Abstract
Free full text

Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast
Abstract
The Tup1–Ssn6 corepressor complex regulates the expression of several sets of genes, including genes that specify mating type in the yeast Saccharomyces cerevisiae. Repression of mating-type genes occurs when Tup1–Ssn6 is brought to the DNA by the Matα2 DNA-binding protein and assembled upstream of a- and haploid-specific genes. We have determined the 2.3 Å X-ray crystal structure of the C-terminal domain of Tup1 (accesion No. 1ERJ), a 43 kDa fragment that contains seven copies of the WD40 sequence motif and binds to the Matα2 protein. Moreover, this portion of the protein can partially substitute for full-length Tup1 in bringing about transcriptional repression. The structure reveals a seven-bladed β propeller with an N-terminal subdomain that is anchored to the side of the propeller and extends the β sheet of one of the blades. Point mutations in Tup1 that specifically affect the Tup1–Matα2 interaction cluster on one surface of the propeller. We identified regions of Tup1 that are conserved among the fungal Tup1 homologs and may be important in protein–protein interactions with additional components of the Tup1-mediated repression pathways.
Introduction
In the yeast Saccharomyces cerevisiae, the transcriptional corepressor complex Tup1–Ssn6 regulates genes responsible for a variety of cellular functions including mating-type determination, glucose repression, oxygen repression and DNA repair. The Tup1 tetramer forms a tight complex with Ssn6, with a stoichiometry of four Tup1 molecules per Ssn6 molecule (Varanasi et al., 1996; Redd et al., 1997). Since neither Tup1 nor Ssn6 binds DNA directly, the ability of the corepressor to recognize this broad array of promoters depends on protein–protein interactions with sequence-specific DNA-binding proteins, each of which is specific to a set of target genes. For example, in the regulation of mating-type genes, the DNA-binding protein, Matα2, binds with MCM1 to operator sites upstream of the promoters for a-specific genes (in α and a/α cells) and with Mata1 to the upstream regions of haploid-specific genes (in a/α cells), recruiting the Tup1–Ssn6 corepressor complex (Keleher et al., 1992). Several genetic and biochemical experiments have uncovered direct protein–protein interactions required for the recruitment of the Tup1–Ssn6 corepressor by the Matα2 protein. The homeodomain of Matα2, located at the C-terminus of the protein, binds both DNA (Hall and Johnson, 1987) and the tetratricopeptide repeats (TPR) of Ssn6 (Smith et al., 1995) while the N-terminal domain of Matα2 binds Tup1 (Komachi et al., 1994; Komachi and Johnson, 1997). Other DNA-binding proteins involved in Tup1–Ssn6-mediated repression pathways include Mig1 and Nrg1, which mediate glucose repression (Treitel and Carlson, 1995; Park et al., 1999), Rox1, which controls hypoxic repression (Balasubramanian et al., 1993), and Crt1, a regulator of DNA repair genes (Huang et al., 1998). Although the Tup1–Ssn6 corepressor complex must interact with DNA-binding proteins in order to be targeted to specific promoters, Tup1–Ssn6 can be artificially tethered to DNA by fusion with LexA and, under these conditions, bring about transcriptional repression (Keleher et al., 1992). LexA–Ssn6 without Tup1 represses weakly, but LexA–Tup1 represses well in the absence of Ssn6 (Tzamarias and Struhl, 1994). These results suggest that, once the DNA-binding protein recruits the Tup1–Ssn6 corepressor complex to the correct promoter, Tup1 makes one of the most important interactions with the downstream component(s) in the repression pathway. However, it should be noted that repression by the LexA derivatives of Ssn6 and Tup1 is considerably weaker than that of the active repression complexes, and it is possible that the DNA-binding proteins make additional contacts with downstream components.
Tup1 from S.cerevisiae is a 713 amino acid protein containing three functional domains. The N-terminal domain (residues 1–91) mediates tetramerization of Tup1 (Williams et al., 1991; Varanasi et al., 1996; Jabet et al., 2000) and is necessary for interaction with the TPR motifs of Ssn6 (Tzamarias and Struhl, 1994, 1995). The C-terminal domain of Tup1 (residues 334–713) interacts with the Matα2 protein and contains seven copies of the WD40 repeat (Williams and Trumbly, 1990; Komachi et al., 1994). Present in many proteins with diverse functions, the WD40 repeat is a degenerate sequence repeat that is ~40 amino acids in length and was first identified in the protein Gβ (Fong et al., 1986). WD40 repeats have been characterized as containing a variable region, which is variable in both length and composition, and a core region, which is more uniform in length and contains certain conserved amino acids including GH at the N-terminus and WD at the C-terminus (Neer et al., 1994). The C-terminal WD40 repeat domain of Tup1 binds Matα2 in vitro and, when overexpressed in cells lacking full-length Tup1 and Ssn6, can interact with Matα2 and cause repression of a-specific genes in α cells (Komachi et al., 1994). The presence of all seven WD40 repeats is required for biological activity, with the deletion of even a single WD40 repeat disrupting Tup1-mediated repression (Williams and Trumbly, 1990; Komachi et al., 1994; Tzamarias and Struhl, 1994). The ‘central domain’ of Tup1 (residues 92–316) connects the N-terminal and C-terminal domains and may also play a role in repression (Tzamarias and Struhl, 1994). Deletion studies have provided evidence for direct interactions between this central domain and the N-terminal tails of histones H3 and H4 (Edmondson et al., 1996), and these interactions have been implicated in the mechanism of repression. Transcriptional repression assays have implicated the central domain of Tup1 and part of the C-terminal domain (residues 92–386) as important for repression (Tzamarias and Struhl, 1994).
