Abstract
Free full text

Hot-spot mutants of p53 core domain evince characteristic local structural changes
Abstract
Most of the oncogenic mutations in the tumor suppressor p53 map to its DNA-binding (core) domain. It is thus a potential target in cancer therapy for rescue by drugs. To begin to understand how mutation inactivates p53 and hence to provide a structural basis for drug design, we have compared structures of wild-type and mutant p53 core domains in solution by NMR spectroscopy. Structural changes introduced by five hot-spot mutations (V143A, G245S, R248Q, R249S, and R273H) were monitored by chemical-shift changes. Only localized changes are observed for G245S, R248Q, R249S, and R273H, suggesting that the overall tertiary folds of these mutant proteins are similar to that of wild type. Structural changes in R273H are found mainly in the loop–sheet–helix motif and the loop L3 of the core domain. Mutations in L3 (G245S, R248Q, and R249S) introduce structural changes in the loop L2 and L3 as well as terminal residues of strands 4, 9, and 10. It is noteworthy that R248Q, which is often regarded as a contact mutant that affects only interactions with DNA, introduces structural changes as extensive as the other loop L3 mutations (G245S and R249S). These changes suggest that R248Q is also a structural mutant that perturbs the structure of loop L2–L3 regions of the p53 core domain. In contrast to other mutants, replacement of the core residue valine 143 to alanine causes chemical-shift changes in almost all residues in the β-sandwich and the DNA-binding surface. Long-range effects of V143A mutation may affect the specificity of DNA binding.
The tumor suppressor p53 responds to DNA damage by activating the transcription of regulatory genes that arrest cell growth until the damage is repaired and by activating additional factors that induce apoptosis. When p53 is inactivated, most commonly by a single missense mutation, cell growth can proceed without regulation, thus causing tumor growth; this fact accounts for the observation that mutations in p53 are found in over half of all human cancers.
p53 is a transcription factor of 393 amino acids divided into three major regions: the 200-residue DNA-binding core domain that folds into a common immunoglobulin motif with a DNA-binding surface formed by two β-turn loops (L2 and L3) and a loop–sheet–helix motif (1), the relatively unstructured transactivation domain located N-terminally to the core and the C-terminal tetramerization domain (2–5). The core domain of p53 is the most highly conserved between species and confers DNA-binding specificity to the protein. Substitutions in the core domain make up 98% of the transforming mutations in p53, and 40% of these involve just six “hot spots” near the DNA-binding surface (6).
p53 is a potential target for cancer therapy by drugs designed to bind to a mutant and restore its stability or wild-type conformation. Characterization at the molecular level of how individual mutations inactivate p53 may help in the design of such drugs. Structural information about p53 mutants is scarce and mainly inferred from gross studies with monoclonal antibodies. Here, we monitored structural changes of five of the most common mutants of p53: V143A, G245S, R248Q, R249S, and R273H.
MATERIALS AND METHODS
Sample Preparation.
The core domain of human p53 encoding residues 94–312 was subcloned into pRSETA (Invitrogen) as described (7). The vector was transformed into Escherichia coli strain C41 (8) for overexpression. To produce 80% 2H, uniformly 13C,15N-labeled p53 core domain, 750 ml of l-broth with 50 μg/ml ampicillin was grown at 37°C until A600 reached 0.8, at which point the cells were collected by centrifugation (7 min at 8,000 × g). The cells were resuspended in 1.5 liters of Bio-Express-1000 medium (Cambridge Isotope Laboratories, Cambridge, MA) and grown at 28°C. When A600 reached 0.8, zinc sulfate was added to give a final concentration of 100 μM, and protein expression was induced with 200 μM of isopropyl β-d-thiogalactoside. The cells were grown for a further 16 h and then harvested by centrifugation (7 min at 8,000 × g). The cells were lysed by sonication in 20 mM sodium phosphate buffer (pH 7.2; with 10 mM β-mercaptoethanol/1 mM phenylmethylsulfonyl fluoride). The extract was centrifuged, and the resulting supernatant was loaded onto a 5-ml SP-HiTrap ion exchange column (Amersham Pharmacia) that was preequilibrated with 20 ml of sonication buffer and washed with 0.1 M NaCl in the same buffer. p53 was eluted in 20 mM sodium phosphate buffer (pH 7.2; with 10 mM β-mercaptoethanol) by using a 0.1–0.3 M NaCl gradient over 60 ml. Eluted protein was immediately diluted to avoid aggregation and dialyzed against 50 mM sodium phosphate buffer (150 mM NaCl/2.5 mM DTT). The sample was concentrated to 500 μM, and 2H2O was added to 5% (vol/vol) concentration, making the final NMR sample in 150 mM NaCl/2.5 mM DTT/50 mM sodium phosphate buffer, pH 7.2.
