Abstract
Free full text
A comprehensive proteomics and genomics analysis reveals novel transmembrane proteins in human platelets and mouse megakaryocytes including G6b-B, a novel ITIM protein
Associated Data
Summary
The platelet surface is poorly characterized due to the low abundance of many membrane proteins and the lack of specialist tools for their investigation. In this study we have identified novel human platelet and mouse megakaryocyte membrane proteins using specialist proteomic and genomic approaches. Three separate methods were used to enrich platelet surface proteins prior to identification by liquid chromatography and tandem mass spectrometry: lectin affinity chromatography; biotin/NeutrAvidin affinity chromatography; and free flow electrophoresis. Many known, abundant platelet surface transmembrane proteins and several novel proteins were identified using each receptor enrichment strategy. In total, two or more unique peptides were identified for 46, 68 and 22 surface membrane, intracellular membrane and membrane proteins of unknown sub-cellular localization, respectively. The majority of these were single transmembrane proteins. To complement the proteomic studies, we analysed the transcriptome of a highly purified preparation of mature primary mouse megakaryocytes using serial analysis of gene expression in view of the increasing importance of mutant mouse models in establishing protein function in platelets. This approach identified all of the major classes of platelet transmembrane receptors, including multi-transmembrane proteins. Strikingly, 17 of the 25 most megakaryocyte-specific genes (relative to 30 other SAGE libraries) were transmembrane proteins, illustrating the unique nature of the megakaryocyte/platelet surface. The list of novel plasma membrane proteins identified using proteomics includes the immunoglobulin superfamily member G6b, which undergoes extensive alternate splicing. Specific antibodies were used to demonstrate expression of the G6b-B isoform, which contains an immunoreceptor tyrosine-based inhibition motif. G6b-B undergoes tyrosine phosphorylation and association with the SH2-containing phosphatase, SHP-1, in stimulated platelets suggesting that it may play a novel role in limiting platelet activation.
Introduction
Platelets are small anucleate cells that circulate in the blood in a quiescent state. Their primary physiological function is to stop bleeding from sites of vascular injury by adhering to and forming aggregates on exposed extracellular matrix proteins following blood vessel damage (19, 38). The platelet aggregate or “primary hemostatic plug” is consolidated by fibrin polymers produced by thrombin generated on the platelet surface (46).
Platelets express a diverse repertoire of surface receptors that allow them to respond to different stimuli and adhere to a variety of surfaces. The expression levels of platelet surface receptors varies widely, with the most abundant being the integrin αIIbβ3, which is essential for platelet aggregation. Quiescent human platelets express 40,000 to 80,000 copies of αIIbβ3 on their surface, which increases by 30% to 50% upon platelet activation (45). In contrast, the ADP receptor P2Y1 is among the least abundant, with quiescent human platelets expressing approximately 150 copies on their surface (5).
In order to fully understand how platelets respond to vessel wall damage we require a comprehensive knowledge of the receptors expressed on their surface. Several novel platelet receptors have been identified in recent years, including the lectin receptor, CLEC-2 (43); CD40L (21); Eph kinases and their counter receptors ephrins (36, 37); cadherins (14); Toll receptors-2,-4 and -9 (1, 2); and the single-pass transmembrane natriuretic peptide receptor type C (NPR-C) (40). These findings suggest that platelets may express additional receptors that have important roles in modulating their function.
Proteomics-based approaches have been used to explore the platelet proteome in its entirety (16, 27, 30), as well as sub-proteomes, including the phosphoproteome of thrombin activated platelets (17, 26, 28) and the platelet releasate (9). One class of proteins conspicuously under-represented in the early platelet proteomics studies were transmembrane proteins. This reflects the relatively low abundance of these proteins and also technical difficulties associated with solubilizing and resolving transmembrane proteins in some of the above techniques, most notably two-dimensional gel electrophoresis (2-DE). More recently, the group of Sickmann have characterised the platelet membrane proteome using a combination of density gradient centrifugation and 1-dimensional gel electrophoresis (1-DE), and 16-benzyldimethyl-n-hexadecylammonium chloride (BAC)/sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (31). This group reported the identification of 83 plasma membrane proteins and 48 proteins localized to other membrane compartments.
The application of molecular techniques to analyse expressed genes in platelets is fraught with difficulties because of the lack of a nucleus and the very low levels of mRNA that are carried over from the megakaryocyte. Thus contamination with mRNA from other cell types is a major issue of concern. Further, only 11% of platelet mRNA appears to be derived from genomic DNA, with the majority being derived from mitochondrial genes, as demonstrated by serial analysis of gene expression (SAGE) (20). These problems can be overcome to a large extent by use of a highly purified, mature population of the platelet precursor cell, the megakaryocyte. These cells contain very high levels of mRNA that includes transcripts for all platelet proteins, as illustrated by Kim et al who used SAGE to analyse mRNA in megakaryocytes derived from human cord blood CD34+ cells (24).
In this study, we have used several membrane protein enrichment techniques, namely lectin and biotin/NA affinity chromatography and free flow electrophoresis, in combination with liquid chromatography and tandem mass spectrometry (LC-MS/MS) to identify novel receptors in human platelets. We have also performed LongSAGE on a population of well characterised, highly purified mature murine megakaryocytes (12). The 21 base pair long LongSAGE sequence tags have the advantage over the 14 base pair tags of standard SAGE in providing more reliable detection of greater than 99% of all expressed genes (39). Moreover, SAGE provides a quantitative measure of mRNA expression, unlike DNA microarrays (44). We chose to use megakaryocytes rather than platelets as the source of RNA in order to minimise contamination from other cells and to limit the contribution of mitochondrial-derived mRNA (see above). A major advantage of using mouse rather than human megakaryocytes is with regard to the widespread use of mouse models for functional studies, especially as SAGE analysis of mouse megakaryocytes has not been reported. In this study, >80% of transmembrane proteins identified in human platelets using proteomics were also present in the mouse megakaryocyte LongSAGE library, thereby validating this approach. In total, the present study reports the identification of 136 transmembrane proteins in human platelets based on the identification of two or more unique peptide hits, of which just under 100 have yet to be studied in platelets using biochemical or functional means. Determination of the functional roles of these proteins will enable the further understanding of platelet regulation and may identify novel targets for development of new types of anti-platelet agents.
Experimental Procedures
Materials
N-acetyl-D-glucosamine and propidium iodide (Sigma-Aldrich Company Ltd, Gillingham, UK). Wheat germ agglutinin (WGA) conjugated to Sepharose 4B and unconjugated Sepharose 4B beads (Amersham Biosciences UK Ltd, Little Chalfont, UK). Amicon Centriprep YM-10 and Ultrafree 0.5 centrifugal filter devices (Millipore Corp., Bedford, MA, USA). EZ-link sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate (sulfo-NHS-SS-biotin) and immobilized NA-beads were supplied with the Cell Surface Protein Biotinylation and Purification Kit (Pierce Biotechnology, Inc, Rockford, IL). Colloidal Coomassie G-250 stain (Geneflow, Staffordshire, UK). Rabbit anti-SHP-1 (C-19) polyclonal antibody (Santa Cruz Biotechnology, Inc, Santa Cruz, CA). Ammonium chloride potassium buffer (BioWhittaker, Rockland, ME). Immunomagnetic sheep anti-rat IgG beads (Dynal, Oslo, Norway). Rat-anti mouse antibodies for immunodepletion experiments (BD Biosciences, Oxford, UK). Recombinant murine Stem Cell Factor (SCF) (Peprotech, Rocky Hill, NJ). Human thrombopoietin was a generous gift from Genentech (San Francisco, CA). Tris-glycine SDS-PAGE gels (4-20%), serum free-medium, L-glutamine, penicillin/streptomycin, I-SAGE Long Kit and SAGE2000 4.5 Analysis Software (Invitrogen Ltd, Paisley, UK). RNeasy Miniprep Kit (Qiagen, Crawley, UK). Rabbit anti-G6b-B polyclonal antibody was generated by Eurogentec (Seraing, Belgium) using KLH-conjugated peptides (amino acids 184-198, VKTEPQRPVKEEEPK; and amino acids 220-235, SRPRRLSTADPADAST) from the cytoplasmic tail of G6b-B. Plasmid pCDNA3-G6bB was a generous gift from Dr. R. D. Campbell (MRC Rosalind Franklin Centre for Genomics Research, Cambridge, UK). All other reagents were obtained as previously described (6, 41).
Preparation of washed platelets
Washed human platelets were prepared from blood collected from healthy drug-free volunteers, as previously described (41). Briefly, 9 volumes of blood were collected into 1 volume of 4% (w/v) sodium citrate solution. One volume of ACD solution (1.5% [w/v] citric acid, 2.5% [w/v] sodium citrate and 1% [w/v] glucose) was added to the anti-coagulated blood before being centrifuged at 200g for 20 min at room temperature. Platelet rich plasma (PRP) was collected, to which 2 nM prostacyclin was added, before being centrifuged at 1000g for 10 min. Platelets were washed in 25 mL of modified Tyrode’s-HEPES buffer pH 7.3 (134 nM NaCl, 2.9 mM KCl, 20 mM HEPES, 12 mM NaHCO3, 1 mM MgCl2, 5 mM glucose) containing 3 mL of ACD and 1 nM prostacyclin. Platelets were centrifuged at 1000g for 10 min and resuspended at 5 × 108/mL in modified Tyrode’s-HEPES buffer. Platelets were counted with a Coulter Z2 Particle Count and Size Analyzer (Beckman Coulter Ltd, High Wycombe, UK).
WGA affinity chromatography
Washed platelets (10 mL at 5 × 108/mL) were lysed with an equal volume of 2 × lysis buffer (2% NP-40, 300 mM NaCl, 20 mM Tris, 10 mM ethylenediaminetetraacetic acid pH 7.4), containing protease inhibitors (1 mM AEBSF, 10 μg/mL leupeptin, 10 μg/mL aprotinin and 1 μg/mL pepstatin A). The platelet lysate was pre-cleared with 2 mL of Sepharose 4B beads for 30 min at 4°C and centrifuged at 10,000g for 15 min at 4°C. WGA conjugated to Sepharose 4B (2 mL) was added to the supernatant. The sample was incubated overnight at 4°C with mixing. The WGA resin was transferred to a column and washed three times with 1 × lysis buffer. Bound proteins were eluted from the WGA resin with 3 mL of 0.3 M N-acetyl-D-glucosamine and concentrated to 200 μL using an Amicon Centriprep YM-10 and Ultrafree 0.5 centrifugal filter devices. A fifth of the volume of 5 × SDS-PAGE sample buffer was added to samples and heated to 100°C for 5 min. Samples were prepared in this way in three separate experiments.
