Abstract
Free full text

The PDZ Ligand Domain of the Human Papillomavirus Type 16 E6 Protein Is Required for E6's Induction of Epithelial Hyperplasia In Vivo
Abstract
Human papillomaviruses (HPVs) are the causative agent of warts. Infections with high-risk HPVs are associated with anogenital and head and neck cancers. One of the viral genes responsible for HPV's oncogenic activity is E6. Mice expressing the HPV-16 E6 protein in their epidermis (K14E6WT) develop epithelial hyperplasia and squamous carcinomas. Numerous cellular proteins interact with E6, some of which can be grouped based on common amino acid motifs in their E6-binding domains. One such group, the PDZ partners, including hDLG, hSCRIBBLE, MUPP1, and MAGI, bind to the carboxy-terminal four amino acids of E6 through their PDZ domains. E6's interaction with the PDZ partners leads to their degradation. Additionally, E6's binding to PDZ proteins has been correlated with its ability to transform baby rat kidney cells in tissue culture and to confer tumorigenicity onto cells in xenograft experiments. To address whether the ability of E6 to bind PDZ domain partners is necessary for E6 to confer epithelial hyperproliferation in vivo, we generated transgenic mice that express in stratified squamous epithelia a mutant of E6 lacking the last six amino acids at its carboxyl terminus, E6Δ146-151, from the human keratin 14 (K14) promoter. The K14E6Δ146-151 mice exhibit a radiation response similar to that of the K14E6WT mice, demonstrating that this protein, as predicted, retains an ability to inactivate p53. However, the K14E6Δ146-151 mice fail to display epithelial hyperplasia. These results indicate that an interaction of E6 with PDZ partners is necessary for its induction of epithelial hyperplasia.
Human papillomaviruses (HPVs) are the causative agent of warts. Infections with high-risk HPVs are associated with anogenital (27) and head and neck cancers (18, 25). HPVs encode two oncogenes, E6 and E7, which are thought to contribute to cervical cancers because they are selectively induced in their expression in those cancers. E6 and E7 are multifunctional proteins best known for their abilities to bind and inactivate the p53 and pRb cellular tumor suppressors, respectively. E6 binds in a ternary complex with p53 and a cellular ubiquitin ligase, E6AP (10). This complex results in ubiquitination and degradation of p53 through the proteosome pathway (11). Thus, E6 abrogates p53 function. However, E6's oncogenic activities cannot be explained entirely by its effects on p53. E6's ability to transform cells does not always correlate with its ability to degrade p53; mutants of E6 that cannot induce p53 degradation are still able to immortalize mammary epithelial cells (13, 19), transform 3Y1 rat fibroblasts, and confer tumorigenicity onto ψ2 mouse fibroblast cells (14). Conversely, some E6 mutations that retain the ability to induce p53 degradation are unable to transform cells (5, 13). Studies with K14E6WT mice also have identified p53-independent activities. Mice expressing the HPV type 16 (HPV-16) E6 protein in their epidermis (K14E6WT) develop epithelial hyperplasia (31). However, this phenotype has not been observed in p53-null mice of the same genetic background. These observations indicate that E6 must confer epithelial hyperplasia through mechanisms other than or in addition to its inactivation of p53.
Numerous cellular proteins have been found to interact with the E6 protein. Many interactors can be classified into groups due to the presence of a specific amino acid motif in their E6-binding region. One group is the α-helix partners, named for the α-helix domain in their E6-binding region (1, 4, 6, 32). In a previous study we investigated the role of the E6's interaction with the α-helix partners in epithelial hyperplasia. We found that expression of a mutant of E6, E6I128T, which was deficient in binding α-helical partners in the skin of transgenic animals, was still able to induce epithelial hyperplasia, albeit to a lesser extent than wild-type E6 (22). Thus, although α-helix partner binding is involved in induction of hyperproliferation, it is not solely responsible for it. Furthermore, the intermediate level of epithelial hyperplasia seen in the K14E6I128T strain was not rescued to the level seen in K14E6WT mice by placing the K14E6I128T transgene on the p53-null background. Therefore, inactivation of p53 does not contribute to E6's induction of epithelial hyperplasia.
These results led us to ask which E6 activities other than α-helix binding are contributing to E6's induction of epithelial hyperplasia. Another group of E6 interactors that may mediate epithelial hyperplasia contain a PDZ domain in their E6-binding region. PDZ domains are conserved protein elements named for the proteins PSD-95, Dlg, and ZO-1. HPV-16 E6 contains a protein motif in its carboxy terminus known to bind PDZ domains (PDZ ligand). E6's PDZ ligand motif, consisting of its carboxy-terminal four amino acids (ETQL), mediates E6's interaction with the PDZ domains of the human homologue of Drosophila discs large (hDlg) (14, 17), the human homologue of Drosophila Scribble (hScrib) (21), multi-PDZ-containing protein (MUPP1) (16), and membrane-associated guanylate kinase with an inverted arrangement of protein-protein interaction domains (MAGI-1) (7). The PDZ domain containing proteins shown to bind E6 will be referred to here as the PDZ partners of E6. E6's interaction with the PDZ partners leads to their degradation (7, 15, 16, 21). Additionally, E6's binding to PDZ proteins has been correlated with its ability to transform baby rat kidney cells in tissue culture and to confer tumorigenicity to cells in xenograft experiments (14). In Drosophila embryos, dlg (9, 24, 34) or scrib (2) mutants exhibit hyperplastic overgrowth and loss of cell polarity in epithelial tissues such as the imaginal discs. To address whether the ability of E6 to bind PDZ domain partners is necessary for E6 to confer epithelial hyperproliferation on transgenic mice, we generated transgenic mice that express a mutant of E6 lacking the six amino acids at the carboxy terminus, E6Δ146-151. Comparisons of the phenotypes of the mice expressing E6Δ146-151 and wild-type E6 demonstrate that the PDZ ligand domain is necessary for E6's induction of epithelial hyperplasia in vivo.
