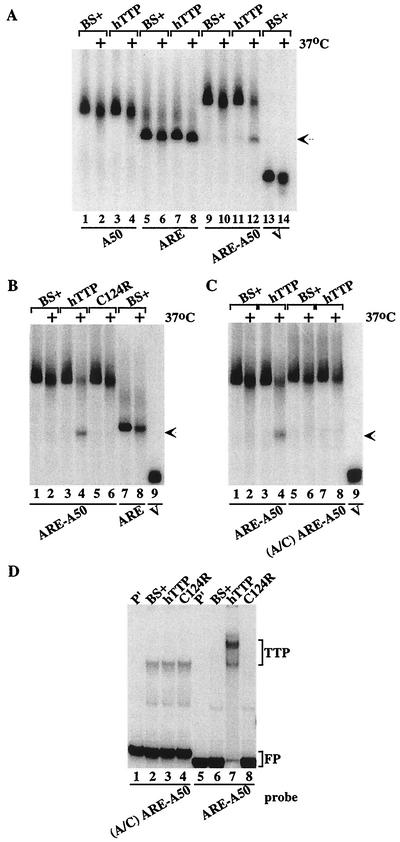
Cell-free deadenylation of polyadenylated, ARE-containing RNA probes. In panels A to C, 293 cell extracts were incubated with 32P-labeled RNA probes on ice (no symbol) or at 37°C (+) for 60 min, and EDTA (final concentration, 20 mM) was added to stop the reaction. RNA was then isolated and subjected to electrophoresis on urea-polyacrylamide gels, followed by autoradiography. The arrow in panels A to C indicates the migration position of the ARE probe and the deadenylated product of probe ARE-A50. (A) Incubation of the RNA probes A50 (lanes 1 to 4), ARE (lanes 5 to 8), ARE-A50 (lanes 9 to 12), and V (lanes 13 and 14) with extracts from 293 cells transfected with vector alone (BS+) or CMV.hTTP.tag (hTTP). (B) Incubation of the RNA probes ARE-A50 (lanes 1 to 6) and ARE (lanes 7 and 8) with extracts from 293 cells transfected with vector alone (BS+), CMV.hTTP.tag (hTTP), or the TTP zinc finger mutant (C124R). The position of probe V migration is shown in lane 9. (C) Incubation of the RNA probe ARE-A50 (lanes 1 to 4) and the mutant probe (A/C) ARE-A50 (lanes 5 to 8) with extracts from 293 cells transfected with vector alone (BS+) or CMV.hTTP.tag (hTTP). The position of probe V migration is shown in lane 9. (D) An electrophoretic mobility shift assay was performed by using extracts from 293 cells transfected with vector alone (BS+), CMV.hTTP.tag (hTTP), or the TTP zinc finger mutant (C124R) with probes (A/C) ARE-A50 (lanes 2 to 4) or ARE-A50 (lanes 6 to 8). The final reaction products were separated on an 8% nondenaturing polyacrylamide gel followed by autoradiography. Lanes 1 and 5 (P′) were loaded with probe alone (RNase T1 digested). The TTP-RNA complexes formed (TTP) and the migration position of the free probe (FP) are indicated.
Effect of nonbinding TTP mutants on deadenylation.
We have shown previously that the increased levels of TNF-α and GM-CSF mRNAs in cells derived from TTP-deficient mice were due to increased stability of those mRNAs (7, 8). Conversely, TTP promoted the instability of ARE-containing mRNAs in a 293 cell cotransfection system, apparently by first degrading the poly(A) tail of the mRNA (19, 20). This mRNA destabilizing effect of TTP required its binding to the ARE of these mRNAs through its TZF domain, since TTP lost both ARE-binding and mRNA-destabilizing activities when key amino acids in the TZF domain were mutated (19, 20, 22). We therefore evaluated the importance of the TZF domain to TTP's ability to promote RNA deadenylation in the cell-free deadenylation assay. As shown in Fig. Fig.2B,2B, an extract containing the TTP zinc finger mutant C124R caused the same minimal degradation of the probe ARE-A50 as seen with extracts from cells transfected with vector alone (Fig. (Fig.2B,2B, compare lanes 1 and 2 to lanes 5 and 6). The mutant TTP also did not cause accumulation of the lower-molecular-weight band, as seen with extracts containing wild-type TTP (Fig. (Fig.2B,2B, lanes 3 and 4), which again migrated to approximately the same position as the probe ARE (Fig. (Fig.2B,2B, lanes 7 and 8; arrow). Previous studies have documented that this mutant TTP protein is expressed at least as well as the wild-type TTP protein under these transfection conditions (see, for example, Fig. Fig.88 in reference 20). These studies demonstrate that the ability of TTP to bind to the ARE was required to promote the deadenylation of ARE-containing, polyadenylated RNA probes.

Attempted cross-linking of coexpressed PARN-FLAG and TTP-HA. Extracts were prepared from 293 cells expressing vector alone (BS+) or CMV-hTTP-tag (hTTP), CMV.hPARN.flag (hPARN), or both together. The extracts were then incubated without (−) or with (+) 3 mM MgCl2 for 20 min at 25°C as indicated. DSS (0.1 mM final concentration) was then added to some of the extracts (+, as indicated), and the extracts were rotated gently at 4°C for 45 min. The reactions were then stopped by the addition of Tris buffer (pH 8.0) to a final concentration of 0.1 M. Equivalent amounts of protein were then loaded onto three SDS-polyacrylamide gels, transferred to nitrocellulose, and blotted with antibodies to the FLAG epitope tag (A), PARN itself (B), or the HA epitope tag (C). Chemiluminescence autoradiography was then performed. The migration positions of molecular weight standards are indicated to the left of each blot. The brackets to the right of panels A and B indicate high-molecular-weight protein species that reacted with both the FLAG (A) and the PARN (B) antibodies. Immunoreactive PARN (A and B) and TTP (C) are also indicated. The faint immunoreactive species that occurred at Mr ~100,000 in panel C may be TTP dimer. See the text for additional details.
Effect of a mutant ARE on TTP-induced deadenylation.
We have shown previously that a mutant ARE, in which all of the A's of the core AUUUA pentamer were changed to G's, did not bind TTP (21). We tested whether the deadenylation of a similar mutant probe could be stimulated by wild-type TTP. The probe used was identical to probe ARE-A50 except that the flanking A's of the AUUUA motif in the ARE had been mutated to C's. In this case, TTP did not stimulate the deadenylation of the mutant probe A/C ARE-A50, despite evidence that TTP promoted the deadenylation of the normal ARE-A50 probe (Fig. (Fig.2C,2C, compare lanes 3 and 4 to lanes 7 and 8). In a gel shift assay with the same mutant probe, A/C ARE-A50, extracts from cells transfected with vector alone, wild-type TTP, or the C124R mutant formed similar complexes that migrated in similar patterns (Fig. (Fig.2D,2D, lanes 2 to 4). When probe ARE-A50 was incubated with extract from cells transfected with CMV.hTTP.tag, there was formation of the usual TTP-probe complex (Fig. (Fig.2D,2D, lane 7), which was not seen with extracts of cells transfected with the TTP mutant C124R or vector alone (Fig. (Fig.2D,2D, lanes 6, 8). The specificity of this TTP-probe complex has been validated in several previous studies (21, 22). These results indicated that a probe competent to bind TTP was necessary for deadenylation to occur in this cell-free assay.