To produce 15N-labeled wild-type and the mutant p53 core domains, a 500-ml starter culture of l-broth (50 μg/ml ampicillin) was grown at 37°C until A600 reached 0.8. The cells were collected by centrifugation and resuspended in 1 liter of M9 minimal medium with 1 g/liter [15N]H4Cl and 4 g/liter glycerol. The culture was allowed to grow at 25°C until A600 reached 0.8, at which point protein expression was induced with 100 μM of isopropyl β-d-thiogalactoside. The cells were grown for another 12 h and then harvested by centrifugation. The cells were lysed in 200 ml of 20 mM Tris buffer (pH 7.2; 0.5 mM phenylmethylsulfonyl fluoride/5 mM DTT) by a EmulsiFelx-C5 high pressure homogenizer (Avestin, Ottawa). The cell debris was removed by centrifugation, and the resulting supernatant was loaded onto a SP-Sepharose ion exchange column (Amersham Pharmacia) that was preequilibrated with 100 ml of 0.1 M NaCl in 20 mM Tris buffer (pH 7.2; 5 mM DTT). The p53 core domain was eluted with a 0.1–0.3 M NaCl gradient over 240 ml. Eluted protein was diluted three times and loaded onto a 5-ml Hi-Trap heparin affinity column (Amersham Pharmacia) preequilibrated with 100 ml of 20 mM Tris buffer (pH 7.2; 5 mM DTT). p53 core domain was eluted with a 0.1–0.3 M NaCl gradient over 240 ml. Eluted protein was concentrated to 7 ml and then loaded onto an Amersham Pharmacia Superdex 75 HR26/60 column equilibrated with 25 mM sodium phosphate buffer (150 mM KCl/10 μM chloramphenicol/5% (vol/vol) 2H2O, pH 7.1). The sample was concentrated to 500 μM, making the final NMR sample in 150 mM KCl/5 mM DTT/5% (vol/vol) 2H2O/25 mM sodium phosphate buffer, pH 7.1.
NMR Spectroscopy.
1H–15N heteronuclear single quantum coherence (HSQC) and Nuclear Overhauser enhancement spectroscopy–HSQC spectra for wild-type p53 core domain were acquired on a Bruker DRX-800 spectrometer equipped with a 5-mm inverse detection triple resonance probe with x, y, z gradient coils. All other NMR experiments were acquired on a Bruker DRX-600 spectrometer equipped with a 5-mm inverse detection triple resonance probe with x, y, z gradient coils. Three pairs of triple resonance experiments were acquired at 298 K on an 80% 2H, uniformly 13C,15N-labeled sample to yield sequential connectivities via Cα [HNCA, HN(CO)CA], Cβ [HN(CA)CB, HN(COCA)CB], and CO [HNCO, HN(CA)CO] resonances (9, 10). 1H–15N HSQC spectra for wild-type and mutant p53 core domain were acquired at 293 K. In all experiments, selective flip-back pulses were applied to minimize attenuation of signals caused by solvent exchange (11). Suppression of the residual water signal was achieved by WATERGATE (water suppression by gradient-tailored excitation; see ref. 12).
RESULTS
p53 Core Domain in Solution Has Similar Secondary Structure to That in the Crystal Structure.
Backbone 1H, 13C, and 15N resonances were assigned by using triple resonance experiments. Short-ranging nuclear Overhauser enhancements were used to confirm the assignment and to resolve any ambiguity when necessary. Of 200 backbone amide signals, 176 were assigned (Fig. (Fig.1).1). Deviations of 13C Cα (ΔCα) and Cβ (ΔCβ) chemical shifts from random-coil values are good indicators for identification of secondary structure elements (13), with residues in helices and strands having positive and negative values of ΔCα − ΔCβ, respectively. Fig. Fig.22 illustrates that the trend of ΔCα − ΔCβ agrees well with secondary structure elements as seen in the crystal structure, suggesting the structure of p53 core domain in solution is similar to that in crystal structure (1). Residues near the C terminus (294–312) have random-coil chemical shifts, suggesting that the C-terminal tail is unstructured. This finding is consistent with the lack of electron density observed for this region in the crystal structure.