MATERIALS AND METHODS
Mice.
The generation of K14E6WT (29) and K14E6I128T (22) transgenic mice were previously described. The K14E6Δ146-151 mice were generated similarly to the K14E6WT mice and will be described in more detail elsewhere (M. M. Nguyen and A. E. Griep, unpublished data). Briefly, a recombinant copy of the HPV-16 E6 gene containing an 18-nucleotide deletion at the 3′ end of the gene was generated by PCR with the primers ML-4 (5′-CATCAAGAACATAATCATGCATGGA-3′) and ML-5 (5′-TCCATGCATGATTATGTTCTTGATG-3′), and the product was cloned into pK14 to generate pK14E6Δ146-151. Subsequently, PCR was used to amplify the E6Δ146-151E7TTL DNA fragment with the primers 709-1 (5′-GGCGGATCCTTTTATGCACCAAAAGAGAACTG-3′) and 709-4 (5′-CCCGGATCCTACCTGCAGGATCAGCCATG-3′) containing BamHI sites. This fragment was restricted with BamHI and inserted into the unique BamHIsite between the hK14 promoter and hK14 polyadenylation sequences in pG3Z-K14 (33) to generate pK14E6Δ146-151B. This recombinant plasmid was digested with HindIII and EcoRI to release a 3.2-kb DNA fragment containing the hK14 promoter, HPV-16 sequences, and the K14 polyadenylation sequence. The fragment was purified by gel electrophoresis and microinjected into fertilized mouse FVB/N eggs as described previously (29). Offspring were screened for the transgene by Southern analysis and PCR. Several K14E6Δ146-151 lines were propagated and housed as described for the K14E6WT mice (29).
In situ hybridization.
Nine-day-old mice were sacrificed, and skin and ear samples were collected. These were fixed in 10% buffered formalin, embedded in paraffin, and cut into 5-μm sections. In situ hybridization was performed as described earlier (31) by using an antisense E6 cRNA probe transcribed in the presence of [35S]UTP and [35S]CTP from a plasmid containing the HPV-16 E6 open reading frame upstream of the T7 promoter. Slides were coated with photographic emulsion and kept at 4°C for 2 weeks before development. The hybridization signal was examined by using dark-field microscopy.
Real-time PCR analysis for transgene expression.
Real-time PCR analysis was performed as described previously (22). Briefly, 5 μg of total cellular RNA extracted from the skin of 9-day-old animals was reverse transcribed by using the You-Prime-Ready-To-Go beads (Amersham Pharmacia; catalog no. 27-9264-01) and NotI-(dT)18 primer (Amersham Pharmacia; catalog no. 27-7806-01). Real-time PCR analysis was performed with the primers 5′-TGGAAGACCTGTTAATGGGCA-3′ and 5′-TGCAGGATCAGCCAATGGTAG-3′ and the Taqman probe 6-FAM-ACTAGGAATTGTGTGCCCCATCTGTTCTCAG-TAMARA. Amplification by using the Perkin-Elmer sequence detector 7700 was performed with the 2× Universal Amplification mix (Perkin-Elmer; catalog no. 4304437). The amplification profile was 45 cycles of 94°C for 15 s and 60°C for 1 min. The threshold (CT) values for serial (1/2, 1/4, and 1/8) dilutions of RNA from K14E6WT line 5737 were used to generate a linear standard curve. CT values of the samples were compared to this standard curve to calculate the relative levels of transgene expression. The signal obtained for undiluted RNA from the K14E6WT line 5737 was set to 100%, and the signal obtained for other mouse lines was reported as a fraction of that amount. RNA samples from three mice in each line were analyzed in parallel within each real-time PCR experiment.
Radiation response analysis.