Ability of TTP to promote deadenylation of a GM-CSF ARE probe.
We also tested a probe derived from an mRNA containing a second class II ARE that was contained in the GM-CSF mRNA. This probe was important to test because its ARE ends ca. 54 bases 5′ of the beginning of the poly(A) tail, as occurs in the natural GM-CSF mRNA; this is in contrast to the ARE-A50 probe used for most of these experiments, in which the core ARE from TNF-α mRNA was linked directly to a poly(A) tail, separated only by the XbaI cloning site. When the GM-CSF probe was incubated with 293 cell extracts, TTP caused degradation of the polyadenylated GM-CSF probe (Fig. (Fig.3A,3A, compare lanes 5 and 6 with lanes 7 and 8), again accompanied by the appearance of a smaller, deadenylated species (lane 8, arrow); however, TTP had no effect on the nonpolyadenylated probe (lanes 1 to 4). As in the case of the TNF-α probe, the TTP mutant C124R was without effect on degradation of the polyadenylated GM-CSF probe (lanes 9 and 10). As expected, native but not C124R mutant TTP could bind to the GM-CSF ARE probe in a gel shift assay (Fig. (Fig.3B3B).
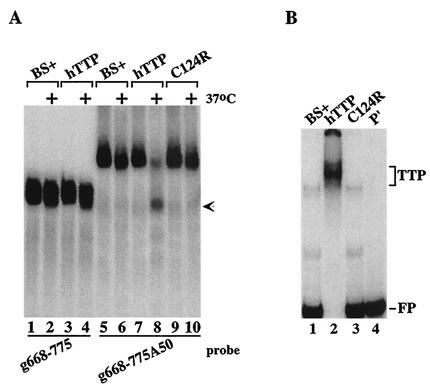
Ability of TTP to promote deadenylation of a GM-CSF ARE probe. (A) Extracts from 293 cells transfected with vector alone (BS+), CMV.hTTP.tag (hTTP), or the TTP zinc finger mutant (C124R) were mixed with the GM-CSF ARE probes g668-775 (lanes 1 to 4) or g668-775A50 (lanes 5 to 10) as described in the legend to Fig. Fig.2.2. The arrow indicates the migration position of the ARE probe (lanes 1 to 4), as well as the deadenylated product of probe g668-775A50 (lane 8). (B) Cell extracts as described above were incubated with probe g668-775A50 (lanes 1 to 3), and a gel shift assay was performed as described in the legend of Fig. Fig.2.2. Lane 4 (P′) was loaded with probe alone (RNase T1 digested). The TTP-RNA complexes formed (TTP) and the migration position of the free probe (FP) are indicated.
Effect of TTP-related proteins on probe deadenylation.
Besides TTP, mammals express two additional CCCH TZF proteins, ZFP36L1 and ZFP36L2 (5). Although the physiological functions of these two proteins are unknown, they have been shown, like TTP, to bind to ARE probes and stimulate the breakdown of ARE-containing mRNAs when coexpressed in cells (21). To examine whether these proteins caused the deadenylation of the ARE-containing polyadenylated probes in this cell-free system, we performed similar assays with extracts from 293 cells transfected with CMV.cMG1.tag (the rat orthologue of ZFP36L1) or CMV.xC3H-3.tag (the Xenopus orthologue of ZFP36L2). When probe ARE-A50 was incubated with extracts from cells transfected with TTP, cMG1, or xC3H-3 expression plasmids, the probe was degraded in a characteristic fashion in the presence of all three proteins (Fig. (Fig.4A,4A, lanes 4, 6, and 8) compared to extracts from vector alone-transfected cells (Fig. (Fig.4A,4A, lanes 1 and 2). The binding of these proteins to this probe could be readily demonstrated by gel shift analysis (Fig. (Fig.4B).4B). Thus, representatives of the two TTP-related proteins behaved like TTP in this cell-free deadenylation assay.
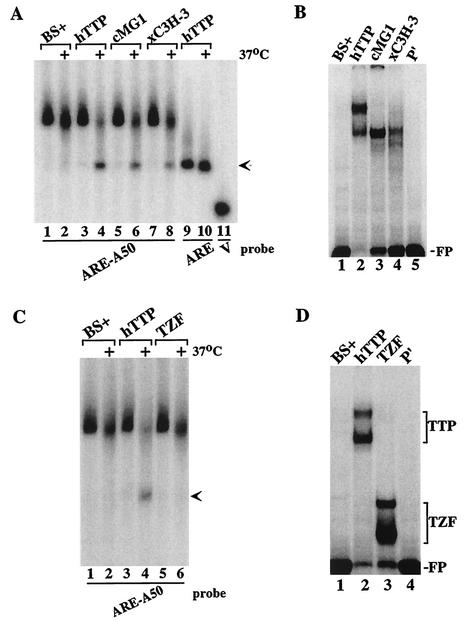
Effect of TTP-related proteins on probe deadenylation. (A) Extracts from 293 cells transfected with vector alone (BS+), CMV.hTTP.tag (hTTP), CMV.cMG1.tag (cMG1), or CMV.xC3H-3 (xC3H-3) were incubated with probe ARE-A50 (lanes 1 to 8) or ARE (lanes 9 and 10). The reaction mixtures were incubated on ice or at 37°C (+) for 60 min and then processed as described for Fig. Fig.2.2. The arrow indicates the migration position of the deadenylated product of probe ARE-A50 (lanes 4, 6, and 8) and the ARE probe (lanes 9 and 10). The position of probe V migration is shown in lane 11. (B) The cell extracts described in panel A were incubated with probe ARE-A50 (lanes 1 to 4) and used in a gel shift analysis. Lane 5 (P′) contained probe alone (RNase T1 digested). The migration position of the free probe (FP) is indicated. (C) Extracts from 293 cells transfected with vector alone (BS+), CMV.hTTP.tag (hTTP), or the TTP TZF domain vector CMV.hTTP(97-173).tag (TZF) were incubated with probe ARE-A50, and the deadenylation assay and analysis of the products were performed as described in panel A. The arrow indicates the migration position of the deadenylated product of probe ARE-A50 (lane 4). (D) The extracts described in panel C were incubated with probe ARE-A50 (lanes 1 to 3) and used in a gel shift assay. Lane 5 (P′) was loaded with probe alone (RNase T1 digested). The migration positions of the TTP-RNA (TTP) and TZF-RNA (TZF) complexes and of the free probe (FP) are indicated.