Biotinylation of surface proteins and isolation by NeutrAvidin affinity chromatography
Platelet surface proteins were biotinylated according to the manufacturer’s instructions, with a few minor modifications. Platelets (10 mL at 5 × 108/mL) were washed twice with 25 mL PBS pH 7.4 containing 1 μM prostacyclin. They were then resuspended in 10 mL of 412 μM EZ-link sulfosuccinimidyl-2-(biotinamido) ethyl-1,3-dithiopropionate (sulfo-NHS-SS-biotin) in PBS pH 7.4 for 30 min at room temperature. Unreacted biotinylation reagent was quenched by adding Tris pH 8.0 to a final concentration of 50 mM; platelets were pelleted at 1000g for 10 min at room temperature; washed twice in 10 mL 0.025 M Tris, 0.15 M NaCl (TBS) pH 7.4 containing 1 μM prostacyclin; and lysed in 500 μL lysis buffer (proprietary) by sonicating on low power at 10 min intervals for 30 min on ice. Lysates were centrifuged at 10,000g for 2 min at 4°C to remove cell debris. Clarified supernatants were incubated with 250 μL NeutraAvidin (NA)-beads for 1 hr at room temperature then centrifuged for 1 min at 1,000g. The gel was washed 3 × 500 μL wash buffer (proprietary). Proteins were eluted in 2 × sample buffer containing 50 mM DTT and heated to 100°C for 5 min. Samples were prepared in this way in three separate experiments.
Preparation of platelet plasma membranes (PM) and intracellular membranes (IM) by free flow electrophoresis
Platelet PM and IM were prepared as described in detail previously (3). Briefly, platelets were separated from freshly obtained platelet concentrates (National Blood Service, Tooting, London, UK) and treated with neuraminidase (type X, 0.05 U/mL) for 20 min at 37°C. After 2 washings, platelets were disrupted by sonication and the platelet homogenate layered on a linear (1-3.5 M) sorbitol density gradient followed by centrifugation at 42,000g for 90 min, to obtain a mixed membrane fraction (free of granular contamination). This membrane fraction was separated into PM and IM by free-flow electrophoresis using an Octopus electrophoresis apparatus (Dr. Weber Gmbh, Germany) running at 750 V, 100 mA. Two discrete peaks comprising PM and IM (more electronegative) were obtained. Tops of peaks were pooled, centrifuged (100,000g for 60 min) and resuspended in 0.4 M sorbitol, 5% glycerol and 10 mM triethanolamine pH 7.2 and kept at −80°C until further analysis. The purity of fractions was checked by analyzing by SDS-PAGE and western blotting for the absence of actin in IM and of SERCA2 Ca2+ATPase in PM fractions, as previously described (3). Samples were prepared in this way on two separate occasions.
Protein preparation for MS/MS
Proteins were resolved on 4-20% Tris-glycine SDS-PAGE gels and stained with Colloidal Coomassie G-250 stain. Twelve to 32 gel slices each with a width of 1-2 mm were manually excised with a razor for subsequent in-gel trypsinization and LC-MS/MS analysis. Bands were excised from three separate WGA affinity purification experiments; three biotin/NA affinity purification experiments and two FFE experiments. Proteins were trypsinized within gel slices and peptides extracted using the method described by Shevchenko (42).
LC-MS/MS and data anlysis
Tryptic peptides were analyzed by LC-MS/MS, using a ThermoFinnigan LCQ Deca XP Plus ion-trap (Thermo Electron Corporation, Hemel Hempstead, UK) coupled to a Dionex/LC Packings nanobore HPLC system (Dionex/LC Packings, Sunnyvale, CA, USA), configured with a 300 μm id/1 mm C18 Pepmap pre-column (LC Packings, San Francisco, CA, USA) and a 75 μm id/15 cm C18 PepMap analytical column (LC Packings). Tryptic peptides were eluted into the ion-trap mass spectrometer using a 45 min 5-95% acetonitrile gradient containing 0.1% formic acid at a flow rate of 200 nL/min. Spectra were acquired in an automatic data dependent fashion using a full MS scan (400-2000 m/z) to determine the 5 most abundant ions which were sequentially subjected to MS/MS analysis. Each precursor ion was analyzed twice before it was placed on an exclusion list for 1 min. MS/MS spectra were converted into dta-format files by Bioworks Browser (3.1) and searched against the NCBInr database (released April 2004) using the TurboSequest (3.1) search algorithm (ThermoFinnigan). Both the precursor mass tolerance and the fragment mass tolerance were set at 1.4 Da. Two missed tryptic cleavages and carbamidomethylation of cysteine residues as a fixed modification were allowed. Positive peptide hits using TurboSequest had a minimum cross-correlation factor of 2.5, a minimum delta correlation value of 0.25 and a preliminary ranking of one. The same dta-format files generated with the LC-MS/MS ion-trap and Bioworks Browser set up were also searched against the NCBInr database using the Mascot 1.8 search algorithm (Matrix Science, London, UK). Mascot searches were restricted to the human taxonomy allowing carbamidomethyl cysteine as a fixed modification and oxidized methionine as a potential variable modification. Both precursor mass tolerance and MS/MS tolerance of 1.4 Da, allowing for up to two missed cleavages. Positive identification was only accepted when the data satisfied the following criteria: (i) MS/MS data were obtained for at least 80% y-ions series of a peptide comprising at least eight amino acids and no missed tryptic cleavage sites; (ii) MS/MS data with more than 50% y-ions were obtained for two or more different peptides comprising at least eight amino acids long and no more than two missed tryptic cleavage sites. Swiss-Prot/TrEMBL accession numbers were obtained for all proteins identified.
MS/MS analysis of tryptic fragments was also carried out with a Q-TOF 1 mass spectrometer (Micromass, Manchester, UK) as a means of verifying proteins identified with the ion-trap mass spectrometer, and of improving both protein and proteome coverage by using complementary instruments for the MS/MS analysis (13). The Q-TOF 1 mass spectrometer was coupled to a CapLC HPLC system (Waters, Milford, MA, USA) configured with a 300 μm id/5 mm C18 pre-column (LC Packings) and a 75 μm id/25 cm C18 PepMap analytical column (LC Packings). Tryptic peptides were eluted to the mass spectrometer using a 45 min 5-95% acetonitrile gradient containing 0.1% formic acid at a flow rate of 200 nL/min. Spectra were acquired in an automatic data dependent fashion with a 1 sec survey scan followed by three 1 sec MS/MS scans of the most intense ions. The selected precursor ions were excluded from further analysis for 2 min. MS/MS spectra were converted into pkl-format files using Mass Lynx 3.4 and searched against the NCBInr database with the Mascot search algorithm, as described above.
All proteins identified by both Sequest and Mascot were checked for predicted transmembrane domains (TMDs) with TMHMM v. 2.0 (25).
Construction of decoy database and estimation of the false positive rate of protein identification by LC-MS/MS
A randomized version of the NCBInr database used in this study was generated by a Perl programme downloaded from Matrix Science Ltd. (London, UK), decoy.pl. This programme was run using the random and append command line switches that appended a random set of sequences, with the same average amino acid composition as those in the original dataset, onto the database. The decoy.pl programme was modified to work correctly with the long header format of the NCBInr database. Database searches with all of the dta-format files generated by LC-MS/MS ion-trap and Sequest were searched against the decoy database using the same search parameters described above for the original searches. The percent false positive rate of protein identification was calculated by dividing the number of “random” proteins identified by the sum of “random” and “real” proteins identified and multiplying by 100. The false positive rate was calculated for random proteins identified by two or more peptide hits and for those identified by 1 peptide hit.
Comparison of proteomics datasets
To compare which proteins were common to both our proteomic dataset reported in this study and that of Moebius et al (31), a non-redundant set of peptide sequences were collected from each study. A total of 295 were obtained from the Moebius study and 136 from the present study. All sequences were subsequently BLAST searched against the Reference Sequence Project peptides. Sixty-two proteins were found to be common to both datasets.
Megakaryocyte culture and purification
Bone marrow cells were flushed from femurs and tibias of 3 to 4 month old C57Bl6 mice, as previously described (12). Mature erythrocytes were lysed with ammonium chloride potassium buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM Na2EDTA pH 7.3). CD16/CD32+Gr1+B220+CD11b+ cells were depleted using immunomagnetic sheep anti-rat IgG beads and rat-anti mouse antibodies according to the manufacturer’s instructions. The cell depleted population was then cultured in serum-free medium supplemented with 2 mM L-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin and 20 ng/mL murine SCF at 37°C and 5% CO2 for 2 days and 5 more days under the same conditions, in addition to 200 ng/mL recombinant human thrombopoietin. High density mature megakaryocytes were then isolated in a 0-3% BSA gradient (4 mL 3% BSA/PBS in a 15 mL Falcon tube overlaid with 4 mL of 1.5% BSA/PBS and 4 mL of suspension cells in PBS) (11). After standing for 40 min at room temperature, the cells remaining in the lower 2 mL were collected, washed in PBS and subjected to another 0-3% BSA gradient to obtain a pure population. DNA content of cells was determined by staining with 50 μg/mL propidium iodide and analyzing cells with a FACScan analyzer and CellQuest software (Becton Dickinson), as previously described (12).
Serial analysis of gene expression
Primary mouse megakaryocyte RNA was made using the RNeasy Miniprep Kit. The LongSAGE library was generated from 20 μg RNA using the I-SAGE Long Kit and sequenced by Agencourt Bioscience Corporation (Beverly, MA, USA). LongSAGE sequence tags were identified using SAGE2000 4.5 Analysis Software with reference to the SAGEmap_tag_ug-rel database (http://www.ncbi.nlm.nih.gov/SAGE/). To identify megakaryocyte-specific genes, the resulting SAGE library, of 53,046 sequence tags, was compared to 30 other mouse SAGE libraries, from T lymphocyte (14 SAGE libraries), dendritic cells (6), intra-epithelial lymphocytes (2), embryonic stem cells (2), brain (2), B lymphocyte (1), heart (1), 3T3 fibroblast cell line (1) and P19 embryonic carcinoma cell line (1), with a combined total of 1,031,389 tags. The data analysis was performed using custom written software (!SAGEClus) as described in Cobbold et al. (8). Genes with predicted TMDs were identified using TMHMM v. 2.0 (25).