Eight-day-old animals were irradiated with 4 Gy of gamma irradiation from a cesium source. After 23 h, irradiated mice and control, unirradiated mice were injected with 100 μg of 5′-bromo-2′-deoxyuridine (BrdU) and 83.75 μg of fluorodeoxyuridine/g (body weight). One hour later, mice were sacrificed, and ear and skin samples were collected, fixed in 10% formalin, paraffin embedded, and cut into 5-μm sections. BrdU was detected by using the BrdU immunohistochemistry kit (Oncogene HCS24) according to the manufacturer's directions with the following exceptions. The trypsin digestion was done with 2 drops of trypsin concentrate for every 3 drops of dilution buffer, the primary antibody incubation was for 3 h, and diaminobenzidine (DAB) incubation time was 3 min. The percentage BrdU-positive cells was calculated by dividing the percentage of BrdU-positive cells by the total number of cells in 10 ×40 microscopic frames per skin section. The levels of BrdU incorporation in the irradiated groups were normalized to the BrdU incorporation in the unirradiated mice of the same genotype. Analysis was performed on three mice per treatment group, except for the irradiated K14E6Δ146-151 group, in which two mice were analyzed. For p53 immunohistochemistry, deparaffinized sections were treated for 15 min with 3% hydrogen peroxide to quench endogenous peroxidase activity, heated for 20 min in boiling 0.01 mol of citrate buffer (pH 6.0) per liter by using a microwave to unmask antigens, blocked with 5% nonfat dry milk-phosphate-buffered saline-5% normal goat serum for 30 min, and incubated for 3 h at room temperature with a 1:500 dilution of the rabbit anti-mouse p53 antibody (CM5, catalog no. NCL-p53-CM5p; Novocastra Laboratories). After incubation with a secondary antibody (30 min) and then with Vectastain ABC reagents (30 min), the slides were exposed to DAB substrate.
Immunohistochemical analysis for BrdU, PCNA, and Dlg.
Nine-day-old or seven-week-old animals were injected with BrdU and fluorodeoxyuridine as described above 1 h prior to sacrifice. Skin and ear sections were prepared and stained for BrdU as described above. To quantify hyperproliferation, the total number of cells, as well as the number of BrdU-positive suprabasal cells, in 10 ×40 microscopic frames per section were counted. Suprabasal cells were defined as any cell not directly attached to the basement membrane. The percentage of suprabasal BrdU-positive cells was calculated by dividing the number of BrdU-positive suprabasal cells by the total number of suprabasal cells. Analysis was performed on at least three mice per genotype. For proliferating cell nuclear antigen (PCNA) and Dlg immunohistochemistry, tissue sections were blocked in 5% nonfat milk and 5% normal horse serum in PBS for 30 min. Tissue sections were exposed to (i) antibody raised against the rat homologue of Dlg, SAP97, at a 1:1,000 dilution overnight at 4°C or (ii) anti-PCNA antibody (Roche catalog no. 1486772) at a 1:200 dilution and for 1 h at room temperature in a humidified chamber. Slides were then incubated with secondary antibody for 30 min. For PCNA staining, tissue sections were exposed to Vectastain ABC reagent (Vector PK-6200) for 30 min and DAB substrate for 2 min. The tissue sections were counterstained with hematoxylin and dehydrated through a series of alcohols. Slides were then coverslipped by using Vectastain mounting medium.
Genotyping the K14E6I128T K14E6Δ146-151 double transgenic mice.
To distinguish between the K14E6I128T singly transgenic and the K14E6I128T K14E6Δ146-151 double transgenic mice, we made use of the MslI site introduced into the E6 gene by the E6Δ146-151 mutation. PCR was used to amplify the sequences encompassing the E6I128T E7TTL and E6Δ146-151 E7TTL regions of the transgenes. The amplified products were then digested with MslI and subjected to gel electrophoresis. Ethidium bromide was used to visualize the DNA fragments. The singly transgenic mice gave two DNA fragments of 760 and 62 bp. The doubly transgenic mice yielded four DNA fragments of 750, 395, 346, and 62 bp.
RESULTS
Generation of the K14E6Δ146-151 mice.
To examine the in vivo function of E6's PDZ ligand domain, mice were generated expressing a mutant version of the E6 protein referred to as E6Δ146-151 that is deleted of the C-terminal six amino acids. This deletion removes the PDZ ligand domain of E6 and renders it defective for binding hDlg, a PDZ domain partner of E6 (13). The E6Δ146-151 mutant gene was cloned into a K14 expression plasmid containing the human K14 promoter and polyadenylation sequences. The K14E6Δ146-151 recombinant DNA was injected into the male pronuclei of fertilized mouse eggs, and the injected eggs transplanted into pseudopregnant female mice. Transgene-positive offspring were identified by PCR and Southern analyses. Positive founders were bred to generate multiple independent lines of K14E6Δ146-151 mice. Transgene expression in each line was quantified by real-time PCR by using a Taqman probe specific for the E6 transgene. Representative results from these assays are displayed in Fig. Fig.1A.1A. Line 513 expressed its transgene at a level 5% that of the reference K14E6WT line 5737. One line of K14E6Δ146-151 mice (i.e., line 512) was found to express the transgene at a level equivalent to the reference K14E6WT line 5737. All other K14E6Δ146-151 lines tested displayed transgene expression at or below that of line 513. We chose K14E6Δ146-151 line 512 for further analysis because it expresses the mutant E6 gene at levels commensurate with that seen in for wild-type E6 in the reference K14E6WT line 5737.