Effect of the TZF domain alone on deadenylation.
The members of this CCCH TZF protein family share a highly conserved TZF region that comprises the ARE-binding domain (21). To determine whether the TZF region alone was sufficient to induce the deadenylating activity in 293 cell extracts, we evaluated extracts from cells transfected with either full-length TTP or the epitope-tagged TZF domain, consisting of amino acids 97 to 173 of human TTP (GenBank RefSeq accession no. NP_003398). As expected, extracts from cells expressing full-length TTP caused degradation of the probe and accumulation of the deadenylated ARE band (Fig. (Fig.4C,4C, lanes 3 and 4, arrow). In contrast, the TZF-containing extract caused minimal degradation of the probe, and no accumulation of the deadenylated probe, after 60 min of incubation (Fig. (Fig.4C,4C, lanes 5 and 6), similar to extracts from cells transfected with the vector alone (Fig. (Fig.4C,4C, lanes 1 and 2). Despite the lack of activity in the deadenylation assay, the TZF domain polypeptide could readily bind to the ARE-A50 probe, as demonstrated by gel shift analysis (Fig. (Fig.4D).4D). These data indicated that the TZF domain peptide was unable to mimic full-length TTP in this cell-free deadenylation system.
Characterization of the TTP-induced deadenylating activity.
As described more fully below, we found that the activity within the 293 cell extracts that acted with TTP to stimulate deadenylation of ARE-containing polyadenylated probes was sensitive to boiling and could be extracted with phenol-chloroform, both of which are characteristics of proteins. Since the TTP-inducible deadenylating activity was present in a non-detergent-containing, 100,000 × g supernatant from 293 cells and required magnesium but not ATP (see below), we focused on the human enzyme PARN. Mammalian PARN activity has been shown to depend on Mg2+ but not on ATP (17). To investigate whether PARN might be involved in the TTP-dependent deadenylation of ARE-containing probes in this cell-free system, we prepared cell extracts in MgCl2-free buffer and then added back various concentrations of MgCl2. In the presence of 3 mM MgCl2 at 37°C, probe ARE-A50 was slightly degraded in extracts prepared from cells transfected with vector alone (Fig. (Fig.5A,5A, lane 2); however, in the absence of added MgCl2, the probe was completely stable at 37°C (Fig. (Fig.5A,5A, lane 3). When the probe was incubated with extracts from cells transfected with TTP, both the usual deadenylation of the probe and the appearance of the deadenylated probe decreased with decreasing concentrations of MgCl2 (Fig. (Fig.5A,5A, lanes 4 to 7). These findings suggested that the ARE-containing, polyadenylated RNA probe was degraded in the presence of TTP by a Mg2+-dependent activity present in the 293 cell extracts. The presence or absence of ATP had no effect on the TTP-dependent deadenylating activity (data not shown).
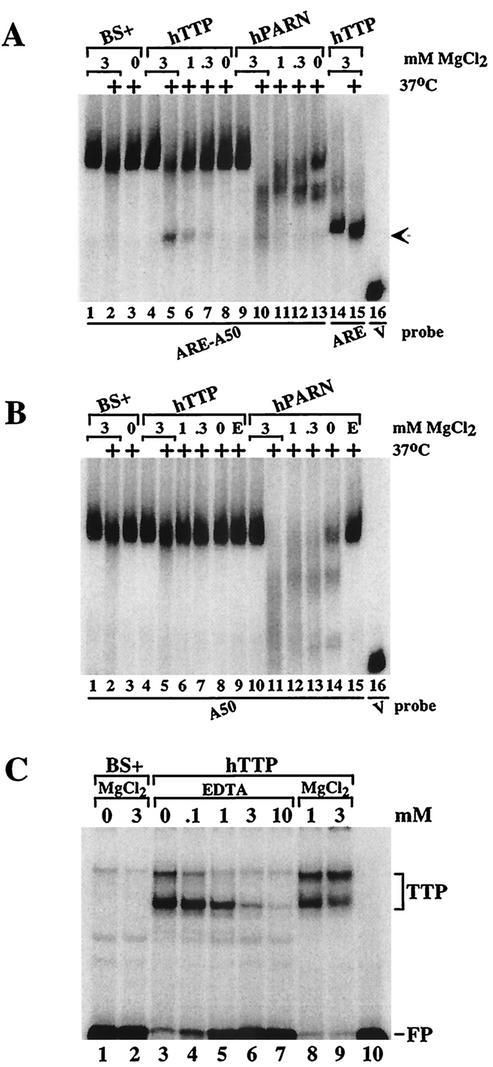
Characterization of the TTP-induced deadenylating activity. Extracts from 293 cells transfected with vector alone (BS+), CMV.hTTP.tag (hTTP), and CMV.hPARN.flag (hPARN) were incubated on ice or at 37°C (+) for 60 min and processed as described for Fig. Fig.2.2. (A) MgCl2 was included in the reaction mixture to final concentrations of 3 mM (lanes 1 and 2, lanes 4 and 5, lanes 9 and 10, and lanes 14 and 15), or 1 mM (lanes 6 and 11), 0.3 mM (lanes 7 and 12) or was not included (lanes 3, 8, and 13), and the mixtures were incubated and processed as described for Fig. Fig.2.2. The arrow indicates the migration positions of the deadenylated product of probe ARE-A50 (lanes 5 to 7) and the ARE probe (lanes 14 and 15). The position of probe V migration is shown in lane 16. (B) MgCl2 was included in the reaction mixture to final concentrations of 3 mM (lanes 1 and 2, lanes 4 and 5, and lanes 10 and 11), 1 mM (lanes 6 and 12), or 0.3 mM (lanes 7 and 13) or was not included (lanes 3, 8, and 13), and the mixtures were incubated and processed as described for Fig. Fig.2.2. EDTA (1 mM) was present during the incubation in lanes 9 and 15. The position of probe V migration is shown in lane 16. (C) Extracts from 293 cells transfected with vector alone (BS+) or CMV.hTTP.tag (hTTP) were incubated with probe ARE-A50 in the absence (lanes 1 and 3) or presence of MgCl2 (3 mM, lanes 2 and 9; 1 mM, lane 8) or with increasing concentrations of EDTA (0 to 10 mM, lanes 3 to 7); the reactions were then used in a gel shift assay. Lane 10 (P′) was loaded with probe alone (RNase T1 digested). The migration positions of the TTP-RNA complexes (TTP) and the free probe (FP) are indicated.
We also examined the effect of Mg2+ on the degradation of probe ARE-A50 in extracts from 293 cells that overexpressed human PARN. In the presence of 3 mM MgCl2, the PARN-containing extracts caused complete disappearance of the polyadenylated probe (Fig. (Fig.5A,5A, lanes 9 and 10). This activity decreased with decreasing concentrations of MgCl2 (Fig. (Fig.5A,5A, lanes 9 to 13), although there was no accumulation of the deadenylated RNA (arrow) as seen with TTP (lane 5). There was some PARN-induced degradation of the probe in the absence of added MgCl2 (Fig. (Fig.5A,5A, lane 13), possibly due to trace amounts of Mg2+ in the cell extracts (see below).