Platelet activation, immunoprecipitations and western blotting
Washed platelets (8 × 108/mL) were stimulated with 10 μg/mL CRP or 5 U/mL thrombin for 90 seconds with constant mixing at 1,200 rpm and 37°C, as previously described (41). Platelets were lysed in 2 × lysis buffer containing 5 mM sodium vanadate in addition to the protease inhibitors described above. Proteins were immunoprecipitated from platelet lysates with 2 μg rabbit anti-SHP-1 antibody and 10 μL rabbit anti-G6b-B serum. Ten microlitres of rabbit pre-immune serum was used as a negative control for immunoprecipitations. Membranes were immunoblotted with 1 μg/mL anti-phosphotyrosine antibody, 0.2 μg/mL anti-SHP-1 antibody and 1/1000 rabbit anti-G6b-B antibody, as previously described (29, 41).
Transient tranfections
Human embryonic kidney (HEK) 293T cells were transfected with 5 μg of either pCDNA3.1 plasmid or pCDNA3-G6bB plasmid by the calcium phosphate technique. Cells were lysed in 2 × lysis buffer containing protease and phosphatase inhibitors; proteins were resolved on 4-20% SDS-PAGE gels and western blotted with either 1/1000 rabbit anti-G6b-B serum or 1/1000 pre-immune serum from the same rabbit in which the anti-G6b-B antibody was raised.
Results
Enrichment of platelet PM proteins by affinity chromatography and free flow electrophoresis
Three different techniques were used to enrich platelet transmembrane proteins, namely WGA affinity chromatography, biotin/NA affinity chromatography and FFE. Proteins were subsequently resolved by 1-DE, stained with Colloidal Coomassie blue, and bands were manually excised and identified by LC-MS/MS. Fragmentation spectra generated by the ion-trap and Q-TOF mass spectrometers were searched against the NCBInr database using the Sequest search algorithm and against the NCBInr and Swiss-Prot/TrEMBL databases using the Mascot search algorithm. The use of two different search algorithms and databases increased the number of identified proteins and also helped to safeguard against erroneous identifications (13). All proteins that met the search criteria outlined in the Experimental Procedures, which includes identification of two or more unique peptides, were investigated for transmembrane domains using TMHMM v. 2.0 (25).
The proteins that have been identified in this study are divided into plasma membrane (PM), intracellular membrane (IM) and proteins of unknown sub-cellular distribution, in accordance with data from NCBI, Swiss-Prot/TrEMBL and PubMed (Table 1, and Supplementary Tables 1 and 2). The techniques and search algorithms which were used in their identification are also shown in Table 1, and Supplementary Tables 1 and 2. Proteins that are found in PM and IMs such as integrin αIIbβ3 are classified as PM proteins. Ten of the proteins of unknown distribution are hypothetical proteins and have not been previously identified in any cell type. Tryptic peptides identified by Sequest are listed in Supplementary Table 3 and those identified only by Mascot are listed in Supplementary Table 4. Selected MS/MS spectra identified by Sequest and Mascot are included as Supplementary Data 1 and Supplementary Data 2, respectively. All raw MS/MS data generated as part of this study can be accessed from the Molecular and Cellular Proteomics website (Supplementary Data 3 and 4).
Table 1
Transmembrane proteins localized to the plasma membrane identified by tandem mass spectrometry in human platelets and SAGE analysis in mouse megakaryocytes. Proteins are arranged according to families. Information is given on general function or specific function in platelets, where known. Several proteins are predominantly expressed on intracellular membranes and platelet α-granules and are translocated to the plasma membrane on activation. General information was obtained from NCBI, Swiss-Prot/TrEMBL and PubMed databases. The number of transmembrane domains (No. of predicted TMDs) in each protein was predicted with TMHMM v.2.0 (25). The highest number of unique peptides (No. of unique peptides) identified in a single mass spectrometry experiment are shown. The search algorithm (Mascot and/or Sequest) used to identify each protein is indicated, as is the method used to enrich transmembrane proteins (biotin/NA, biotinylation and NeutrAvidin affinity chromatography; FFE-IM, free flow electrophoresis-intracellular membrane fraction; FFE-PM, free flow electrophoresis-plasma membrane fraction; WGA, wheat germ agglutinin affinity chromatography). All proteins were identified by two or more peptide hits with at least one of the search algorithms. The number of SAGE tags (No. of SAGE tags) identified in mouse megakaryocytes is indicated in the final columns. Transcripts for 37 out of 44 proteins (84%) identified by MS/MS analysis of human platelets were identified in mouse megakaryocytes by SAGE. A question mark in the SAGE tag column indicates that the transcript could be present, but that there is currently not enough sequence information in the public databases for the identification of SAGE tags.
| Family name (Protein name) | Swiss-Prot/TrEMBL accession | Function | No. of predicted TMDs | No. of unique peptides | Search algorithm | Enrichment | No. of SAGE tags |
|---|---|---|---|---|---|---|---|
| Cadherin superfamily | |||||||
| protocadherin FAT 2 | Q9NYQ8 | Calcium-dependent cell-adhesion protein | 1 | 2 | Mascot | WGA, FFE-PM | ? |
| Ig superfamily and associated proteins | |||||||
| basigin | P35613 | Associates with carboxylate transporters; undergoes homophilic binding; function in platelets is not known | 2 | 3 | Sequest | WGA | 26 |
| CD226 | Q15762 | Involved in intercellular adhesion and modulation of signaling; supports platelet adhesion to endothelial cells | 1 | 5 | Mascot, Sequest | biotin/NA, WGA | 3 |
| CD84 | O15430 | Member of the SLAM family of homophilic adhesion receptors; stabilizes platelet-platelet interactions during thrombosis | 1 | 4 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 3 |
| endothelial cell-selective adhesion molecule | Q96AP7 | Found at tight junctions in endothelial cells; function in platelets is not known | 1 | 3 | Mascot, Sequest | biotin/NA, FFE-PM | 9 |
| FcR γ-chain | P30273 | Present as a complex with the collagen receptor, GPVI; the ITAM in the FcR γ-chain is critical for GPVI signaling | 1 | 2 | Mascot, Sequest | biotin/NA | 23 |
| G6f | Q7Z5H2 | G6f gene is found within the MHC class III; interacts with Grb2 when phosphorylated; function in platelets is not known | 1 | 4 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM | 7 |
| GPVI | Q9UIF2 | Major signaling receptor for collagen; present as a complex with FcR γ-chain | 1 | 3 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 6 |
| MHC class I antigen (HLA-A) | Q8MHP8 | Presentation of antigens to the immune system | 1 | 6 | Mascot, Sequest | biotin/NA | 80 (total) |
| MHC class I antigen B-52 | P30490 | Presentation of antigens to the immune system | 1 | 4 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM | 80 (total) |
| MHC class I antigen Cw-15 | Q07000 | Presentation of antigens to the immune system | 1 | 8 | Mascot, Sequest | biotin/NA, FFE-PM | 80 (total) |
| ICAM 2 | P13598 | Ligand for β2-integrins; function in platelets is not known | 1 | 3 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 2 |
| JAM1 | Q9Y624 | Plays a role in tight junction formation and transmigration; function in platelets is not known | 1 | 4 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM | 5 |
| JAM3 | Q9BX67 | May participate in cell-cell adhesion distinct from tight junctions; function in platelets is not known | 1 | 3 | Mascot, Sequest | biotin/NA, FFE-PM | 0 |
| FcγRIIA | P12318 | Low affinity IgG receptor; mediates platelet activation by immune complexes via an ITAM regulated pathway | 1 | 2 | Mascot, Sequest | biotin/NA | Not in mouse |
| PECAM-1 | P16284 | Major platelet receptor; undergoes homotypic binding; inhibits platelet activation by collagen and VWF | 1 | 12 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 7 |
| TLT-1 | Q8IWY2 | Found on platelet α-granules and translocates on activation; contains an ITIM; supports late stage platelet activation | 1 | 2 | Mascot, Sequest | biotin/NA, FFE-IM | 13 |
| Integrin family | |||||||
| α2 integrin subunit | P17301 | α2β1 mediates platelet adhesion to collagen | 1 | 8 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 0 |
| α6 integrin subunit | P23229 | α6β1 mediates platelet adhesion to laminin | 1 | 12 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 47 |
| αIIb integrin subunit | P08514 | αIIbβ3 is the major platelet adhesion and aggregation receptor | 1 | 15 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 136 |
| β1 integrin subunit | P05556 | associates with α2, α5 and α6 integrin subunits in platelets | 1 | 8 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 14 |
| β3 integrin subunit | P05106 | associates with αIIb and αv integrin subunits in platelets | 1 | 17 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 41 |
| Leucine-rich repeat family | |||||||
| GPIX | P14770 | Subunit of the GPIb-IX-V complex that binds VWF | 1 | 5 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 11 |
| GPV | P40197 | Subunit of the GPIb-IX-V complex that binds VWF | 1 | 11 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 9 |
| GPIbα | P07359 | Subunit of the GPIb-IX-V complex that binds VWF | 1 | 5 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 21 |
| GPIbβ | P13224 | Subunit of the GPIb-IX-V complex that binds VWF | 1 | 4 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 31 |
| LRRC32 protein | Q14392 | Not known | 1 | 2 | Mascot, Sequest | biotin/NA | 0 |
| Peptidase family | |||||||
| ADAM 10 | O14672 | Proteolytic release of cell surface proteins, including TNFα and ephrin-A2; function in platelets is not known. | 1 | 5 | Mascot, Sequest | biotin/NA, FFE-PM | 11 |
| ECE-1 | P42892 | Metabolism of big endothelin-1 to endothelin-1 | 1 | 2 | Mascot, Sequest | FFE-IM, WGA | 0 |
| Protein tyrosine phosphatase family | |||||||
| DEP-1 | Q12913 | Regulates contact inhibition of cell growth; function in platelets is not known | 1 | 11 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 1 |
| Selectin family | |||||||
| P-selectin | P16109 | Found on platelet α-granules and translocates on activation; mediates interaction with microparticles and leukocytes | 1 | 11 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 13 |
| Steroid receptor family | |||||||
| membrane associated progesterone receptor component 1 | O00264 | Receptor for progesterone; function in platelets is not known | 1 | 2 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM | 9 |
| membrane associated progesterone receptor component 2 | O15173 | Receptor for progesterone; function in platelets is not known | 1 | 4 | Mascot, Sequest | FFE-IM, FFE-PM | 1 |
| type I sigma receptor | Q99720 | Interacts with endogenous steroid hormones (progesterone and testosterone); function in platelets is not known | 1 | 2 | Mascot, Sequest | FFE-IM | 1 |
| Tetraspanin family | |||||||
| CD9 | P21926 | Major platelet tetraspanin; associates with platelet glycoproteins | 4 | 4 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM | 34 |
| Tspan-9 | O75954 | Function in platelets is not known | 4 | 2 | Mascot, Sequest | biotin/NA, FFE-PM | 5 |
| Tspan-33 | Q86UF1 | Function in platelets is not known | 4 | 3 | Mascot, Sequest | FFE-PM | 3 |
| Tyrosine protein kinase family | |||||||
| EphB1 | P54762 | Supports late stage aggregation via interaction with ephrin-B1 | 1 | 2 | Mascot, Sequest | biotin/NA | 0 |
| Miscellaneous | |||||||
| adipocyte plasma membrane-associated protein | Q9HDC9 | May play a role in adipocyte differentiation; function in platelets is not known | 1 | 3 | Mascot, Sequest | FFE-IM, FFE-PM | 2 |
| BAT5 | O95870 | BAT5 gene is found within the MHC class III; function in platelets is not known | 2 | 2 | Mascot, Sequest | FFE-IM | 2 |
| CD36 | P16671 | Putative receptor for collagen and thrombospondin in platelets | 2 | 5 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 0 |
| CD92 | Q96KU3 | Probable choline transporter; function in platelets is not known | 9 | 2 | Sequest | FFE-PM | 5 |
| sodium/potassium transporting, β 3 polypeptide | P54709 | Non-catalytic component of a Na+/K+-ATPase; responsible for establishing and maintaining resting membrane potential | 1 | 3 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM | 3 |
| solute carrier family 2, facilitated glucose transporter member 3 | P11169 | Facilitative glucose transporter; present in platelet PM and α-granules; required for glucose uptake by platelets | 10 | 2 | Mascot, Sequest | FFE-IM, FFE-PM | 11 |
| STIM1 | Q13586 | A Ca2+ sensor that links Ca2+ store depletion from the ER with store-operation Ca2+ influx from the PM | 1 | 5 | Sequest | FFE-IM, FFE-PM | 2 |
| stomatin, isoform a | P27105 | Act as a cytoskeletal anchor in erythrocytes; present in platelet α-granules | 1 | 5 | Mascot, Sequest | biotin/NA, FFE-IM, FFE-PM, WGA | 12 |
Since a large proportion of platelet surface proteins are glycosylated, we initially used the lectin WGA to purify platelet glycoproteins followed by elution with N-acetylglucosamine (Fig. 1A), as illustrated for the platelet glycoproteins GPIbα and PECAM-1 (Fig. 1B). The distinct staining pattern of the WGA-purified sample relative to that of the whole cell lysate confirms that a substantial level of protein purification has been achieved, a result that is further supported by comparing the αIIbβ3:actin ratio before and after enrichment (Fig. 1A, WCL versus WGA lanes). In total, 21 PM proteins and 2 IM proteins were identified by two or more peptide hits using this approach (Table 1, and Supplementary Table 1). This approach also identified a similar number of cytosolic and granule proteins, possibly because of association with the cytoplasmic regions of transmembrane proteins or because of their glycosylation (data not shown).