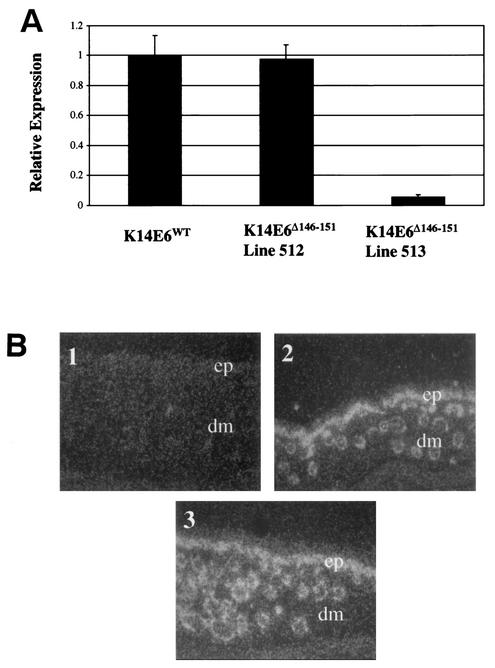
Analysis of transgene expression. (A) Quantification of transgene expression by real-time PCR. Total RNA from the skin of each transgene line was reverse transcribed into cDNA. The abundance of cDNA was then quantified by real-time PCR analysis with a FAM-labeled Taqman probe specific for the transgene. Shown are the relative transgene expression levels for K14E6WT and K14E6Δ146-151 mice. (B) In situ hybridization. Transgene mRNA was examined in skin sections from 9-day-old mice with an antisense probe. Dark-field images (magnification, ×20) of skin sections from nontransgenic (subpanel 1), K14E6WT (subpanel 2), and K14E6Δ146-151 (subpanel 3) animals are shown. The epidermal (ep) and dermal (dm) regions of the skin are indicated. The transgene hybridization signal is localized to the epidermis and hair follicles in the dermis.
The tissue localization of the transgene expression in K14E6Δ146-151 line 512 was examined by in situ hybridization in comparison to that seen in the reference K14E6WT line 5737. Skin sections were exposed for an equal time to a cRNA probe specific for the transgene transcript. Photomicrographs of in situ hybridizations performed in parallel on nontransgenic, K14E6Δ146-151 line 512 and and on K14E6WT line 5737 skin sections are displayed in Fig. Fig.1B.1B. The transgene expression was localized to the epidermis and epithelial appendages (hair follicles and sebaceaous glands) in the skin of both the K14E6WT and K14E6Δ146-151 mice, as expected for genes under the regulation of the K14 promoter. This result demonstrated that the K14E6Δ146-151 transgene in line 512 was expressed in an appropriate, tissue-specific manner. Furthermore, the transgene-specific in situ signal seen in the skin of K14E6Δ146-151 line 512 mice was similar to that seen in skin of K14E6WT line 5737 mice. This finding corroborated the more quantitative RNA analysis performed by real-time PCR (Fig. (Fig.1A1A).
E6Δ146-151 abrogates DNA damage response.
We have previously reported that the wild-type E6 can abrogate responses to DNA damage in vivo (29). This response is primarily due to E6's inactivation of p53 (22, 29). The E6Δ146-151 mutant has been shown to inactivate p53 in vitro. To determine whether expression of the K14E6Δ146-151 transgene inhibits the function of the p53 protein in vivo, we monitored the DNA damage response of the line 512 K14E6Δ146-151 mice. Mice were exposed to 4 Gy gamma radiation 24 h prior to sacrifice. 1 h prior to sacrifice, mice were injected with the nucleotide analog BrdU. Skin was then harvested, and paraffin-embedded histologic sections were stained immunohistochemically for BrdU. The percentages of BrdU-positive epidermal cells in unirradiated and irradiated animals were calculated. The level of BrdU incorporation in the unirradiated group was set to 1 for each genotype. The BrdU incorporation in the irradiated group is reported in Table Table11 as a fraction of the BrdU incorporation in the unirradiated group. The irradiated nontransgenic animals incorporated BrdU at a level 0.2 that of the unirradiated group, a finding consistent with prior studies that demonstrated an inhibition of DNA synthesis at 24 h postirradiation (29). The K14E6WT mice incorporated BrdU at a level 0.9 that of the unirradiated indicating, as previously shown (29), that E6 abrogates the normal DNA damage response in the skin. After irradiation, the K14E6Δ146-151 mice and p53-null animals incorporated BrdU at a level of about 0.7 that of unirradiated animals. This result is consistent with E6Δ146-151 inactivating p53 in vivo. When we performed immunohistochemistry on the skin from the irradiated K14E6Δ146-151 and K14E6WT mice, we failed to detect staining for the p53 protein (Fig. (Fig.2)2) or a protein induced by p53 in response to DNA damage, p21 (data not shown). These results further demonstrate the inactivation of p53 by E6Δ146-151. The observations that K14E6Δ146-151 mice display an altered DNA damage response and can inactivate p53 demonstrate that functional E6Δ146-151 protein is expressed in the epidermis of these transgenic mice.