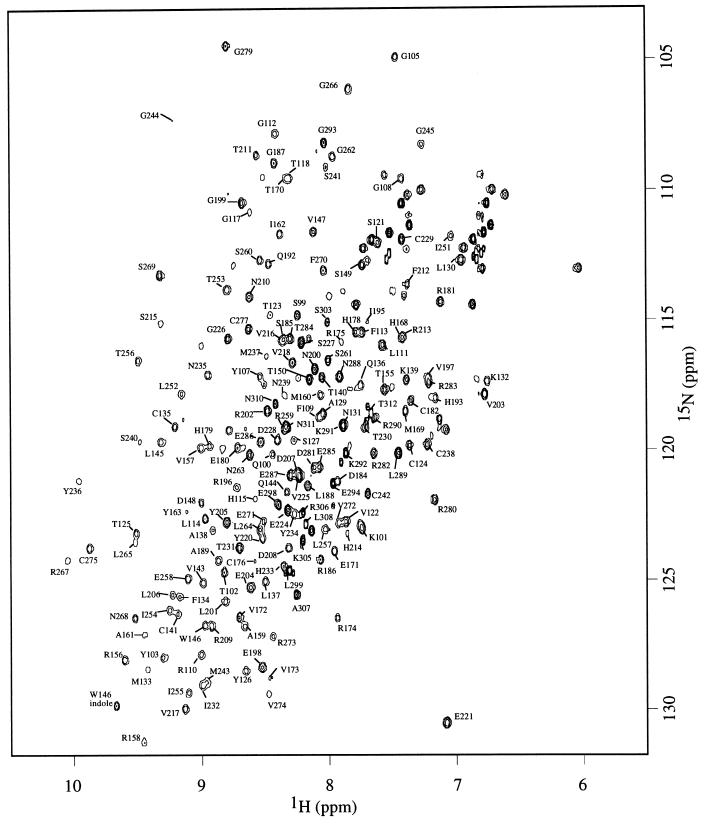
1H–15N HSQC spectra of wild-type p53 core domain (residues 94–312) acquired at 293 K. Of 200 (excluding proline and N terminus) backbone amide signals, 176 were assigned. Amide signal from the side-chain indole group of Trp-146 is indicated.
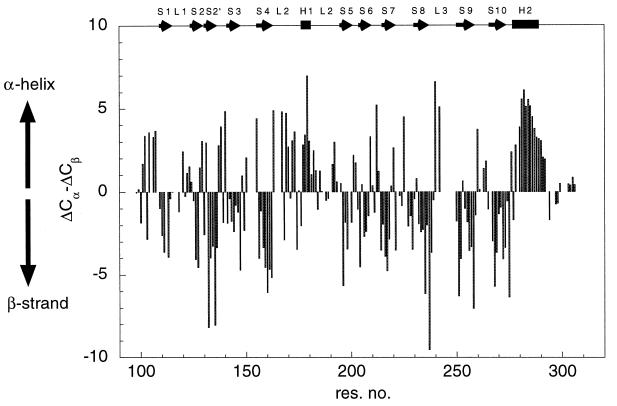
Secondary structure of p53 core domain in solution. Deviation of ΔCα and ΔCβ shifts from random-coil values are plotted against sequence position. Random-coil chemical shifts for Cα and Cβ resonances were obtained from ref. 22. Positive values of ΔCα − ΔCβ suggest helical structure, whereas negative values of ΔCα − ΔCβ are indicative of strands. Helices and strands, according to the definition in the crystal structure, are shown as boxes and arrows at the top of the figure.
Structural Changes Caused by Mutation of p53 Core Domain.
1H–15N HSQC spectra for five mutants (R273H, R248Q, R249S, G245S, and V143A) of the p53 core domain were acquired to monitor structural changes introduced by the mutations. Backbone 1H,15N resonances of the mutant were assigned by comparison of the mutant 1H–15N HSQC spectra to that of the wild-type. Residues that have similar 1H and 15N chemical shifts in the wild-type and the mutant spectra are color-coded green in Fig. Fig.33B. Most of the residues in the G245S, R248Q, R249S, and R273H mutants fall into the later category. Because chemical shifts are highly sensitive to structural changes in the environment, these mutants should adopt overall fold similar to wild-type. Chemical-shift changes (color-coded red in Fig. Fig.33B) for these four mutants are found predominantly near the site of mutation.