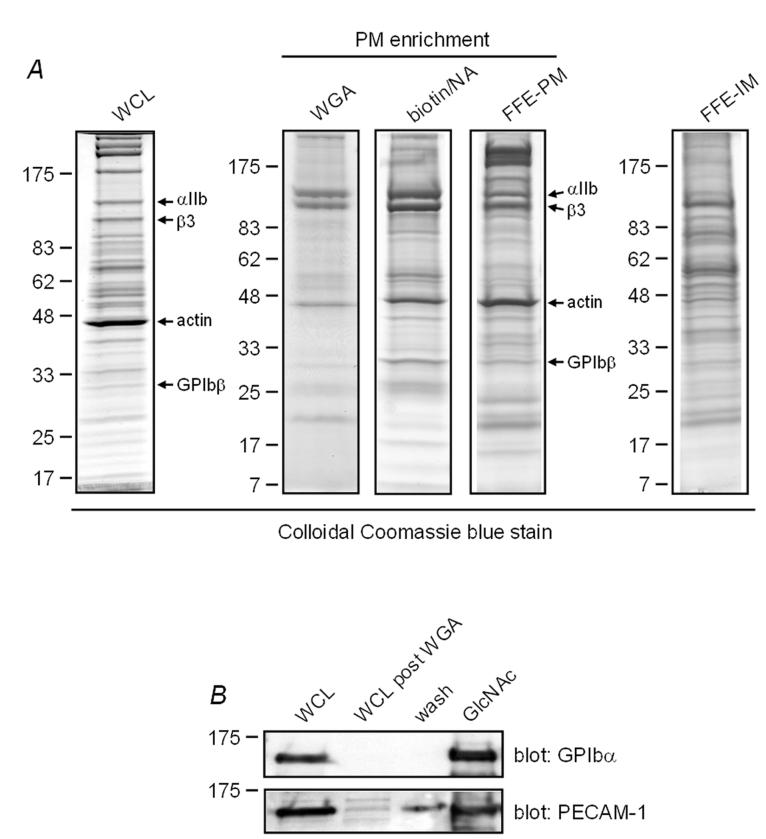
Comparison of proteins isolated by WGA affinity chromatography, biotin/NeutrAvidin (NA) affinity chromatography and free flow electrophoresis (FFE). A, platelet whole cell lysate (WCL) and proteins isolated by the three enrichment techniques were resolved on 4-20% SDS-PAGE gels and stained with Colloidal Coomassie blue. Bands corresponding to αIIb, β3, actin and GPIbβ were identified by tandem mass spectrometry and are shown to the left of the panels. WGA, wheat germ agglutinin affinity chromatography; biotin/NA, biotin/NeutrAvidin affinity chromatography; FFE-PM, free flow electrophoresis-plasma membrane fraction; FFE-IM, free flow electrophoresis-intracellular membrane fraction. Images shown are representative of three WGA, three biotin/NA, and two FFE enrichment experiments. B, aliquots taken at various stages of the WGA affinity chromatography procedure, including elution by N-acetylglucosamine (GlcNac), were western blotted for PECAM-1 and GPIbα.
As an alternative approach, exposed lysine residues of platelet surface proteins were labelled with biotin prior to affinity purification with NA-beads. The membrane insoluble biotinylating reagent sulfo-NHS-SS-biotin was used to biotinylate surface proteins and thereby limit labelling of intracellular proteins (35). NA-beads were used rather than avidin- or streptavidin-beads in order to facilitate removal of bound proteins through the reducing agent DTT. An estimate of the amount of enrichment of transmembrane proteins can be obtained by comparing the αIIbβ3:actin and GPIbβ:actin ratios before and after enrichment (Figure 1A, WCL versus biotin/NA lanes). This approach detected a greater number of proteins than that using WGA chromatography as shown by the increased number of bands in Figure 1A. This is most likely due to the higher proportion of transmembrane proteins with free lysine residues compared with those that are precipitated by the lectin. Furthermore, the high affinity of NA for biotin enables the use of more stringent wash conditions, thereby removing a greater proportion of cytosolic proteins which would interfere with detection of membrane proteins. Thirty-five PM, 14 IM and 5 transmembrane proteins of unknown localization were identified by two or more peptide hits using biotin/NA (Table 1, and Supplementary Tables 1 and 2).
FFE was used to separate PM and IM proteins on the basis of a charge difference generated by treatment of platelets with neuraminidase, which selectively removes sugar residues from the outer, plasma membrane (3). The purity of the two FFE fractions was estimated by western blotting for the absence of actin in IM and of SERCA2 Ca2+ATPase in PM fractions. The presence of actin in the PM fraction is a consequence of its association with surface glycoproteins, including the GPIb-IX-V complex. The results demonstrate a level of contamination of less than 5% of PM in the IM fraction, which is consistent with our experience of this technique (3). The purity of the two membrane fractions was further supported by the distinct banding pattern of the PM and IM samples, with the banding pattern of the former being similar to that obtained using biotin labelling, but with a greater number of bands (Fig. 1A). A total of 35 PM, 30 IM and 10 transmembrane proteins of unknown location were found in the FFE-generated PM sample by a minimum of two peptide hits (Table 1, and Supplementary Tables 1 and 2), compared with 31 PM, 66 IM and 20 transmembrane proteins of unknown location in the FFE-generated IM sample (Table 1, and Supplementary Tables 1 and 2). Significantly, only two of the 44 proteins identified only in the FFE-IM fraction were known PM proteins, further illustrating the successful separation of plasma and intracellular membranes (Table 2). The presence of IM proteins in the PM fraction, and vice versa, is therefore most likely due to the presence of proteins in both membrane regions, as well as a degree of cross contamination. The majority of the IM proteins are expressed in the endoplasmic reticulum (ER) (Supplementary Table 1).
Table 2A
Number of transmembrane proteins identified by each enrichment method. Proteins identified using each enrichment technique were pooled from three WGA affinity chromatography experiments, three biotin/NeturAvidin affinity chromatography experiments and two free-flow electrophoresis experiments. Samples from each experiment were analyzed once by LC-MS/MS.
| Enrichment method | Number of transmembrane proteins identified | |||
|---|---|---|---|---|
| PM proteins | IM proteins | Proteins of unknown compartment | Total | |
| biotin/NA | 35 | 15 | 5 | 55 |
| FFE-IM | 31 | 65 | 20 | 116 |
| FFE-PM | 35 | 29 | 10 | 74 |
| WGA | 21 | 2 | 0 | 23 |
PM, plasma membrane; IM, intracellular membrane; biotin/NA, biotin/NeutrAvidin affinity chromatography; FFE-IM, free flow electrophoresis-intracellular membrane fraction; FFE-PM, free flow electrophoresis-plasma membrane fraction; WGA, wheat germ agglutinin affinity chromatography
In total, these three approaches identified 46 PM, 68 IM and 22 transmembrane proteins of unknown compartmentalization on the basis of identification of two or more unique peptides by MS/MS. A summary of the number of transmembrane proteins identified by each enrichment method and the overlap between the different enrichment methods is provided in Tables Tables2A2A and andB.B. Eighty-three percent of the proteins were identified by both Mascot and Sequest search algorithms and 60% were identified by more than one enrichment method. Strikingly, the 17 proteins identified by all of the enrichment techniques are well known platelet surface transmembrane proteins that are present at high levels (see Table 1). Interestingly, only a small number (17%) of the identified PM proteins had more than one predicted transmembrane domain, including the three tetraspanin proteins, CD9, Tspan-9 and Tspan-33. On the other hand, there are no seven transmembrane G protein-coupled receptors (GPCR) in this list, a result which was also found by Moebius and coworkers who used a combination of density gradient centrifugation, 1-DE, and 16-BAC/SDS-PAGE to purify platelet membranes (31). Significantly, a greater proportion of IM (58%) and proteins of an undefined membrane distribution (59%) are predicted to contain more than one transmembrane domain, suggesting that the lack of identification of multi-spanning proteins in the PM fraction may be due, in part, to their low abundance. We estimate that just under 100 of the identified proteins have not been previously described in platelets on the basis of biochemical and functional data. Of this list, 10 are hypothetical proteins in that they have not been identified in any cell type. Together, these results illustrate the power of using all three approaches to identify platelet membrane proteins.