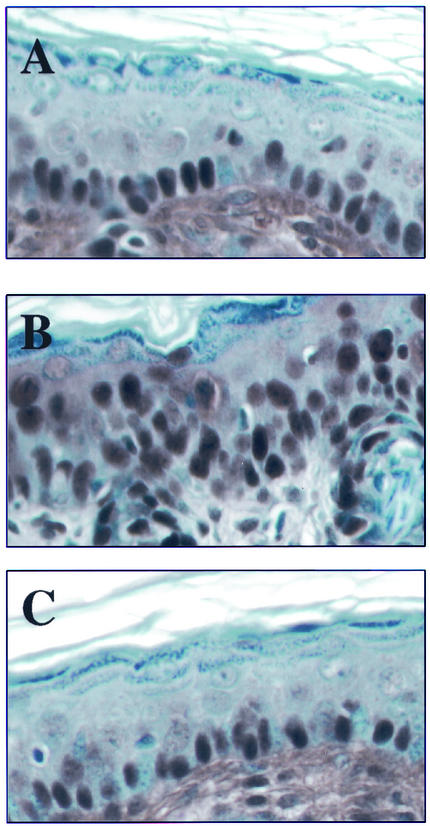
p53 induction in the mouse epidermis after irradiation. Shown are cross sections of skin from unirradiated K14E6Δ146-151 (A), irradiated nontransgenic (B), irradiated K14E6WT (C), and irradiated K14E6Δ146-151 (D) mice stained for p53. Note the frequent positively stained nuclei in the epidermis of the irradiated nontransgenic mouse compared to their absence in the epidermis of both the irradiated K14E6WT mice and the K14E6Δ146-151 mice. No p53 staining was detected in unirradiated mice of all genotypes (representative staining is shown only for unirradiated K14E6Δ146-151 mice [see panel A]).
TABLE 1.
DNA damage responses in the skin of wild-type or mutant HPV-16 E6 transgenic mice
| Mouse genotype | Mean fraction of BrdU-positive cells in irradiated epidermis ± SDa |
|---|---|
| Nontransgenic | 0.2 ± 0.2 |
| K14E6WT | 0.91 ± 0.2 |
| P53 null | 0.72 ± 0.3 |
| K14E6Δ146-151 | 0.7b |
Epidermal hyperproliferation is absent in the K14E6Δ146-151 mice.
The expression of wild-type E6 in the skin of mice induces epidermal hyperplasia, which leads to the overt thickening of the skin, which is most evident in the ears of the mice. We did not detect any overt thickening of the skin in any of the lines of K14E6Δ146-151 mice. To characterize the epithelial phenotype in the K14E6Δ146-151 mice further, we measured the proliferative indices in the epidermis of line 512 K14E6Δ146-151 mice. To accomplish this, 9-day-old and 7-week-old mice of the nontransgenic, K14E6WT (line 5737) and K14E6Δ146-151 (line 512) mouse lines were injected with BrdU 1 h prior to sacrifice. Paraffin-embedded histologic sections from the ears of these mice were subjected to immunohistochemistry specific for BrdU. Expression of wild-type E6 led to BrdU incorporation in the suprabasal layers of the epidermis in K14E6WT mice, a compartment of the epidermis that normally does not efficiently support DNA synthesis (Fig. (Fig.3B).3B). This unscheduled DNA synthesis, which is a hallmark of epithelial hyperplasia in the K14E6WT mice, was not evident in the epidermis of K14E6Δ146-151 (Fig. (Fig.3C)3C) or nontransgenic (Fig. (Fig.3A)3A) mice. The percentage of BrdU-positive suprabasal cells was calculated for 9-day-old and 7-week-old K14E6WT, K14E6Δ146-151, and nontransgenic animals (Table (Table2).2). There was no increase in the percentage of suprabasal cells staining positive for BrdU in the skin of K14E6Δ146-151 transgenic mice compared to that in nontransgenic animals at either time point.
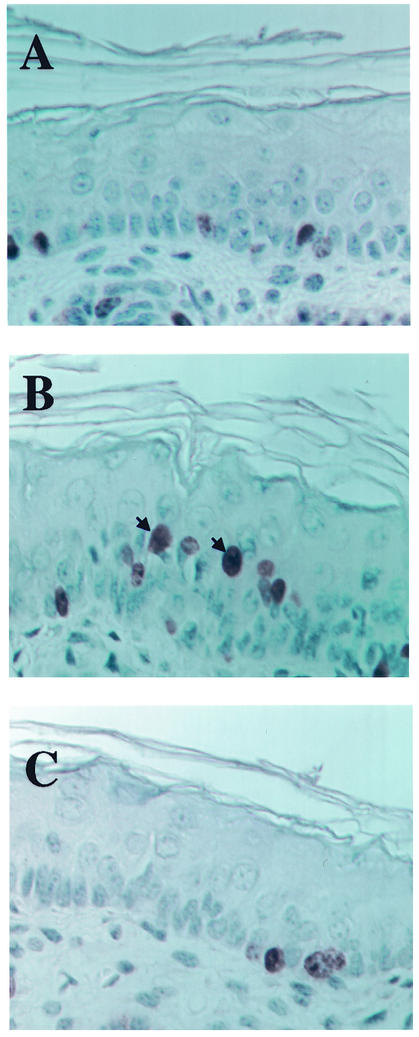
Hyperproliferation in the skin of nontransgenic (A), K14E6WT (B), and K14E6Δ146-151 (C) mice. Shown are cross sections of skin from the ear of 9-day-old nontransgenic, K14E6WT, and K14E6Δ146-151 mice stained immunohistochemically for BrdU incorporation. BrdU was injected (100 μg/g [body weight]) 1 h prior to sacrifice. Examples of BrdU-positive suprabasal cells are denoted by arrows.