A polyadenylated probe (A50) that did not contain the ARE was minimally degraded by the 293 cell extracts from vector alone-transfected cells, both in the presence or absence of added 3 mM MgCl2 (Fig. (Fig.5B,5B, lanes 1 to 3). This probe was also minimally degraded in extracts from TTP-transfected cells, either when various concentrations of Mg2+ were present or in the presence of 1 mM EDTA (Fig. (Fig.5B,5B, lanes 4 to 9). However, when extracts from PARN-transfected 293 cells were exposed to the A50 probe, there was dramatic, MgCl2-dependent degradation of the probe that did not occur in the presence of 1 mM EDTA (Fig. (Fig.5B,5B, lanes 10 to 15).
Although the experiments described above demonstrate that the TTP-dependent deadenylating activity present in 293 cell extracts on ARE-containing, polyadenylated RNA substrates was dependent on Mg2+, it remained possible that the association of TTP with the ARE was itself Mg2+ dependent. However, neither the gel shift patterns of endogenous 293 cell proteins forming complexes with probe ARE-A50 (Fig. (Fig.5C,5C, lanes 1 and 2) nor TTP expressed in 293 cells (Fig. (Fig.5C,5C, compare lanes 3, 8, and 9) required added MgCl2. The formation of both TTP-ARE complexes decreased with increasing concentrations of EDTA (Fig. (Fig.5C,5C, lanes 3 to 7), perhaps due to chelation of the zinc ions within TTP's zinc fingers. These data indicate that the lack of TTP-induced deadenylation seen in the absence of Mg2+ was not due to inhibited TTP binding to the ARE under these conditions.
Effects of TTP and PARN together to promote deadenylation.
To evaluate the possible synergistic activation of deadenylation caused by TTP and PARN, cells were transfected with cDNAs expressing PARN and TTP, singly and together. Extracts containing TTP alone caused a time-dependent degradation of the ARE-A50 probe and accumulation of the deadenylated probe (Fig. (Fig.6A,6A, compare lanes 5 to 8 with lanes 3 and 4). Transfection of PARN alone caused a time-dependent deadenylation of the probe, but no accumulation of the deadenylated band (Fig. (Fig.6A,6A, lanes 9 to 12). However, when the effects of PARN and TTP together were evaluated under these conditions, there was complete probe degradation, and marked accumulation of the deadenylated probe (Fig. (Fig.6A,6A, lanes 13 and 14). Note that lane 14 (TTP plus PARN) was from only a 15-min incubation and is thus directly comparable to lane 6 (TTP alone) and lane 10 (PARN alone) at this time point. Thus, the two proteins together produced a dramatic and synergistic stimulation of probe ARE-A50 deadenylation under these conditions.
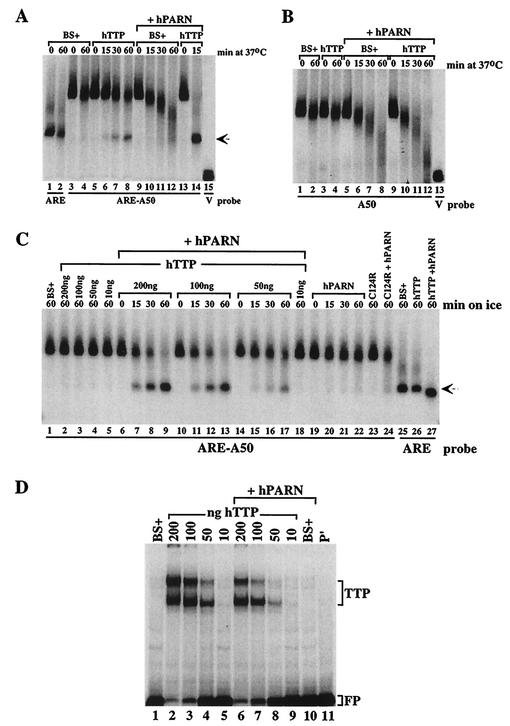
Effects of TTP and PARN expressed together to promote deadenylation. For panels A and B, extracts from 293 cells transfected with vector alone (BS+), CMV.hTTP.tag (hTTP), CMV.hPARN.flag (hPARN), or either BS+ or CMV.hTTP.tag together with CMV.hPARN.flag were incubated with the probes ARE-A50, ARE, or A50 as indicated. The deadenylation assays and sample processing were performed as described in the legend to Fig. Fig.2.2. (A) Extracts were incubated with the probes ARE or ARE-A50 at 37°C for the times indicated, except that reactions labeled “0” were incubated on ice for 60 min. The arrow indicates the migration position of the ARE probe (lanes 1 and 2) and the deadenylated product of probe ARE-A50 (lanes 6 to 8). The position of probe V migration is shown in lane 15. (B) Extracts were incubated with probe A50 at 37°C for the times indicated, except that reactions labeled “0” were incubated on ice for 60 min. The position of probe V migration is shown in lane 13. (C) Extracts from 293 cells transfected with vector alone (BS+), CMV.hPARN.flag (hPARN), or different amounts of CMV.hTTP.tag DNA (hTTP) extract (expressed as nanograms of TTP vector DNA added per 10-cm plate of cells) were mixed with CMV.hPARN.flag extracts and then were incubated on ice for the times indicated and processed for the deadenylation assay. The arrow indicates the migration positions of the deadenylated product of probe ARE-A50 (lanes 7 to 9, lanes 11 to 13, and lanes 15 to 17), as well as that of probe ARE (lanes 25 to 27). (D) The extracts described in panel C were incubated with ARE-A50 and used in the gel shift assay. Lane 11 (P′) was loaded with probe alone (RNase T1 digested). The migration positions of the TTP-RNA complexes and the free probe (FP) are indicated.
When similar experiments were performed with the A50 probe that lacked an ARE, there was no effect of TTP on probe degradation compared to extracts from cells transfected with vector alone (Fig. (Fig.6B,6B, compare lanes 3 and 4 to lanes 1 and 2). In extracts from cells cotransfected with PARN, the time courses of the probe degradation profiles were essentially the same when vector alone or TTP was cotransfected (Fig. (Fig.6B,6B, lanes 5 to 12). Thus, TTP had no apparent effect on the ability of PARN to cause deadenylation of a poly(A) probe that lacked an ARE.