Table 2B
Overlap in transmembrane proteins identified by different enrichment methods. The number of transmembrane proteins identified by each enrichment technique individually (rows 1-4) or by multiple enrichment techniques (rows 5-15), are shown.
| Enrichment methods | Number of transmembrane proteins identified | ||
|---|---|---|---|
| PM proteins | IM proteins | Proteins of unknown compartment | |
| 1) biotin/NA | 5 | 0 | 2 |
| 2) FFE-IM | 2 | 32 | 10 |
| 3) FFE-PM | 2 | 0 | 0 |
| 4) WGA | 1 | 0 | 0 |
| 5) biotin/NA, FFE-IM | 1 | 6 | 0 |
| 6) biotin/NA, FFE-PM | 5 | 1 | 0 |
| 7) biotin/NA, WGA | 1 | 1 | 0 |
| 8) FFE-IM, FFE-PM | 4 | 20 | 7 |
| 9) FFE-IM, WGA | 1 | 0 | 0 |
| 10) FFE-PM, WGA | 1 | 0 | 0 |
| 11) biotin/NA, FFE-IM, FFE-PM | 6 | 7 | 3 |
| 12) biotin/NA, FFE-IM, WGA | 0 | 0 | 0 |
| 13) biotin/NA, FFE-PM, WGA | 0 | 0 | 0 |
| 14) FFE-IM, FFE-PM, WGA | 0 | 1 | 0 |
| 15) biotin/NA, FFE-IM, FFE-PM, WGA | 17 | 0 | 0 |
| Total | 46 | 68 | 22 |
The false positive rate of protein identification was determined by re-analyzing all of the Sequest dta-format files against a decoy database consisting of the original NCBInr database with a randomized version of the same database appended to the end of it. Scrambled peptides were marked “random” so that they could be easily distinguished from real proteins. The estimated false positive identification rate was 0.025% for proteins identified by 2 or more peptide hits, reflecting the stringent settings used in the study and thereby giving increased confidence to the data.
As part of this study, we also identified 45 proteins on the basis of a single unique peptide using the above techniques. These proteins are listed in Supplementary Table 5. The estimation of the false positive rate for this group of proteins was 5% thereby demonstrating the need for supporting biochemical or functional data to confirm their expression in platelets. Nevertheless, it is emphasised that several of these proteins are already known to be expressed in platelets, including the α5 integrin subunit and the C-type lectin-like receptor, CLEC-2.
Identification of G6b-B in human platelets: a novel tyrosine phosphorylated ITIM-bearing protein
One of the novel platelet PM proteins is the immunoglobulin superfamily member G6b, which is reported to have seven splice variants, G6b-A to G6b-G (10). Two of these splice variants, G6b-A and G6b-B, have transmembrane domains and have been shown to be expressed on the surface of transiently transfected cells (10). The main difference between these two splice variants is in their cytoplasmic tails. The G6b-A isoform lacks any tyrosine residues in this region, whereas the G6b-B isoform contains an ITIM and therefore has the potential to selectively inhibit signalling by the platelet immunoreceptor tyrosine-based activation motif (ITAM)-receptors, GPVI and FcγRIIA. Three unique peptides were identified for different isoforms of G6b by MS/MS. MS/MS spectra for all three peptides are shown in Figure 2. One of the peptides (TVLHVLGDR) could have come from any of the seven splice variants. A second peptide (LPPQPIRPLPR) could only have come from G6b-A, whereas the third peptide (IPGDLDQEPSLLYADLDHLALSR) could have come from either G6b-B, -C or -E. However, neither G6b-C nor G6b-E are predicted to contain transmembrane domains. In order to clarify the ambiguity of the MS/MS result and determine whether G6b-B is expressed in human platelets, we raised a rabbit polyclonal antibody to peptides found in a portion of the cytosolic tail of G6b-B that is absent from G6b-A, and used these to confirm expression of the ITIM-bearing isoform of G6b in platelets by western blotting (Fig. 3A). Whole cell lysate prepared from HEK 293T cells transiently transfected with G6b-B was used as a positive control (Fig. 3A). The specific antibody identified two bands at 32 and 38 kDa on a 4-20% SDS-PAGE gel in platelets, which are most likely to represent differentially glycosylated isoforms of G6b-B, as similar bands were also seen in G6b-B-transfected, but not mock-transfected HEK 293T cells (Fig. 3A). Multiple forms of G6b-B that can be separated by SDS-PAGE have been described in transfection studies in other cell types (10).
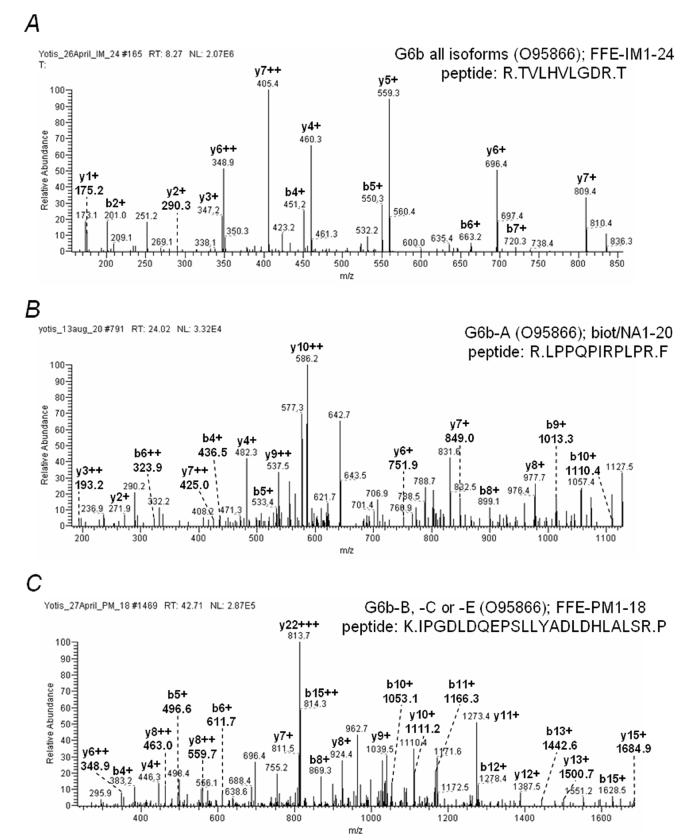
MS/MS spectra of G6b peptides. Peptides corresponding to each MS/MS spectra are shown in the top right corner of each panel, along with the G6b isoforms from which each peptide may have been derived, the Swiss-Prot/TrEMBL accession number (in parenthesis), and the experiment and band slice identification numbers. The start and end of each peptide are indicated by dots. Adjacent amino acids to the peptide identified are also included (outside dots). Selected b- and y-ions identified are indicated. (A) Peptide TVLHVLGDR is present in all seven isoforms of G6b. (B) Pepetide LPPQPIRPLPR is only present in G6b-A. (C) Peptide IPGDLDQEPSLLYADLDHLALSR is present in G6b-B, -C and -E.
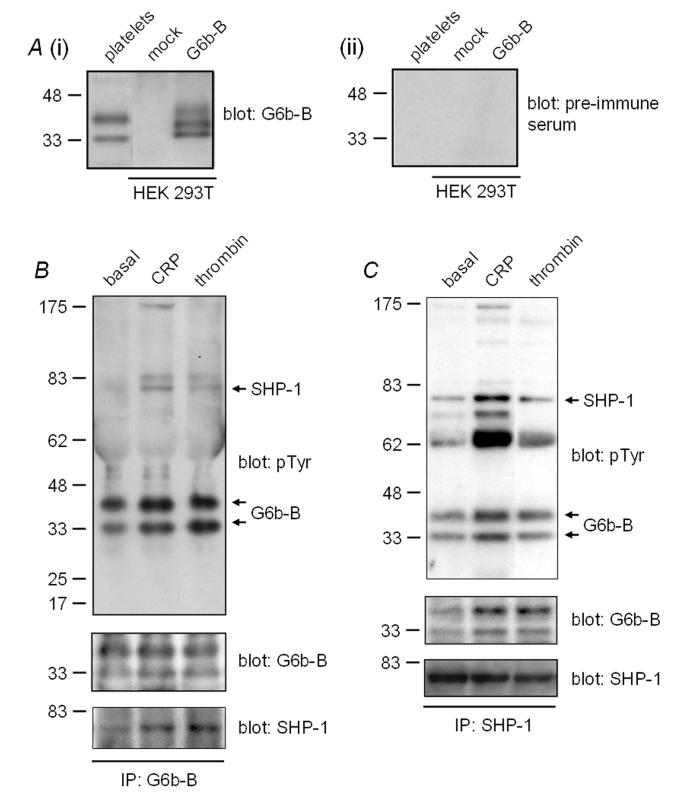
Expression of G6b in human platelets. A, (i) whole cell lysates were prepared from human platelets and HEK 293T cells transiently transfected with either plasmid alone (mock) or a G6b-B expression plasmid (G6b-B) were western blotted for G6b-B using a rabbit anti-G6b-B polyclonal antibody raised against two peptides from the cytoplasmic tail of the protein; (ii) as a control, the same samples western blotted in (i) were blotted with pre-immune serum from the same rabbit in which the G6b-B anti-serum was raised. B, G6b-B undergoes an increase in tyrosine phosphorylation in response to CRP and thrombin stimulation, and interacts with SHP-1 in human platelets. G6b-B was immunoprecipitated (IP) from whole cell lysates prepared from resting platelets and platelets stimulated with either 10 μg/mL CRP or 5U/mL thrombin. Samples were western blotted for tyrosine phosphorylated proteins, then stripped and blotted for G6b-B, followed by SHP-1. C, G6b-B is tyrosine phosphorylated in resting and CRP and thrombin activated platelets and interacts with SHP-1. SHP-1 was immunoprecipitated from whole cell lysates prepared from resting platelets and platelets stimulated with either 10 μg/mL CRP or 5 U/mL thrombin. Samples were western blotted for tyrosine phosphorylated proteins, then stripped and blotted for G6b, followed by SHP-1. Results are representative of three experiments.