TABLE 2.
Epithelial hyperplasia in wild-type or mutant HPV-16 E6 transgenic mice
| Mouse genotypea | % Suprabasal BrdU-positive cellsb in:
| |
|---|---|---|
| 9-day-old mice | 7-wk-old mice | |
| Nontransgenic | 1.7 ± 0.5 | 0.1 ± 0.3 |
| K14E6WT | 4.3 ± 1.4 | 6.8 ± 2.0 |
| K14E6Δ146-151 | 2.0 ± 0.7 | 0.0 ± 0.0 |
| K14E61128T | 2.9 ± 1.1c | 2.0 ± 1.4 |
| K14E61128T/K14E6Δ146-151 | ND | 1.9 ± 0.8 |
PCNA immunostaining was used as an independent means of monitoring epidermal hyperplasia. PCNA is a component of the DNA synthesis machinery. The PCNA antibody used in our analyses is specific for an epitope exposed only in proliferating cells. Ear cross sections prepared as described above were stained for PCNA by immunohistochemistry. Photomicrographs of this staining are presented in Fig. Fig.4.4. PCNA staining was evident in the basal, parabasal, and supraparabasal layers of the K14E6WT ear (Fig. (Fig.4B).4B). PCNA staining was seen in the basal and parabasal layers but not in the supraparabasal layers of the nontransgenic (Fig. (Fig.4A)4A) and K14E6Δ146-151 (Fig. (Fig.4C)4C) mice. This result, along with the BrdU staining results, indicates that the K14E6Δ146-151 mice lack the epidermal hyperproliferation phenotype evident in the reference K14E6WT line 5737.
K14E6Δ146-151 K14E6I128T double-transgenic mice.
In a prior study we generated and analyzed transgenic mice in which we expressed in the mouse epidermis a mutant of E6, E6I128T, that is deficient in binding α-helix binding partners of E6. Mice expressing E6I128T displayed a reduced level of epithelial hyperplasia compared to that seen in mice expressing wild-type E6 in the epidermis (22). This finding indicated that E6's interactions with one or more of its α-helix binding partners contribute to the induction of epithelial hyperplasia. Because the E6I128T mutant protein retains the PDZ ligand domain, it is predicted to retain an ability to interact with PDZ binding partners of E6. Conversely, we have shown in vivo (Table (Table11 and Fig. Fig.2)2) and others have shown in vitro (13) that the E6Δ146-151 mutant protein must be able to bind at least one α-helix binding partner, E6AP, since it retains an ability to functionally inactivate p53. Thus, two E6 mutant proteins, which appear to be defective in binding distinct subsets of cellular proteins, show reduced ability or no ability to induce epithelial hyperplasia. This led us to question whether E6 induces epithelial hyperplasia through a coupled interaction between α-helix and PDZ partners. Functional interactions between α-helix and PDZ partners of E6 have been proposed. E6 induces the degradation of hScrib, a PDZ domain partner, via E6AP, an α-helix partner (21). Moreover, E6-TP1, another α-helix partner, interacts with hDLG, another PDZ domain partner, although it is not known whether this interaction is affected by E6 (23). An alternative explanation for the fact that E6I128T and E6Δ146-151 proteins are reduced or absent in their ability to induce epithelial hyperplasia is that E6 works to alter epithelial growth through two (or more) independent pathways that are not functionally coupled. To distinguish between these possibilities, we sought to determine whether E6Δ146-151 and E6I128T can complement each other to induce wild-type levels of epithelial hyperplasia. Evidence for complementation would argue that the partner interactions affected by each mutation contribute to independent (uncoupled) pathways. A lack of complementation would be consistent with E6 inducing epithelial hyperplasia through some coupled interaction between α-helix and PDZ domain protein partners. Lines of K14E6I128T (line 6061) and K14E6Δ146-151 (line 512) mice expressing their transgenes at matched levels to that in the reference K14E6WT line (line 5737) were bred and tissues harvested from 7-week-old single- or double-transgenic offspring after exposure for 1 h to BrdU (injected intraperitoneally). Histologic sections from ears were subjected to immunohistochemical staining for BrdU. The percentages of BrdU-positive cells observed in the suprabasal layers of the ears are presented in Table Table2.2. The presence of the K14E6Δ146-151 transgene did not affect the level of proliferation in the K14E6I128T mice. The lack of complementation is consistent with E6 inducing epithelial hyperplasia through a coupled interaction between α-helix and PDZ domain protein partners.