We next performed similar experiments on ice, in an attempt to slow the reaction that was essentially complete by 15 min at 37°C in the presence of both PARN and TTP (see lane 14 in Fig. Fig.6A).6A). At 0°C, extracts from cells transfected with vector alone or various amounts of the TTP expression plasmid did not promote destabilization of the ARE-A50 probe after 60 min (Fig. (Fig.6C,6C, lanes 1 to 5). In the extract from cells transfected with PARN alone, there was barely detectable degradation of the ARE-A50 probe on ice, even after 60 min of incubation (Fig. (Fig.6C,6C, lanes 19 to 22). However, in extracts from cells expressing both exogenous PARN and TTP, there was a time-dependent degradation of the probe ARE-A50 at 0°C, accompanied by a gradual increase in the accumulation of the deadenylated ARE band (arrow; Fig. Fig.6C,6C, lanes 6 to 18), and the deadenylating activity was dependent on the amount of transfected TTP DNA used. At the lowest concentration of TTP DNA used, 10 ng of CMV.hTTP.tag, there was no apparent degradation of the probe after 60 min on ice (Fig. (Fig.6C,6C, lane 18). The TTP zinc finger mutant C124R alone did not induce any endogenous deadenylating activity (Fig. (Fig.6C,6C, lane 23); after 60 min of incubation with extract from cells cotransfected with the PARN vector, the probe was found to be degraded to approximately the same extent as occurred with extracts from cells transfected with PARN alone (Fig. (Fig.6C,6C, compare lanes 22 and 24).
The possible effects of PARN on the association of various concentrations of expressed TTP with the ARE-A50 probe were evaluated in a gel shift assay (Fig. (Fig.6D).6D). The presence of PARN in the extract did not increase the binding of TTP to the RNA probe, nor did it result in the formation of a “supershifted” complex, as might be expected if there were direct physical association between TTP and PARN (Fig. (Fig.6D).6D). PARN alone did not shift this probe into the gel (Fig. (Fig.6D,6D, compare lanes 1 and 10). At the concentration of TTP DNA, 10 ng, at which no detectable synergistic activation of deadenylation with PARN occurred at 0°C (Fig. (Fig.6C,6C, lane 18), there was barely detectable formation of a TTP-probe complex (Fig. (Fig.6D,6D, lanes 5 and 9).
Effects of affinity-purified TTP and PARN on deadenylation.
To begin to address the question of whether PARN plus TTP alone could promote the ARE-dependent deadenylation of poly(A) probes, the fusion proteins hTTP-FLAG or hPARN-FLAG were isolated by affinity chromatography from 293 cells transfected with the expression plasmids (Fig. (Fig.7).7). The fusion proteins were eluted from the affinity matrix with FLAG epitope peptide in an attempt to decrease nonspecific elution of contaminating proteins. When the probe ARE-A50 was incubated at 37°C with either the TTP eluate alone (Fig. (Fig.7A,7A, lane 1) or the PARN eluate alone (P) (Fig. (Fig.7A,7A, lane 2), there was minimal degradation of the intact probe compared to the effects of extract from cells transfected with vector alone (Fig. (Fig.7A,7A, lane 5). However, the probe was almost completely degraded, along with formation of the characteristic lower band, when both FLAG eluates were combined (Fig. (Fig.7A,7A, lane 3). When the FLAG-TTP eluate was added to the vector-transfected extract, the ARE-A50 probe degradation profile was virtually identical to that seen when extract from TTP-transfected cells was used (Fig. (Fig.7A,7A, compare lanes 6 and 10). The addition of PARN-FLAG in the absence of TTP caused more degradation of the probe than the presence of the endogenous deadenylating activity (Fig. (Fig.7A,7A, compare lanes 4 and 7). When both eluates were added, the probe was almost completely degraded (Fig. (Fig.7A,7A, lane 8), with formation of the lower band (Fig. (Fig.7A,7A, lane 8) that migrated to the same position as probe ARE (arrow; Fig. Fig.7A,7A, lanes 11 to 14). Thus, under these conditions, the individual TTP and PARN eluates each had minimal deadenylating activity; however, the combination of the two had marked deadenylating activity toward the ARE-A50 probe. Similar results were seen when the TTP and PARN eluates were incubated together with probe ARE-A50 on ice (results not shown).

Effects of affinity-purified TTP and PARN on deadenylation. Deadenylation assays were performed with the fusion proteins hTTP-FLAG or hPARN-FLAG that had been isolated by affinity chromatography from 293 cells transfected with the appropriate expression plasmids; in some cases, these were mixed with extracts from 293 cells transfected with vector alone (BS+) or CMV.hTTP.tag (hTTP). (A) Effects of affinity-purified hTTP-FLAG (T) or hPARN-FLAG (P) alone or together (TP) on the deadenylation of probe ARE-A50 in the absence (lanes 1 to 3) or presence of extracts from 293 cells transfected with vector alone (BS+) (lanes 4 to 8), either on ice (no legend) or after 60 min at 37°C (+). Lanes 9 and 10 show deadenylation of the probe in extracts from cells transfected with CMV.hTTP.tag (hTTP). The arrow indicates the migration positions of the deadenylated product of probe ARE-A50 (lanes 3, 6, 8, and 10) and the ARE probe (lanes 11 to 14). The position of probe V migration is shown in lane 15. (B) Similar extracts prepared from 293 cells were either untreated (C), extracted with phenol-chloroform (E), or boiled (B), after which they were incubated with probe ARE-A50 (lanes 1 to 4) either in the presence of FLAG peptide (F; lanes 5 to 8) or of affinity-purified hTTP-FLAG (T; lanes 9 to 12). The effects of 293 cell extracts from cells expressing transfected CMV.hTTP.tag (hTTP) on probes ARE-A50 (lanes 13 and 14) or ARE (lanes 15 and 16) are also shown. The arrow indicates the migration positions of the deadenylated product of probe ARE-A50 (lanes 10 and 14) and the ARE probe (lanes 15 and 16). The position of probe V migration is shown in lane 17.
To characterize further the endogenous factor(s) in 293 cells whose deadenylating activity was stimulated by TTP in the degradation of the ARE-containing poly(A) probes, we treated 293 cell extracts by phenol-chloroform extraction or by boiling. When the phenol-chloroform extracted or boiled 293 extracts were incubated with the ARE-A50 probe at 37°C, there was little degradation of probe compared to untreated extract (Fig. (Fig.7B,7B, lanes 1 to 4). Likewise, the ARE-A50 probe stability in the FLAG peptide eluate added to untreated, or extracted, or boiled extracts at 37°C was comparable to that seen in the absence of the eluate (Fig. (Fig.7B,7B, lanes 5 to 8). When the eluted FLAG-TTP was incubated with the probe, there was a slight degradation of the probe but no change in the size of the probe was seen (Fig. (Fig.7B,7B, lane 9). However, in the presence of the untreated 293 extract, almost all of the probe was degraded, with the formation of the smaller band (Fig. (Fig.7B,7B, lane 10) that migrated to the same position as that seen when extracts from 293 cells transfected with TTP were used (Fig. (Fig.7B,7B, lane 14). FLAG-TTP added to phenol-chloroform or heat-treated 293 extracts did not cause the probe to degrade (Fig. (Fig.7B,7B, lanes 11 and 12), suggesting that the deadenylation factor(s) effectively activated by TTP was a protein and was heat labile. Probe ARE incubated with the TTP-containing extract was also shown in this experiment (Fig. (Fig.7B,7B, lanes 15 and 16).