To investigate a possible functional role for G6b-B in platelets, the protein was immunoprecipitated from resting and stimulated platelets and analysed for tyrosine phosphorylation. Platelets were stimulated with the GPVI-specific peptide, CRP, and the G protein coupled receptor agonist, thrombin. G6b-B was constitutively phosphorylated on tyrosine residues under resting conditions and underwent a small increase in tyrosine phosphorylation upon stimulation by both agonists (Fig. 3B). The tyrosine phosphatase SHP-1, which is regulated by ITIM receptors, was weakly precipitated with G6b-B under basal conditions and more strongly following stimulation by the two agonists. Importantly, G6b-B was also precipitated by an antibody to SHP-1, with the level of G6b-B in the immunoprecipitate increasing upon stimulation with CRP and thrombin (Fig. 3C). Taken together, these results demonstrate that G6b-B associates with SHP-1 in resting and stimulated platelets, consistent with the idea that the immunoglobulin superfamily protein may function as a novel ITIM receptor in platelets.
Identification of transmembrane proteins in mouse megakaryocytes by SAGE
To complement the proteomics studies, LongSAGE was performed on a highly enriched population of primary mouse bone marrow-derived megakaryocytes that had been allowed to fully differentiate, as indicated by the fact that over 95% of cells had ploidy values of 64n or 128n (Fig. 4). The characteristics of this highly purified preparation have been previously described (12). Sequencing of 53,046 SAGE tags identified 8,316 expressed genes of which approximately 1,200 contain transmembrane domains as predicted by TMHMM v. 2.0 (25). Strikingly, the total number of transmembrane proteins identified by SAGE was greater than eight times that identified by proteomics on the basis of two or more unique peptides. Importantly, however, 81% of the proteins identified in the proteomics studies in human platelets were also identified in mouse megakaryocytes by SAGE (Table 1, and Supplementary Tables 1 and 2), suggesting a high degree of similarity in the membrane proteomes of human platelets and mouse megakaryocytes. Further, the high purity of the SAGE library was verified by the absence of tags for many well-known markers of other haematopoietic lineages, including CD3δ, CD3ε, CD3γ, CD4 and CD8α (T cells), CD19, Igα and Igβ (B cells), F4/80 (macrophages) and CD16 (macrophages, NK cells, neutrophils and myeloid precursors).
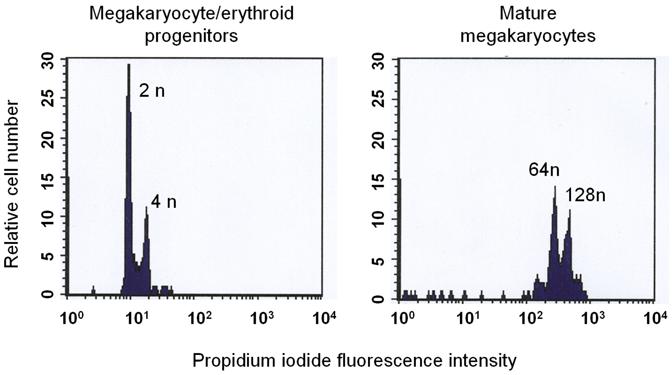
The ploidy of bone marrow-derived megakaryocytic cells was assessed by flow cytometry in the presence of propidium iodide. Mature megakaryocytes (>64n) were used to generate the SAGE library.
The list of membrane proteins that were identified by SAGE includes nearly all of the known platelet surface proteins and, moreover, for the majority of these, there was a good agreement between the number of SAGE tags and their reported levels of expression (Table 1, Supplementary Tables 1 and 2, and data not shown). For example, the major platelet PM protein, integrin αIIb (80,000 copies per platelet), was the most abundant PM protein identified by SAGE (136 SAGE tags). The tetraspanin CD9 (45,000 copies; 34 tags) and the GPIb-IX-V complex (25,000 copies; 21, 31, 11 and 9 tags for GPIbα, GPIbβ, GPIX and GPV, respectively) were intermediate, whereas GPVI (4,000 copies; 6 tags) and P2Y1 (150 copies; 2 tags) had relatively few tags. The near comprehensive coverage of the SAGE library is illustrated by the identification of 20 class I G protein-coupled receptors, of which 18 have been previously reported in platelets (Supplementary Table 6), and the presence of 15 tetraspanins, each of which was verified in mouse megakaryocytes by RT-PCR (Tomlinson and Watson, unpublished). Moreover, the 2 novel class I G protein-coupled receptors are orphans and so have evaded discovery through functional means. Significantly, however, a small number of platelet proteins were not detected by SAGE, including the α2- and α5-integrin subunits and the P2Y12 G protein-coupled ADP receptor, suggesting that the mRNA levels for these genes are relatively low in megakaryocytes. A list of the top 50 transmembrane proteins with the greatest number of SAGE tags is shown in Table 3.
The megakaryocyte SAGE library was compared with 30 other mouse SAGE libraries to identify megakaryocyte-specific expressed genes (Table 4). As anticipated, this identified the integrin αIIb subunit as the major megakaryocyte-specific gene. Strikingly, however, seventeen of the 25 most megakaryocyte-specific expressing genes encoded transmembrane proteins, emphasizing the unique nature of the megakaryocyte surface. This includes all of the proteins that make up the GPIb-IX-V complex, as well as the recently identified type II C-type lectin-like receptor CLEC-2 and the ITIM-containing protein, TREM-like transcript 1 (TLT-1) (4, 43, 47).
Table 3
Fifty most abundant megakaryocyte transmembrane proteins. Each of the 53,046 sequence tags in the mouse megakaryocyte LongSAGE library were identified by comparison to a reference sequence database (SAGEmap tag ug-rel.zip) from SAGEmap at the NCBI website. Genes with predicted transmembrane domains (TMDs) were identified using TMHMM version 2.0 (25).
| NCBI accession number | Gene symbol | Protein name | TMDs | SAGE tags |
|---|---|---|---|---|
| NM_010575 | Itga2b | αIIb integrin subunit | 1 | 136 |
| NM_008410 | Itm2b | Integral membrane protein 2B | 1 | 128 |
| NM_029478 | Tmem49 | Transmembrane protein 49 | 6 | 116 |
| NM_018882 | Gpr56 | G protein-coupled receptor 56 | 7 | 79 |
| NM_007653 | Cd63 | CD63 | 4 | 71 |
| NM_012032 | Serinc3 | Tumor differentially expressed 1 | 11 | 65 |
| NM_001001892 | H2-K1 | MHC Class I | 1 | 63 |
| NM_175015 | Atp5g3 | Mitochondrial H+transporter | 2 | 57 |
| NM_009941 | Cox4i1 | Cytochrome c oxidase subunit IV | 1 | 55 |
| AF201457 | Clec-2 | C-type lectin-like receptor 2 | 1 | 51 |
| NM_007750 | Cox8a | Cytochrome c oxidase VIIIa | 1 | 47 |
| NM_008397 | Itga6 | α6 integrin subunit | 1 | 47 |
| NM_025650 | Uqcr | Ubiquinol-cytochrome c reductase | 1 | 45 |
| NM_026432 | Tmem66 | Transmembrane protein 66 | 2 | 44 |
| NM_133668 | Slc25a3 | Solute carrier family 25 member 3 | 2 | 43 |
| NM_010686 | Laptm5 | Lysosomal-associated TM5 | 4 | 42 |
| NM_011216 | Ptpro | Protein tyrosine phosphatase RO | 1 | 41 |
| NM_026617 | Tmbim4 | Transmembrane BAX inhibitor | 6 | 41 |
| NM_026124 | 1110008 F13Rik | RAB5-interacting protein | 3 | 37 |
| NM_019699 | Fads2 | Fatty acid desaturase 2 | 1 | 37 |
| NM_020258 | Slc37a2 | Solute carrier family 37 member 2 | 1 | 37 |
| NM_009128 | Scd2 | Stearoyl-Coenzyme A desaturase 2 | 4 | 36 |
| NM_007657 | Cd9 | CD9 | 4 | 34 |
| NM_009663 | Alox5ap | 5-lipoxygenase-activating protein | 3 | 33 |
| NM_010581 | Cd47 | CD47 | 5 | 33 |
| NM_053272 | Dhcr24 | 24-dehydrocholesterol reductase | 1 | 32 |
| NM_015747 | Slc20a1 | Solute carrier family 20 member 1 | 8 | 32 |
| NM_028608 | Glipr1 | GLI pathogenesis-related 1 | 1 | 31 |
| NM_010327 | Gp1bb | GPIbβ | 1 | 31 |
| NM_016741 | Scarb1 | Scavenger receptor class B1 | 2 | 31 |
| NM_025378 | Ifitm3 | Interferon-induced TM protein 3 | 2 | 30 |
| NM_025509 | 2310008 M10Rik | DC2 protein | 3 | 29 |
| NM_026155 | Ssr3 | Signal sequence receptor γ | 4 | 29 |
| NM_013532 | Lilrb4 | gp49B | 1 | 28 |
| NM_016906 | Sec61a1 | Sec61α1 subunit | 10 | 28 |
| NM_009775 | Bzrp | Peripheral benzodiazepine receptor | 5 | 27 |
| NM_007806 | Cyba | Cytochrome b-245α | 3 | 27 |
| NM_009768 | Bsg | Basigin | 1 | 26 |
| NM_030694 | Ifitm2 | Interferon-induced TM protein 2 | 2 | 25 |
| NM_008562 | Mcl1 | Myeloid cell leukemia sequence 1 | 1 | 24 |
| NM_133933 | Rpn1 | Ribophorin I | 1 | 24 |
| NM_025468 | Sec11l3 | Sec11-like 3 | 1 | 24 |
| NM_010185 | Fcer1g | Fc receptor γ | 1 | 23 |
| NM_008147 | Gp49a | gp49A | 1 | 22 |
| NM_010326 | Gp1ba | GPIbα | 1 | 21 |
| NM_008640 | Laptm4a | Lysosomal-associated TM4A | 4 | 21 |
| NM_022995 | Tmepai | Nedd4 WW binding protein 4 | 1 | 21 |
| NM_026820 | Ifitm1 | Interferon-induced TM protein 1 | 2 | 20 |
| NM_009842 | Cd151 | CD151 | 4 | 19 |
| AK035304 | P2rx1 | P2X1 ATP receptor | 2 | 19 |
These findings demonstrate that the mouse megakaryocyte SAGE library represents a powerful bioinformatics source for analysis of expression of transmembrane proteins in mature murine megakaryocytes with clear implications for their expression in platelets. The SAGE data has been deposited in the NCBI SAGEmap database (http://www.ncbi.nlm.nih.gov/SAGE/).
Discussion
The main objective of this study was to identify novel receptors expressed on the surface of human platelets using proteomics and to determine which of these proteins are likely to be expressed on mouse platelets using a megakaryoctye SAGE library. The latter information is important because the mouse is the model system of choice for functional studies of novel platelet proteins. Megakaryocytes rather than platelets were chosen as they contain a considerably greater level of mRNA and the application of SAGE to these cells is not hampered by the presence of mitochondrial DNA (20).