(A) Stereo diagram of ribbon representation of wild-type p53 core domain. Secondary structure elements loops L1, L2, L3, helix H2, and strands S6 and S7 are indicated in the diagram. Residues involved in DNA binding in the crystal structure are colored light blue to illustrate the DNA-binding regions of the p53 core domain. (B) Structural changes for hot-spot mutants of the p53 core domain. Five mutants, R273H, R249S, G245S, R248Q, and V143A, were studied. 1H–15N correlation spectra of the five mutants were compared with the wild-type spectrum. Residues with no change in amide chemical shifts are colored green. Those with changes in amide chemical shifts (>0.05 ppm for 1H or >0.25 ppm for 15N) are colored red. Residues are colored grey when quantitative comparison cannot be made (e.g., overlapping in the spectra).
R273H.
Chemical-shift changes are mainly found in the loop–sheet–helix motif and the L3 loop of core domain (Fig. (Fig.33B). Introduction of an aromatic residue perturbs the local chemical environment, which results in chemical-shift changes in surrounding residues. In addition, the guanidinium group of Arg-273 is salt-bridged to the carboxylate group of D281 in helix 2. The R273H mutation breaks this interaction, and structural changes in helix-2 were reflected in the chemical-shift changes for residues 281, 282, 285, and 286 (Fig. (Fig.33B).
G245S, R248Q, and R249S.
The three mutations located in L3 introduced chemical-shift changes for residues in L2 and L3 as well as terminal residues of strands 4, 9, and 10 (Fig. (Fig.33B). L3 is responsible for interactions with the minor groove of DNA (1). Both loops L2 and L3 lack regular secondary structure. They are stabilized by a zinc coordination and side-chain interactions. Both G245S and R249S are classified as “structural mutants.” The guanidinium group of Arg-249 is salt-bridged to the carboxylate group of Glu-171 in L2 and backbone oxygens of Gly-245 and Met-246 (1). Replacement of serine at this position removes these interactions, which are important in stabilizing the conformation of both loops. Gly-245 in the wild type adopts unusual backbone dihedral angles (1), which are not favored when the residue is replaced by serine. Extensive chemical-shift changes found for G245S and R249S in regions near L2 and L3 suggest disruption of conformation that is important to DNA binding.
In the crystal structure of p53 core domain complexed with DNA, Arg-248 makes contacts to the minor groove of DNA (1). R248Q was, therefore, referred as “contact mutant.” However, our data suggest that the R248Q is, in fact, both a contact mutant and a structural mutant. R248Q and R249S have similar thermodynamic stability and are both ≈2 kcal/mol less stable than the wild type (7). However, based on the crystal structure, it is not obvious how Arg-248 contributes to the thermodynamic stability of the p53 core domain. It is intriguing to find that pattern of chemical-shift changes for R248Q are as extensive as those for R249S. This finding suggests that the R248Q mutation also perturbs the structure of L2 and L3. Chemical-shifts changes were found for residues in helix 2, loop L1, and strand 2 and 2′, which are distant from the site of mutation. This finding suggests that Arg-248 must play a role in stabilizing the structure in the L2–L3 loop regions.
V143A.
In contrast to other mutants studied, extensive chemical changes are found for almost all residues in the β-sandwich (Fig. (Fig.33B). The side-chain of Val-143 is buried deep inside the hydrophobic core of the β-sandwich. The extensive range of residues affected is a result of a rearrangement of core residues to fill the cavity created by the V143A mutation.
DISCUSSION
Hot-Spot Mutants Have Characteristic Local Structural Changes.
The structure of p53 mutants has mainly been inferred from interaction with monoclonal antibodies (14–17). The monoclonal antibody PAb240 binds to some p53 mutants but not the wild type (15). It was suggested that these mutants adopt a “mutant conformation” that is distinctly different from wild-type conformation (18). The present study of mutants by NMR spectroscopy provides direct structural evidence on the effects of mutations in the p53 core domain. Our NMR data suggest that, at 20°C, R249S and G245S (both PAb240+) fold into structures in a manner similar to the wild type. Structural changes are mapped mainly to loop L2 and L3 (Fig. (Fig.33B).
The crystal structure of p53 core domain shows that the epitope of PAb240, located in strand 7, is inaccessible to antibody binding (1), and at least a partly unfolded form is required to expose the epitope. PAb240 was shown to bind denatured wild-type or mutant p53 (15). However, whether PAb240 could bind to a partly unfolded form is still under active debate. It has been proposed that global unfolding may not always be necessary; a conformational exchange between folded and partly folded structures might be sufficient to expose the epitope to the antibody (18). Studies of backbone dynamics by 15N relaxation measurements (K.-B.W., S.M.V.F., B.S.D., and A.R.F., unpublished data) showed that residues in strands 6 and 7 exhibit chemical exchange processes on the microsecond–millisecond time scale, suggesting intrinsic flexibility in this region. It is conceivable that this flexibility may be enhanced when the L2, which is packed against strand 7, is destabilized by mutation (e.g., G245S/R249S). Work comparing changes in dynamics of wild type and hot-spot mutants of the p53 core domain may prove informative.