Attempted cross-linking of coexpressed FLAG-PARN and hemagglutinin (HA)-TTP.
These studies do not establish the mechanism by which TTP effectively increases PARN activity toward ARE-containing, polyadenylated RNA substrates. One simple model is that TTP acts as a tether or adaptor molecule, physically linking PARN to the RNA by a direct physical interaction between TTP and PARN. To test this possibility, we performed protein cross-linking experiments in cell extracts by using the bifunctional cross-linker DSS. These studies used 293 cell extracts in which one or both proteins were overexpressed as fusions with different epitope tags, followed by cross-linking in the presence or absence of magnesium. Figure Figure88 shows the results of the present study; each panel of Fig. Fig.88 is a similar Western blot of the various 293 cells extracts, in each case probed with a different antibody.
Figure Figure8A8A shows a Western blot of extracts probed with the FLAG antibody to recognize the FLAG epitope on PARN but not on TTP. This antibody identified immunoreactive PARN as an Mr ~80,000 protein (lanes 9 to 16); this band was not seen in extracts from cells transfected with vector alone (lanes 1 to 4) or with HA-tagged TTP alone (lanes 5 to 8). Cross-linking with DSS revealed a new complex of Mr ~190,000 that strongly reacted with the FLAG antibody (bracket in Fig. Fig.8A).8A). This complex was only seen after cross-linking; it was not affected by the presence of cotransfected TTP (lanes 13 to 16) or by the presence or absence of magnesium. The identity of the components of this complex is not known, except that the immunoreactivity identifies one component as PARN-FLAG; this complex may represent the oligomeric form of the activated human enzyme, as shown previously (26). However, the presence of this larger complex confirms that the cross-linking was effective. An identical blot was then probed with a PARN antibody (Fig. (Fig.8B).8B). This showed the expected reactivity with transfected PARN at Mr ~80,000 (lanes 9 to 16) and confirmed that the higher-Mr complex (bracket) also contained immunoreactive PARN. A much smaller amount of immunoreactive PARN was noted in the cells transfected with vector alone (lanes 1 to 4) or with TTP-HA alone (lanes 1 to 8). This endogenous PARN immunoreactivity decreased in amount after cross-linking, and longer exposures of this blot showed that the large complex was also formed between endogenous PARN and unknown partners after cross-linking (not shown).
An identical blot was then probed with the HA antibody to identify TTP-HA (Fig. (Fig.8C).8C). TTP was not detected in the vector alone transfected cells (lanes 1 to 4) or the cells transfected with PARN alone (lanes 9 to 12); however, a broad band of immunoreactivity of the appropriate size was present when TTP was transfected, alone (lanes 5 to 8) or with PARN (lanes 13 to 16). A faint band at Mr ~110,000 is probably the TTP dimer noted in previous studies (20). Neither the amount of TTP nor its larger complex was affected by the presence or absence of the cross-linker, PARN, or magnesium.
If there had been direct complex formation between TTP and PARN, we would have expected formation of a novel Mr ~120,000 complex that reacted to all three of the antibodies used: FLAG, PARN, and HA. However, no such complex was observed with any of the three antibodies used, either in the presence or in the absence of magnesium (Fig. (Fig.8,8, lanes 13 to 16). Therefore, despite abundant expression of both TTP and PARN and despite evidence that cross-linking had occurred, we found no evidence for a direct physical association between the two proteins under these conditions.
Effect of inactive PARN mutants.
To determine whether the TTP effect could be mediated or inhibited by inactive PARN mutants, we relied on previous data demonstrating that mutation of any one of four key amino acids within the primary sequence of human PARN completely inactivated the enzyme (30). We therefore made similar mutations in our PARN expression vector and examined both their enzymatic activity and their ability to influence TTP activity in 293 cell extracts.
First, we examined the activity of the PARN mutants on a poly(A) substrate (Fig. (Fig.9A);9A); in all panels of Fig. Fig.9,9, the reactions were conducted at 30°C in an attempt to slow them somewhat. As noted earlier, cell extracts enriched in transfected TTP were essentially identical to extracts from cells transfected with vector alone in their inability to promote deadenylation of the poly(A) substrate (Fig. (Fig.9A,9A, compare lanes 2 and 3). Cell extracts enriched in native (i.e., nonmutant) PARN exhibited the usual ability to cause shortening of this substrate (Fig. (Fig.9A,9A, lane 4). However, each of three PARN mutants—D28A, E30A, and D382A—when expressed in 293 cells exhibited essentially no deadenylating activity under these conditions (Fig. (Fig.9A,9A, lanes 5 to 7). The expression of TTP plus native PARN had no effect on PARN′s ability to promote deadenylation of this non-ARE-containing probe (Fig. (Fig.9A,9A, lane 8). Similarly, the coexpression of TTP with the three mutant PARN proteins had no effect on their inability to promote probe deadenylation (Fig. (Fig.9A,9A, lanes 9 to 11). Each of the three mutant proteins was expressed at levels comparable to that observed with the native protein, as determined by Western blotting of the same extracts with the HA epitope antibody (Fig. (Fig.9B).9B). The expression of the mutant PARN proteins was modestly increased by the coexpression of TTP in this experiment, whereas the expression of FLAG-tagged TTP was not affected by the coexpression of native or mutant PARN (Fig. (Fig.9B9B).

Effect of inactive PARN on probe deadenylation in the presence or absence of TTP. Extracts (5 μg of protein) from 293 cells transfected with vector alone (BS+), CMV.hTTP.tag (hTTP), CMV.hPARN.flag (hPARN), or its mutants (D28A, E30A, and D382A), or CMV.hTTP.tag together with CMV.hPARN.flag (or its mutants) were incubated with probes ARE-A50, ARE, or A50, as indicated. Deadenylation assays were carried out at 30°C in this experiment to slow the reaction rate. The buffer used in these experiments contained 100 mM KCl, 1 mM MgCl2, and 10 mM HEPES (pH 7.6).(A) Effect of native PARN (Wt) and the three mutant PARN proteins on the deadenylation of the poly(A) probe (A50) in the presence or absence of TTP, as indicated, either at 0°C (−) or after 60 min at 30°C (+). BS+ refers to extracts from cells transfected with vector alone. Probe V is the remnant vector sequence with no attached poly(A) tail. (B) Western blot demonstrating the expression of native and mutant PARN species in these experiments, as indicated. The symbols are the same as in panel A. The expression of FLAG-tagged TTP is also indicated by a Western blot with the FLAG antibody. The positions of molecular weight standards are shown on the left. (C) Effect of the extracts from vector alone (BS+), TTP alone, native and mutant PARN alone, and various combinations on the deadenylation of ARE-containing, polyadenylated probe ARE-A50, as well as the nonpolyadenylated ARE-containing probe ARE. The times of incubation at 30°C are indicated; the position of the completely deadenylated probe is indicated by the arrow. See the text for further details.