In total, 136 transmembrane proteins were identified by proteomics on the basis of identification of two or more unique peptides using three distinct membrane purification procedures, compared with over 1,200 identified by SAGE. While it is likely that the relatively large and more complex megakaryocyte expresses more transmembrane proteins than platelets, the reason for the differences in total numbers may be largely due to a fundamental difference between the two techniques in that genomics detects essentially all expressed genes, but provides no information on protein expression, while proteomics detects protein expression, but preferentially identifies the most highly expressed proteins. In addition, the application of proteomics as used in the present study is critically-dependent on the presence of suitably spaced trypsin-cleavage sites in order to generate peptides of the appropriate size for identification. Such factors may explain why multi-spanning proteins, such as G protein-coupled receptors and tetraspanins, were particularly under-represented in the proteomic study, as was also reported by Moebius et al in their analysis of the platelet membrane proteome (31). This is likely to reflect the low abundance of the majority of these proteins (the tetraspanin CD9, which was detected, is a notable exception with 45,000 copies per platelet) and relatively low number of tryptic cleavage sites, as is typical for small, multispan membrane proteins.
There was, however, a good correlation between reported expression levels of platelet receptors and the number of SAGE tags for a significant number of proteins. Furthermore, the degree of overlap between the genomic and proteomic data was strong, with 81% of the transmembrane proteins identified in human platelets using proteomics being present in the mouse megakaryocyte SAGE library. The remaining 19% may be due to a number of factors, including differences in the levels of expression in the two species, the absence of certain genes from the mouse genome (e.g. FcγRIIA), differential gene expression between the two species (e.g. human but not mouse platelets express PAR1) (23, 32) or differences in expression in megakaryocytes and platelets. We conclude that the combined use of proteomic- and genomic-based approaches represents a powerful way of mapping the platelet membrane proteome.
Our study has also shown that the use of SAGE data alone is a good method for identifying platelet-specific transmembrane proteins. Since SAGE is quantitative, different libraries can be directly compared. Comparison of the megakaryocyte SAGE library to 30 other SAGE libraries, the majority of which are haematopoietic in origin, revealed that transmembrane proteins feature strongly in the list of the most megakaryocyte-specific proteins. Indeed, the 25 most megakaryocyte-specific genes contained 17 with predicted transmembrane domains, including the known platelet marker integrin αIIb and all four components of the GPIb-IX-V complex. The list also included the recently-identified platelet transmembrane proteins CLEC-2 (43), TLT-1 (4, 47) and endothelial cell-selective adhesion molecule (34), for which functions remain to be elucidated. The results of this SAGE analysis suggest that cell specificity is governed to a large extent by the receptors expressed on the cell surface. Similar analyses will facilitate the identification of cell-specific transmembrane proteins in other cell types. Moreover, given that the NCBI SAGEmap depository now contains over 300 human and 200 mouse SAGE libraries, such experiments can be done entirely in silico.
Three different membrane enrichment techniques were used in this study in combination with LC-MS/MS analysis to identify transmembrane proteins expressed in human platelets. A total of 46 PM, 68 IM and 22 proteins of unknown localization were identified by this approach. Eighty-three percent of these were identified by both Mascot and Sequest search algorithms, which correlates well with the study of Elias et al who reported a figure of >85% when evaluating mass spectrometry platforms used in large-scale proteomics investigations (13). Reproducibility between experiments using the same enrichment technique was high for abundant, known platelet surface proteins (e.g. αIIb and β3 integrin subunits and all of the subunits of the GPIb-IX-V complex) and much lower for novel platelet transmembrane proteins (<50%). This was not surprising as low reproducibility (~70%) between replicate data acquisitions of the same sample has been previously reported (13). The lower reproducibility in our study compared with the Elias study is probably largely due to inter-experimental variation, bearing in mind that each set of samples was only analysed once per experiment, but that either two (FFE) or three (WGA and biotin/NA) purifications were performed.
Additional, biochemical and functional studies were performed on one of the novel proteins that was identified in this study, namely G6b, as this is alternatively spliced to seven different isoforms, one of which contains a transmembrane domain and an ITIM, and is therefore a potential inhibitor of platelet activation. To date, only one inhibitory ITIM-containing receptors has been identified in platelets, PECAM-1, which selectively inhibits platelet activation by GPVI (7, 15, 22). A second platelet ITIM receptor, TLT-1, has been reported to support weak platelet activation (4, 47). Biochemical evidence using a G6b-B specific polyclonal antibody confirmed the presence of G6b-B in human platelets and demonstrated that it is constitutively phosphorylated on tyrosine in platelets and that it undergoes a further increase in tyrosine phosphorylation upon stimulation by the GPVI specific agonist CRP and thrombin. Further, the non-receptor protein tyrosine phosphatase SHP-1 is constitutively associated with G6b-B in resting platelets and undergoes an increase in association in parallel with tyrosine phosphorylation. Thus, G6b-B may potentially play an important role in regulating platelet activation by the two ITAM receptors, the collagen receptor GPVI and the low affinity immune receptor, FcγRIIA, through its association with SHP-1. Further work is necessary to determine which other forms of G6b are expressed in platelets and their functional roles.
The initial proteomic studies in platelets used 2-DE in combination with LC-MS/MS (16, 17, 26, 28). These studies reported the presence of a small number of platelet membrane proteins, most likely because many are expressed at low level and because a significant number precipitate during isoelectric focussing. More recently, combined fractional diagonal chromatography technology, a non-gel-based “shot-gun” approach developed by Gevaert and coworkers, was used in combination with MS/MS to study the platelet proteome (30). Sixty-nine platelet transmembrane proteins were identified using this approach, only 12 of which had been previously reported in platelet proteomics studies. Further, Moebius and co-workers used a combination of 1-DE and 16-BAC/SDS-PAGE prior to LC-MS/MS to identify 83 PM and 48 IM proteins (31). However, these investigators report both transmembrane and membrane-associated proteins, such as Gα13 subunit and Rap-1A, which lack transmembrane domains. Taking this into account, the number of proteins predicted to contain transmembrane domains identified by Moebius et al using proteomics was 124 (31), which is similar to that of 136 identified in the present study. The slightly larger number of proteins identified in the present study can be largely attributed to the number of identified IM proteins, which is likely due to the fact that we used FFE to enrich the IM fraction. A direct comparison of the proteomics dataset reported in the present study with that from the Moebius study showed that 62 proteins were identified in both studies, approximately half of which are known platelet PM proteins. This low level of overlap between the two studies is a reflection of the different techniques, but may also be partially inherent to MS/MS studies as pointed out by Elias et al (13). Together, the present study and that from the Moebius group illustrate the requirement for affinity/membrane purification for the identification of platelet membrane proteins using proteomics.
It is beyond the scope of this study to address the question of the functional roles in platelets of novel receptors identified in the study, but it is noteworthy that a number of the identified proteins have either recently been shown to regulate platelet function or to have characteristics which strongly indicate that they may regulate platelet function. Examples of the former group include the immunoglobulin superfamily protein, CD84, which has recently been shown to play an important role in supporting late stage events in platelet aggregation (33); the C-type lectin receptor, CLEC-2, which has been shown to mediate platelet activation through a distinct signalling cascade (43); and the immunoglobulin superfamily protein, G6f, which has been shown to localize Grb2 to the membrane in GPVI-activated platelets (18).
In summary, the present study has illustrated the power of the combined use of proteomic- and genomic-based approaches in identifying proteins in the platelet membrane. It has also highlighted the high degree of similarity in proteins expressed on the surface of human platelets and mouse megakaryocytes, further validating the use of the mouse model for studying the role of platelets in thrombosis. Future studies need to focus on establishing the biological and biochemical functions of the newly identified proteins in the physiological and pathological regulation of platelets, in anticipation that this may lead to the identification of novel targets for anti-thrombotic agents.
Table 4
Twenty-five most megakaryocyte-specific genes. The megakaryocyte SAGE library, of 53,046 sequence tags, was compared to 30 other mouse SAGE libraries, from T lymphocyte (14 SAGE libraries), dendritic cells (6), intra-epithelial lymphocytes (2), embryonic stem cells (2), brain (2), B lymphocyte (1), heart (1), 3T3 fibroblast cell line (1) and P19 embryonic carcinoma cell line (1), using custom written software (!SAGEClus) as described in Cobbold et al. (8). The top 25 most megakaryocyte-specific genes, that had at least 8 megakaryocyte tags, are listed in order of tag number. The number of non-megakaryocyte tags for each gene is not shown, but was between 0 and 5 tags per 1,031,389 total tags. Genes encoding known and predicted transmembrane proteins are shown in bold.
| NCBI accession no. | Gene symbol | Protein name | SAGE tags |
|---|---|---|---|
| NM_010575 | Itga2b | αIIb integrin subunit | 136 |
| NM_011111 | Serpinb2 | Ser/Cys peptidase inhibitor B2 | 71 |
| NM_022029 | Nrgn | Neurogranin | 64 |
| AF201457 | Clec-2 | C-type lectin-like receptor 2 | 51 |
| NM_008397 | Itga6 | α6 integrin subunit | 47 |
| NM_010327 | Gp1bb | GPIbβ | 31 |
| NM_010326 | Gp1ba | GPIbα | 21 |
| AK035304 | P2rx1 | P2X1 ATP receptor | 19 |
| NM_026018 | Pdzk1ip1 | PDZK1-interacting protein 1 | 16 |
| NM_010823 | Mpl | Thrombopoietin receptor | 15 |
| NM_198028 | Serpinb10 | Ser/Cys peptidase inhibitor B10 | 15 |
| XM_110660 | AI427122 | Hypothetical protein LOC102502 | 14 |
| NM_010484 | Slc6a4 | 5-HT transporter | 14 |
| NM_011347 | Selp | P-selectin | 13 |
| NM_027763 | Treml1 | TLT-1 (ITIM-containing receptor) | 13 |
| BC019416 | Tmem40 | Transmembrane protein 40 | 12 |
| NM_018762 | Gp9 | GPIX | 11 |
| NM_029529 | Slc35d3 | Fringe-like 1 | 10 |
| XM_484044 | BC011467 | Hypothetical protein | 9 |
| NM_027102 | Esam1 | Endothelial adhesion molecule | 9 |
| BC003755 | Eya2 | Eyes absent 2 homolog (Drosophila) | 9 |
| NM_008148 | Gp5 | GPV | 9 |
| AK035425 | Ltb4dh | LeukotrieneB4 hydroxydehydrogenase | 9 |
| NM_025926 | Dnajb4 | DnaJ (Hsp40) homolog B4 | 8 |
| NM_172708 | A930013K19Rik | Hypothetical protein LOC231134 | 8 |
Supplementary Material
supldata1
supldata2
supltab1
supltab2
supltab3
supltab4
supltab5
supltab6
Acknowledgments
We would like to thank Miss Donna Holmes and Mr. Neil Shimwell from the Cancer Research-UK Institute, University of Birmingham, for analyzing samples on the ion trap mass spectrometer; Kath Nolan from the Therapeutic Immunology Group, Sir William Dunn School of Pathology, University of Oxford for SAGE advice; and Majd Protty from the Centre for Cardiovascular Sciences, Institute of Biomedical Research, University of Birmingham for excellent secretarial assistance. AG would like to thank The Oxford Glycobiology Institute Endowment for funding. This research was supported by the BHF, Wellcome Trust and Cancer Research-UK.