DISCUSSION
K14E6Δ146-151 mice lack epithelial hyperplasia.
Our results demonstrate that the PDZ ligand domain of E6 is necessary for the induction of epithelial hyperplasia. Induction of epithelial hyperplasia is an activity of E6 that is likely to contribute to its role in tumorigenesis in vivo (30); therefore, we predict that the K14E6Δ146-151 mice will display reduced tumorigenic phenotypes compared to that of K14E6WT mice. This is in agreement with previous studies in which E6 mutants deficient in binding PDZ partners, unlike wild-type E6, were unable to transform baby rat kidney cells in tissue culture or confer tumorigenicity onto cells in xenograft experiments (14). Long-term tumorigenesis studies, now initiated, comparing the tumorigenic phenotypes of K14E6Δ146-151 mice to those of K14E6WT mice, will allow us to define that contribution. That E6Δ146-151 is capable of immortalizing mammary epithelial cells (13) indicates that PDZ partner binding is not required for this activity and that the activities of E6 required for in vitro immortalization and in vivo hyperplasia are distinct.
Our results indicate that E6 requires interactions with one or more of the PDZ partners to induce epithelial hyperplasia. Evidence from Drosophila studies links two of these PDZ partners, Dlg and Scrib, to epithelial growth regulation. In Drosophila embryos, dlg mutants exhibit hyperplastic overgrowth and loss of cell polarity in epithelial tissues such as the imaginal discs (9, 24, 34). Therefore, E6's degradation of Dlg could potentially mediate the epithelial hyperplasia of the K14E6WT mice. Caruna et al. have identified mice that contain a gene trap insertional mutation in mouse Dlg (3). Mice homozygous for this mutant allele are born with severe cleft palates and die at or close to birth, preventing us from scoring the influence of this Dlg mutation in neonates of the same age at which we detect epithelial hyperplasia in the K14E6WT mice (day 6 or later). We failed to detect epidermal hyperplasia in the skin of mice homozygous for the mutant Dlg allele that were sacrificed at embryonic day 19.5 or as neonates that were less then 24 h old (M. L. Nguyen, M. M. Caruna, A. Bernstein, and P. F. Lambert, unpublished results). The significance of this observation is further limited by the suspicion that this gene trap insertional mutant does not represent a true null allele, a concern raised by the fact that mice engineered by conventional homologous recombination approaches to bear null alleles of Dlg die as embryos (P. Bryant, unpublished data), a finding similar to that seen in Drosophila melanogaster (9, 24). Therefore, a model in which Dlg is specifically inactivated in the epidermis would need to be generated in order to determine whether a lack of Dlg leads to epithelial hyperplasia in mice.
E6 may alter Dlg's activity through means other than inducing its degradation. Dlg binds to the PDZ ligand domain of the cellular tumor suppressor and regulator of the canonical Wnt pathway, adenomatous polyplosis coli (APC) (20). A peptide corresponding to the carboxyl terminus of E6 has been shown to disrupt the APC-Dlg interaction (14). This suggests that the E6 protein may be able to disrupt the Dlg-APC complex. However, what effect this disruption would have on epithelial growth and carcinogenesis is unclear. Overexpression of Dlg abrogates an APC induced cell cycle block in epithelial cells (12), suggesting that Dlg regulates APC's activity in controlling cell growth. However, mice containing a mutant of APC (APC1638T) lacking the Dlg-binding regions did not develop epithelial hyperplasia (M. L. Nguyen, R. Fodde, and P. F. Lambert, unpublished results) or spontaneous tumors (28). Further investigation is needed to determine whether E6's interaction with Dlg is responsible for the induction of epithelial hyperplasia in mice.
E6's interaction with one of the other PDZ partners, Scrib, MAGI, or MUPP1 may also mediate epithelial hyperplasia. Similarly to dlg, Drosophila mutations in scrib lead hyperplastic overgrowth and loss of cell polarity in epithelial tissues (2). Mice with similar mutations in scrib have not been generated; thus, it is not known at this time whether disruption of scrib would lead to comparable epithelial phenotypes in mice. Thus far, there is no direct evidence pertaining to the role of MAGI-1 or MUPP-1 in the control of epithelial cell growth.
E6Δ146-151 and E6I128T cannot complement each other to induce wild-type levels of epithelial hyperplasia.