We then examined the effect of TTP on the ability of cotransfected native and mutant PARN to deadenylate the ARE-containing, polyadenylated substrate. The concentrations of expressed TTP and native PARN were adjusted so that each would have a relatively minor effect alone, making synergy between the two readily detectable. The deadenylation reactions were performed at 30°C for the same reason. As shown in Fig. Fig.9C,9C, there was essentially no probe degradation after the incubation of extracts from cells transfected with vector alone for 60 min at 30°C (compare lane 19 to lane 18). The expression of native PARN led to a modest shortening of the probe under these conditions (Fig. (Fig.9C,9C, compare lane 20 to lane 19). As noted with the poly(A) substrate, there was no difference in deadenylating activity between extracts from cells transfected with vector alone (Fig. (Fig.9C,9C, lane 19) and those from cells transfected with the plasmids encoding the PARN mutants D28A (lane 21), E30A (lane 22), or D382A (lane 23). These data indicate that these PARN mutants had no effect on the ARE-containing, polyadenylated substrate, as noted for the poly(A) substrate.
We next performed time courses of probe degradation with extracts containing TTP alone and then with extracts containing TTP plus either native or mutant PARN proteins. Under these conditions of relatively low protein concentration and 30°C incubation, TTP alone had a modest, time-dependent effect on probe degradation and accumulation of the deadenylated substrate (Fig. (Fig.9C,9C, lanes 2 to 5). Coexpression of native PARN plus TTP resulted in the expected marked increase in time-dependent probe degradation accompanied by accumulation of the deadenylated probe (Fig. (Fig.9,9, lanes 6 to 8). The 60-min time point for TTP plus PARN (lane 8) exhibited markedly increased disappearance of the polyadenylated probe and appearance of the deadenylated probe compared to the control extract (lane 19), native PARN alone (lane 20), or TTP alone (lane 5) at the same time point. When TTP and the mutant PARN were cotransfected, there was no apparent increase in the net effect on deadenylation of this probe compared to TTP alone (compare lanes 3 to 5 with lanes 9 to 11, lanes 12 to 14, and lanes 15 to 17), demonstrating that TTP could not “effectively activate” the mutant enzyme. Importantly, the effects of TTP to promote probe deadenylation and the accumulation of the deadenylated probe were not inhibited by the coexpression of the mutant PARN, suggesting that the endogenous deadenylating activity that was increased in the presence of TTP was not inhibited by the presence of the mutant PARN. This apparent lack of an inhibitory effect of the mutant PARN proteins was more evident at either 37°C or when higher concentrations of TTP were used (data not shown). Figure Figure9C9C also demonstrates the complete lack of effect of TTP alone (lane 26), PARN alone (lane 27), and TTP plus PARN (lane 28) on the stability of the nonpolyadenylated, ARE-containing probe.
DISCUSSION
The most important finding of the present study is that TTP, as well as the other known members of the TTP family of mammalian CCCH TZF proteins, can stimulate the deadenylating activity of PARN, specifically toward ARE-containing RNA substrates, in a cell-free system. This “TTP effect” was not seen with the following controls: with mutant forms of TTP that were unable to bind to the ARE; with mutant forms of PARN that had no intrinsic enzyme acitivity; with transcripts containing a mutant ARE that were unable to bind TTP; with a transcript consisting of poly(A) alone; and with ARE-containing but nonpolyadenylated transcripts. Thus, TTP binding to the ARE appears to specifically target that ARE-containing RNA for PARN-mediated deadenylation and subsequent destruction. This provides a potential cellular mechanism by which the stability of a given transcript can change depending upon the relative abundance of TTP or its family members at that time, even without changes in the abundance or characteristics of the ubiquitous PARN enzyme. This in turn allows specific ARE-containing transcripts to be degraded more rapidly, although the rate of degradation of the vast majority of mature transcripts is unchanged. Modulation of TTP's RNA-binding activity by factors such as phosphorylation (6, 35) or subcellular localization (29, 34), in addition to transcriptional regulation of protein abundance (23-25), thus provide the means for a complex regulatory system that can control the deadenylation and thus the degradation rate of ARE-containing transcripts.
Although these data indicate that TTP and its family members can promote the deadenylation of ARE-containing transcripts by PARN in this cell-free system, this does not mean that PARN is necessarily the enzyme responsible for the analogous effects of the CCCH proteins in intact cells. Indeed, our previous data showing that TTP can promote the destabilization of ARE-containing, nonpolyadenylated histone mRNAs in cells (19) suggest that binding of TTP or its relatives to the ARE can promote the degradation of nonpolyadenylated mRNAs by alternative enzymes. We also cannot conclude that PARN was necessarily the responsible endogenous enzyme “effectively activated by” TTP in 293 cell extracts. Using an antibody that was very effective in Western blots, we were not able to effect complete immunodepletion of PARN from 293 cell extracts, one possible approach to this question. In addition, although the results obtained with catalytically inactive, potential “dominant-negative” mutant forms of PARN indicated that these proteins lacked PARN activity, we saw no evidence that these mutant proteins could inhibit the ability of TTP to activate an endogenous deadenylating activity in 293 cells extracts. There are many potential reasons why this approach was not effective; a major possibility is that, since the mutant PARN was expressed only in the fraction of the cells actually transfected, it would have been synthesized and presumably allowed to oligomerize only with the native PARN molecules in those transfected cells and presumably not with the native PARN molecules in the nontransfected cells with which it was mixed in the final extract. Other approaches to completely depleting the cells of PARN activity, e.g., stable cell lines expressing the mutant proteins, other types of knockout approaches, etc., will be necessary to answer this question. For now, we can conclude that TTP is capable of effectively activating PARN toward ARE-containing, polyadenylated substrates, but whether PARN is responsible for all or most of the TTP-activated deadenylation of ARE-containing substrates that occurs in nontransfected cells remains to be determined.