Abbreviations
| 16-BAC | 16-benzyldimethyl-n-hexadecylammonium chloride |
| CRP | collagen related peptide |
| 1-DE | one-dimensional electrophoresis |
| 2-DE | two-dimensional electrophoresis |
| DTT | dithiothreitol |
| ER | endoplasmic reticulum |
| FFE | free flow electrophoresis |
| GPCR | G protein-coupled receptor |
| HPLC | high performance liquid chromatography |
| IM | intracellular membrane |
| ITAM | immunoreceptor tyrosine-based activation motif |
| ITIM | immunoreceptor tyrosine-based inhibitory motif |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry |
| MS | mass spectrometry |
| NA | NeutrAvidin |
| PM | plasma membrane |
| PGI2 | prostacyclin |
| SAGE | serial analysis of gene expression |
| SCF | Stem Cell Factor |
| SDS-PAGE | sodium dodecylsulfate-polyacrylamide gel electrophoresis |
| SHP-1 | SH2 domain containing protein-tyrosine phosphatase-1 |
| sulfo-NHS-SS-biotin | sulfosuccinimidyl-2-(biotinamido) ethyl-1,3-dithiopropionate |
| TBS | Tris-buffered saline |
| TMD | transmembrane domain |
| WGA | wheat germ agglutinin |
Footnotes
YAS is a BHF Research Fellow; MGT is a MRC New Investigator Award Fellow; ÁG is a Parga Pondal Fellow (Xunta de Galicia, Spain); SPW holds a BHF Chair
References
Full text links
Read article at publisher's site: https://doi.org/10.1074/mcp.d600007-mcp200
Read article for free, from open access legal sources, via Unpaywall:
http://www.mcponline.org/article/S1535947620314055/pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Probing the Interactions of Thiazole Abietane Inhibitors with the Human Serine Hydrolases ABHD16A and ABHD12.
ACS Med Chem Lett, 14(10):1404-1410, 18 Sep 2023
Cited by: 0 articles | PMID: 37849541 | PMCID: PMC10577890
Lost in traffic: consequences of altered palmitoylation in neurodegeneration.
Front Physiol, 14:1166125, 30 May 2023
Cited by: 8 articles | PMID: 37324388 | PMCID: PMC10268010
Review Free full text in Europe PMC
Species Differences in Platelet Protease-Activated Receptors.
Int J Mol Sci, 24(9):8298, 05 May 2023
Cited by: 2 articles | PMID: 37176005 | PMCID: PMC10179473
Review Free full text in Europe PMC
Platelet-derived TLT-1 promotes tumor progression by suppressing CD8+ T cells.
J Exp Med, 220(1):e20212218, 28 Oct 2022
Cited by: 14 articles | PMID: 36305874 | PMCID: PMC9814191
A Novel Morphological Parameter Predicting Fibrotic Evolution in Myeloproliferative Neoplasms: New Evidence and Molecular Insights.
Int J Mol Sci, 23(14):7872, 17 Jul 2022
Cited by: 0 articles | PMID: 35887218 | PMCID: PMC9322985
Review Free full text in Europe PMC
Go to all (96) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Genes & Proteins (Showing 46 of 46)
- (1 citation) UniProt - Q9Y624
- (1 citation) UniProt - P08514
- (1 citation) UniProt - P05106
- (1 citation) UniProt - P12318
- (1 citation) UniProt - O14672
- (1 citation) UniProt - O00264
- (1 citation) UniProt - P11169
- (1 citation) UniProt - Q9UIF2
- (1 citation) UniProt - P13224
- (1 citation) UniProt - P35613
- (1 citation) UniProt - O95870
- (1 citation) UniProt - Q96AP7
- (1 citation) UniProt - Q99720
- (1 citation) UniProt - P23229
- (1 citation) UniProt - P21926
- (1 citation) UniProt - P30490
- (1 citation) UniProt - P16284
- (1 citation) UniProt - Q9NYQ8
- (1 citation) UniProt - Q14392
- (1 citation) UniProt - Q15762
- (1 citation) UniProt - Q8IWY2
- (1 citation) UniProt - P27105
- (1 citation) UniProt - O15173
- (1 citation) UniProt - P05556
- (1 citation) UniProt - P07359
- (1 citation) UniProt - Q7Z5H2
- (1 citation) UniProt - P54762
- (1 citation) UniProt - Q13586
- (1 citation) UniProt - O15430
- (1 citation) UniProt - Q9BX67
- (1 citation) UniProt - O75954
- (1 citation) UniProt - Q9HDC9
- (1 citation) UniProt - P30273
- (1 citation) UniProt - P16109
- (1 citation) UniProt - Q12913
- (1 citation) UniProt - P13598
- (1 citation) UniProt - Q07000
- (1 citation) UniProt - Q96KU3
- (1 citation) UniProt - P17301
- (1 citation) UniProt - P14770
- (1 citation) UniProt - P40197
- (1 citation) UniProt - P16671
- (1 citation) UniProt - Q86UF1
- (1 citation) UniProt - P54709
- (1 citation) UniProt - Q8MHP8
- (1 citation) UniProt - P42892
Show less
Nucleotide Sequences (Showing 7 of 7)
- (2 citations) ENA - AK035304
- (2 citations) ENA - AF201457
- (1 citation) ENA - AK035425
- (1 citation) ENA - BC003755
- (1 citation) ENA - BC019416
- (1 citation) ENA - BC011467
- (1 citation) ENA - AI427122
Show less
RefSeq - NCBI Reference Sequence Database (Showing 64 of 64)
- (2 citations) RefSeq - NM_010575
- (2 citations) RefSeq - NM_010326
- (2 citations) RefSeq - NM_010327
- (2 citations) RefSeq - NM_008397
- (1 citation) RefSeq - NM_010581
- (1 citation) RefSeq - NM_010185
- (1 citation) RefSeq - NM_007806
- (1 citation) RefSeq - NM_133668
- (1 citation) RefSeq - NM_053272
- (1 citation) RefSeq - NM_011347
- (1 citation) RefSeq - XM_484044
- (1 citation) RefSeq - NM_007653
- (1 citation) RefSeq - NM_008148
- (1 citation) RefSeq - NM_008147
- (1 citation) RefSeq - NM_027102
- (1 citation) RefSeq - NM_001001892
- (1 citation) RefSeq - NM_015747
- (1 citation) RefSeq - NM_026018
- (1 citation) RefSeq - NM_012032
- (1 citation) RefSeq - NM_025926
- (1 citation) RefSeq - NM_029529
- (1 citation) RefSeq - NM_026617
- (1 citation) RefSeq - NM_008410
- (1 citation) RefSeq - NM_009941
- (1 citation) RefSeq - NM_011216
- (1 citation) RefSeq - NM_016906
- (1 citation) RefSeq - NM_010686
- (1 citation) RefSeq - NM_009663
- (1 citation) RefSeq - NM_010484
- (1 citation) RefSeq - NM_025650
- (1 citation) RefSeq - NM_198028
- (1 citation) RefSeq - NM_018882
- (1 citation) RefSeq - NM_022029
- (1 citation) RefSeq - NM_019699
- (1 citation) RefSeq - NM_026820
- (1 citation) RefSeq - NM_018762
- (1 citation) RefSeq - NM_016741
- (1 citation) RefSeq - NM_025378
- (1 citation) RefSeq - NM_009768
- (1 citation) RefSeq - NM_028608
- (1 citation) RefSeq - NM_007750
- (1 citation) RefSeq - NM_009775
- (1 citation) RefSeq - NM_008640
- (1 citation) RefSeq - NM_008562
- (1 citation) RefSeq - NM_027763
- (1 citation) RefSeq - NM_026432
- (1 citation) RefSeq - NM_026155
- (1 citation) RefSeq - NM_133933
- (1 citation) RefSeq - NM_022995
- (1 citation) RefSeq - NM_020258
- (1 citation) RefSeq - NM_175015
- (1 citation) RefSeq - NM_025468
- (1 citation) RefSeq - NM_007657
- (1 citation) RefSeq - NM_025509
- (1 citation) RefSeq - NM_009842
- (1 citation) RefSeq - NM_172708
- (1 citation) RefSeq - NM_009128
- (1 citation) RefSeq - NM_013532
- (1 citation) RefSeq - NM_029478
- (1 citation) RefSeq - NM_030694
- (1 citation) RefSeq - NM_011111
- (1 citation) RefSeq - NM_026124
- (1 citation) RefSeq - NM_010823
- (1 citation) RefSeq - XM_110660
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Heparan sulfates are critical regulators of the inhibitory megakaryocyte-platelet receptor G6b-B.
Elife, 8:e46840, 22 Aug 2019
Cited by: 21 articles | PMID: 31436532 | PMCID: PMC6742478
Uncoupling ITIM receptor G6b-B from tyrosine phosphatases Shp1 and Shp2 disrupts murine platelet homeostasis.
Blood, 132(13):1413-1425, 11 Jun 2018
Cited by: 16 articles | PMID: 29891536 | PMCID: PMC6212653
Mice lacking the ITIM-containing receptor G6b-B exhibit macrothrombocytopenia and aberrant platelet function.
Sci Signal, 5(248):ra78, 30 Oct 2012
Cited by: 38 articles | PMID: 23112346 | PMCID: PMC4973664
Targeting platelet inhibition receptors for novel therapies: PECAM-1 and G6b-B.
Platelets, 32(6):761-769, 01 Mar 2021
Cited by: 10 articles | PMID: 33646086
Review
Funding
Funders who supported this work.
British Heart Foundation (1)
The BHF Chair of Cardiovascular Sciences and Cellular Pharmacology
Professor Stephen Watson, University of Birmingham
Grant ID: CH/03/003/15571
Medical Research Council (2)
Tetraspanin superfamily proteins in megakaryocytes and platelets
Dr Michael Tomlinson, University of Birmingham
Grant ID: G0400247
Grant ID: G9818340B