We have shown previously that the α-helix partner binding of E6 contributes to epithelial hyperplasia. In the present study we found that the PDZ partner binding of E6 is necessary for epithelial hyperplasia. Thus, both α-helix and PDZ partner binding play a role in inducing epithelial hyperplasia. When we bred the K14E6I128T mice to the K14E6Δ146-151 mice, the double-transgenic mice did not display the level of epithelial hyperplasia of mice expressing the wild-type E6 (Table (Table2).2). This lack of complementation is consistent with E6 inducing epithelial hyperplasia through some coupled interaction between α-helix and PDZ domain protein partners. One way in which α-helix partners and PDZ partners have been shown to be coupled is through E6AP. The α-helix partner, E6AP, has been shown to mediate the degradation of the PDZ partner hScrib (21), raising the hypothesis that E6 needs not only to bind PDZ partners (mediated by the PDZ ligand domain of E6) but also to induce their degradation (mediated by E6's interaction with E6AP) in order to induce the level of epithelial hyperplasia observed in the K14E6WT mice. However, we have evidence that the ability of E6 to induce epithelial hyperplasia is not altered on an E6AP-null background (M. L. Nguyen and P. F. Lambert, unpublished data). The possibility of another link between α-helix and PDZ partners is suggested by the recent finding that the rat homologue of the α-helix partner, E6TP1, has been shown to bind to the PDZ partner Dlg (23). It is not known what influence E6's interaction with E6TP1 and/or Dlg has on the association of E6TP1 with Dlg. An alternative explanation for the lack of complementation between the K14E6I128T and K14E6Δ146-151 mice is that there is defect common to E6I128T and E6Δ146-151 mutants that is not restored in the double-transgenic animals. Although we cannot formally rule out this possibility, no known functions of E6 are known or predicted to be absent in both the E6I128T and E6Δ146-151 mutants.
We have found that the PDZ ligand domain of HPV-16 E6 is required for the induction of epithelial hyperplasia in mice. Two other viruses, human T-cell leukemia virus (26) and adenovirus 9 (17), encode oncogenes that bind PDZ domain-containing proteins. Interestingly, the PDZ ligand domain of adenovirus 9 oncoprotein, E4 ORF-1, has recently been found to be important for its oncogenic activities (8). These results and ours indicate that interactions between viral proteins and PDZ domain-containing proteins constitute a general mechanism for virus-induced oncogenesis.
Acknowledgments
We are grateful to Norman Drinkwater and Bill Sugden for critical reading of the manuscript. We thank Kathleen Helmuth and the University of Wisconsin Transgenic Animal Facility for generating the transgenic mice, Jane Weeks and Harlene Edwards of the McArdle Laboratory and the University of Wisconsin Comprehensive Cancer Center Histology Core for processing tissue sections, and Megan Poehler for help with immunohistochemistry. We are grateful to Johannes Hell for kindly providing SAP97/Dlg antisera used in the present study, to Georgina Caruna and Alan Bernstein for the Dlg mutant mice, and to Riccardo Fodde for the APC1638T mice.
This study was supported by grants from the National Institute of Health (CA22443, CA09135, and CA07175) and by the American Cancer Society.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.77.12.6957-6964.2003
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc156174?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.77.12.6957-6964.2003
Article citations
Comparative Analysis of Alpha and Beta HPV E6 Oncoproteins: Insights into Functional Distinctions and Divergent Mechanisms of Pathogenesis.
Viruses, 15(11):2253, 14 Nov 2023
Cited by: 0 articles | PMID: 38005929 | PMCID: PMC10674601
Review Free full text in Europe PMC
MmuPV1 E6 induces cell proliferation and other hallmarks of cancer.
mBio, 14(6):e0245823, 31 Oct 2023
Cited by: 1 article | PMID: 37905801 | PMCID: PMC10746199
Additive energetic contributions of multiple peptide positions determine the relative promiscuity of viral and human sequences for PDZ domain targets.
Protein Sci, 32(4):e4611, 01 Apr 2023
Cited by: 2 articles | PMID: 36851847 | PMCID: PMC10022582
Transcriptomic analysis provides insight into the mechanism of IKKβ-mediated suppression of HPV18E6-induced cellular abnormalities.
G3 (Bethesda), 13(4):jkad020, 01 Apr 2023
Cited by: 0 articles | PMID: 36722216 | PMCID: PMC10085804
HPV16 E6 induces chromosomal instability due to polar chromosomes caused by E6AP-dependent degradation of the mitotic kinesin CENP-E.
Proc Natl Acad Sci U S A, 120(14):e2216700120, 29 Mar 2023
Cited by: 7 articles | PMID: 36989302 | PMCID: PMC10083562
Go to all (136) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Two distinct activities contribute to human papillomavirus 16 E6's oncogenic potential.
Cancer Res, 65(18):8266-8273, 01 Sep 2005
Cited by: 47 articles | PMID: 16166303
The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals.
J Virol, 73(7):5887-5893, 01 Jul 1999
Cited by: 140 articles | PMID: 10364340 | PMCID: PMC112649
Role of the PDZ domain-binding motif of the oncoprotein E6 in the pathogenesis of human papillomavirus type 31.
J Virol, 78(22):12366-12377, 01 Nov 2004
Cited by: 96 articles | PMID: 15507623 | PMCID: PMC525055
Structure and function of the papillomavirus E6 protein and its interacting proteins.
Front Biosci, 13:121-134, 01 Jan 2008
Cited by: 20 articles | PMID: 17981532
Review
Funding
Funders who supported this work.
NCI NIH HHS (5)
Grant ID: CA22443
Grant ID: T32 CA009135
Grant ID: CA07175
Grant ID: CA09135
Grant ID: P01 CA022443