These studies were made possible by the development of the cell-free, TTP-dependent, ARE-dependent deadenylation assay described here. This assay in turn depended on the fact that 293 cells, although they apparently do not express TTP or its two known mammalian relatives, nonetheless contain the cellular machinery to degrade class II ARE-containing mRNAs if TTP is artificially transfected into the cells (19, 21). Using cell extracts from these cells, we found that extracts containing TTP expressed from plasmid transfection exhibited markedly increased rates of deadenylation of ARE-containing RNA substrates compared to control extracts. This TTP-dependent deadenylating activity (i) required the presence of a so-called class II ARE in the RNA substrate, (ii) required that the substrate have a poly(A) tail, (iii) required an intact TZF domain in TTP that was capable of RNA binding, (iv) appeared to require additional domains of TTP besides the TZF domain alone, (v) did not require detergents or organelles sedimented by a centrifugation at 100,000 × g for 45 min, (vi) required magnesium but not ATP, and (vi) was heat labile and phenol-chloroform extractable. These properties suggested that the enzyme that was effectively activated was PARN or a similar 3′-to-5′ RNA exonuclease, since PARN is a soluble protein that requires magnesium but not ATP for activity (26). In confirmation of this hypothesis, coexpressed PARN and TTP exhibited dramatically increased activity against ARE-containing, polyadenylated RNA substrates compared to the activity of either protein alone; this was especially striking at 0°C, at which temperature neither TTP alone in 293 cell extracts, nor PARN alone in 293 cell extracts, exhibited significant activity. This synergistic effect of TTP plus PARN was also seen when both proteins, expressed as FLAG fusion proteins, were purified by elution from FLAG affinity columns with the FLAG epitope peptide. It is premature to conclude that only these proteins plus the RNA substrate are necessary for the apparently synergistic effect on deadenylation, since low concentrations of other cellular proteins are likely to have contaminated the FLAG eluates, even when the epitope-tagged proteins were eluted with the epitope peptide. Nonetheless, the affinity-purified proteins had negligible activity on their own but marked activity when mixed, providing some support for the notion that no other components are necessary.
This cell-free deadenylation assay differs in certain respects from that recently described by Wilusz and colleagues (12-14). In contrast to the current system with 293 cells, which lack detectable levels of endogenous TTP, ZFP36L1, and ZFP36L2, these authors used extracts from HeLa cells, which express readily detectable concentrations of TTP. Their assay required the addition of high concentrations of poly(A), presumably to prevent endogenous poly(A)-binding proteins from associating with and protecting the RNA target. In contrast, our assay system did not require added poly(A) for activity. The assay described by Wilusz and colleagues required that the transcripts be capped; in contrast, capping of RNA substrates was not required for the TTP-dependent deadenylation to occur in the 293 cell assay system described here and in fact had minimal effect on deadenylation rates. Other methodological differences may explain some of the different behaviors of the two assays. However, development of the present assay finally allowed for the detection of the “TTP effect” in our hands.
Several types of mechanistic models are possible to explain this effect, including (i) direct physical linking of PARN to the ARE by TTP; (ii) indirect physical linking of these components through intermediary proteins; (iii) displacement of ARE- or poly(A) tail-protecting proteins by TTP, such as other ARE-binding proteins (36) and the poly(A) binding protein (3); and (iv) a TTP-induced change in RNA substrate conformation, including either a local conformational change or disruption of the stable circular conformation formed by the poly(A) tail-cap interaction. At present, we have no evidence for the first of these mechanisms, since TTP and PARN could not be cross-linked with DSS, despite high-level expression of both proteins, and there was no evidence that PARN could “supershift” the TTP-ARE complex in the gel shift assay. In addition, since capped and uncapped RNA probes responded similarly to TTP plus PARN, we found no evidence to support a TTP-induced disruption of the stable circular mRNA proposed to result from the poly(A) tail-cap interaction. Distinguishing among the remaining mechanisms will most likely require an assay containing pure, recombinant components.
It is important to distinguish the TTP-activated deadenylation assay described here from the phenomenon described in two recent studies of the exosomal degradation of the deadenylated mRNA body of ARE-containing mRNAs (9, 27). In the first study (9), the deadenylation of ARE-containing, polyadenylated mRNA substrates was only slightly decreased by prior immunodepletion of exosomal components from the extracts. TTP was found to be loosely associated with the exosome in immunoprecipitation experiments but was not part of the exosomal complex purified by TAP affinity purification. In addition, the exosomal degradation reaction of mRNA bodies described in those studies required ATP as a cofactor, although Ford et al. (12) found that ATP was not required for the deadenylation reaction in the same extracts; these authors also noted that both the deadenylation and mRNA body destruction reactions were inhibited by EDTA, supporting a role for divalent cations in both processes. The authors of these papers concluded that the further degradation of ARE-containing deadenylated mRNA bodies, but not deadenylation per se, occurred in this protein complex; in one case, TTP was found to be involved in the loose association of the ARE-containing mRNA body with the exosome. From these and other data, it seems likely that the Mg2+-dependent, ATP-independent, PARN- and TTP-stimulated deadenylation of ARE-containing mRNAs described here is a prelude to the association of the deadenylated mRNA bodies with the exosome. Nonetheless, as suggested by Chen et al. (9), it is possible that TTP and related proteins also play a role in the further exosomal degradation of ARE-containing deadenylated mRNA bodies.
Acknowledgments
We are grateful to Mike Wormington for the gift of the PARN antibody, to Khalid Khabar for communicating an unpublished paper, and to Dori Germolec and Trevor Archer for helpful comments on the manuscript.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.23.11.3798-3812.2003
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc155217?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
<i>AtC3H3</i>, an <i>Arabidopsis</i> Non-TZF Gene, Enhances Salt Tolerance by Increasing the Expression of Both ABA-Dependent and -Independent Stress-Responsive Genes.
Int J Mol Sci, 25(20):10943, 11 Oct 2024
Cited by: 0 articles | PMID: 39456724 | PMCID: PMC11507560
RNA-Binding Protein-Mediated mRNA Deadenylation in Mammalian Cell Extracts.
Methods Mol Biol, 2723:173-191, 01 Jan 2024
Cited by: 0 articles | PMID: 37824071
RNA localization during early development of the axolotl.
Front Cell Dev Biol, 11:1260795, 19 Oct 2023
Cited by: 0 articles | PMID: 37928901 | PMCID: PMC10620976
Synergistic roles of tristetraprolin family members in myeloid cells in the control of inflammation.
Life Sci Alliance, 7(1):e202302222, 30 Oct 2023
Cited by: 0 articles | PMID: 37903626 | PMCID: PMC10616675
The PARN, TOE1, and USB1 RNA deadenylases and their roles in non-coding RNA regulation.
J Biol Chem, 299(9):105139, 06 Aug 2023
Cited by: 2 articles | PMID: 37544646 | PMCID: PMC10493513
Review Free full text in Europe PMC
Go to all (168) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (3)
- (3 citations) ENA - X02611
- (2 citations) ENA - X03020
- (1 citation) ENA - X02333
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NM_002582.1
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Interactions of CCCH zinc finger proteins with mRNA: tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail.
J Biol Chem, 276(25):23144-23154, 28 Mar 2001
Cited by: 93 articles | PMID: 11279239
Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs.
J Biol Chem, 277(11):9606-9613, 08 Jan 2002
Cited by: 107 articles | PMID: 11782475
Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA.
J Biol Chem, 275(23):17827-17837, 01 Jun 2000
Cited by: 243 articles | PMID: 10751406
Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover.
Biochem Soc Trans, 30(pt 6):945-952, 01 Nov 2002
Cited by: 262 articles | PMID: 12440952
Review