Mutation V143A Introduces Long-Range Effects That Might Affect the Specificity of DNA Binding.
The biological activity of V143A is sensitive to temperature. DNA binding and transcriptional activity of V143A are observed at 32°C, but it is inactive at 37°C. However, even at permissive temperatures (<32°C), the specificity of the DNA binding of V143A is different from that of wild type. For example, V143A can activate transcription of p21 promoter but failed to activate Bax promoter at 30°C, whereas the wild type can activate both (19). This result is in agreement with an earlier report that V143A failed to induce apoptosis in H1299 cells (20). The change in specificity can be understood by the long-range effect of V143A. Chemical-shift changes are found for residues in helix 2 and loop L3 (the DNA-binding region), which are quite far away from the site mutation (Fig. (Fig.33B). Presumably, structural changes are propagated by rearrangement of the core residue to fill the cavity created by the V143A mutation.
Implication in Cancer Therapy.
The NMR spectra presented here are structural fingerprints of mutated p53. Our results show that, for the structural mutants G245S and R249S, the overall scaffold is retained when folded. Structural changes are found predominantly in loops L2 and L3, which probably disrupt the DNA-binding surface. This finding implies that a drug that can rescue the function of these mutants must fulfill two requirements: (i) increasing the thermodynamic stability so that the mutants are folded at physiological temperature and (ii) recovering the conformation in the DNA-binding regions. The finding that V143A, G245S, and R249S mutants were rescued by second-site mutations (21) suggests that it is possible to design small molecules that can alter the mutants to wild-type conformation.
Acknowledgments
We thank Professor I. Bertini for providing access to the 800-MHz spectrometer at the European large-scale facility PARABIO at Florence, Italy. This work was supported by the Cancer Research Campaign of the United Kingdom. B.S.D. is a recipient of a Burroughs–Wellcome Fund Hitchings–Elion Fellowship.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.96.15.8438
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc17534?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.96.15.8438
Article citations
Canonical and non-canonical functions of p53 isoforms: potentiating the complexity of tumor development and therapy resistance.
Cell Death Dis, 15(6):412, 12 Jun 2024
Cited by: 2 articles | PMID: 38866752
Review
p53 biology and reactivation for improved therapy in MDS and AML.
Biomark Res, 12(1):34, 13 Mar 2024
Cited by: 3 articles | PMID: 38481290 | PMCID: PMC10936007
Review Free full text in Europe PMC
Conformational plasticity and allosteric communication networks explain Shelterin protein TPP1 binding to human telomerase.
Commun Chem, 6(1):242, 07 Nov 2023
Cited by: 0 articles | PMID: 37935941 | PMCID: PMC10630336
Multidimensional quantitative phenotypic and molecular analysis reveals neomorphic behaviors of p53 missense mutants.
NPJ Breast Cancer, 9(1):78, 29 Sep 2023
Cited by: 1 article | PMID: 37773066 | PMCID: PMC10541912
Reactivation of mutant p53 in esophageal squamous cell carcinoma by isothiocyanate inhibits tumor growth.
Front Pharmacol, 14:1141420, 24 Apr 2023
Cited by: 1 article | PMID: 37168998 | PMCID: PMC10164965
Go to all (133) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mechanism of rescue of common p53 cancer mutations by second-site suppressor mutations.
EMBO J, 19(3):370-378, 01 Feb 2000
Cited by: 97 articles | PMID: 10654936 | PMCID: PMC305574
Structural basis of restoring sequence-specific DNA binding and transactivation to mutant p53 by suppressor mutations.
J Mol Biol, 385(1):249-265, 30 Oct 2008
Cited by: 39 articles | PMID: 18996393
Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations.
J Biol Chem, 280(16):16030-16037, 09 Feb 2005
Cited by: 102 articles | PMID: 15703170
Structure-function-rescue: the diverse nature of common p53 cancer mutants.
Oncogene, 26(15):2226-2242, 01 Apr 2007
Cited by: 272 articles | PMID: 17401432
Review
Funding
Funders who supported this work.