Several possible mechanisms for Tup1-mediated repression have been proposed. In the case of the regulation of mating-type genes, repression may involve repositioning of nucleosomes (Shimizu, 1991; Cooper, 1994), perhaps involving a direct interaction between Tup1 and the N-terminal tails of histones H3 and H4 (Edmondson, 1996). H3–H4 interactions are probably not a general feature of Tup1–Ssn6-mediated repression because histones H3 and H4 are not implicated in hypoxic-gene repression (Deckert et al., 1998). Moreover, nucleosome positioning per se does not appear to be required for repression of a1/α2-controlled genes or for reporter constructs repressed by α2/MCM1 (Huang et al., 1997; Redd et al., 1997). Some lines of evidence indicate a possible direct interaction between Tup1 and the general transcription machinery. Srb10, Srb11, Sin4 and Rox3 were each identified in genetic screens as being involved in repression by the Tup1–Ssn6 complex (Wahi and Johnson, 1995; Carlson, 1997) and have now been shown to be part of the general transcription machinery (Nonet and Young, 1989; Thompson et al., 1993; Li et al., 1995; Liao et al., 1995). Furthermore, the Srb10/Srb11 kinase is necessary for complete repression by LexA-Tup1–LexA-Ssn6 (Kuchin and Carlson, 1998).
We report here the X-ray crystal structure of a C-terminal 43 kDa fragment of Tup1 from S.cerevisiae (residues 282–713 with an internal deletion of residues 389–431). This C-terminal Tup1 fragment contains all seven of the WD40 repeats and 50 additional amino acids, located N-terminal to the first WD40 repeat, which are conserved among Tup1 proteins from the yeasts S.cerevisiae, Kluyveromyces lactis and Candida albicans. The protein folds into a seven-bladed β propeller with an N-terminal subdomain that extends one of the blades of the propeller. The model of the C-terminal domain of Tup1 presented here provides a framework for understanding the multitude of protein–protein interactions likely to be present in Tup1-mediated repression.
Results and discussion
Crystallization and structure determination
Initial crystallization attempts were carried out with a C-terminal 50 kDa fragment of Tup1, referred to as Tup1c, which contains amino acids 253–713. Tup1c did not crystallize, and analysis of the protein in the crystallization drops by gel electrophoresis revealed a major degradation product (~35 kDa) that resulted from proteolysis in the linker separating WD40 repeats 1 and 2. N-terminal sequencing confirmed that the degradation product corresponded to a fragment of Tup1 containing WD40 repeats 2–7. Since the linker is not required for Tup1 function and is not conserved in other Tup1 family members, a new fragment of Tup1 lacking this protease-sensitive linker region was therefore cloned and expressed (see Materials and methods) and used in the structure determination reported here. This fragment, Tup1cΔ, contains residues 282–713 with a deletion of residues 389–431. The Tup1 crystals belong to the space group P31 with three molecules in the asymmetric unit. The 2.3 Å crystal structure of a 43 kDa C-terminal domain of Tup1 was determined using multiple isomorphous replacement (MIR) phasing techniques with three derivatives: ethylmercurithiosalicylate (EMTS), KAu(CN)2 and di-µ-iodobis(ethylenediamine)diplatinum (II) (PIP). The structure was refined imposing tight non-crystallographic symmetry (NCS) restraints to an Rfree/Rcryst = 26.6/22.8% on all data (see Materials and methods). Data collection and refinement statistics are shown in Table I. The model spans residues 283–710 and contains two disordered regions (Figure 1A–C): one in the loop connecting blades 1 and 2 (residues 385–441) where the internal deletion of residues 389–431 was made; and the other in the loop connecting strands 5C and 5D (residues 607–620). In addition, residues 566–571 in the loop between blades 4 and 5 and residues 306–310 are modeled as alanines because of poor side chain density. The residues have been numbered according to the intact protein, leading to a discontinuity where the protein fragment contains an internal deletion of residues 389–431 (Figure 1C).

Fig. 1. (A) Ribbon representation of the C-terminal domain of S.cerevisiae Tup1 looking down the central axis of the propeller onto the top surface. (B) View rotated 90° about the horizontal axis in (A), looking at the propeller from the side. The blades as well as the ‘top’ and ‘bottom’ surfaces are labeled according to convention and the N- and C-termini are marked. The broken chain between blades 1 and 2 (**) and between strands 5C and 5D in blade 5 (*) represents disordered regions. Side chains important for the Tup1–Matα2 interaction are shown in green as ball-and-stick models. (A) and (B) were generated with RIBBONS (Carson, 1997). (C) Secondary structure elements mapped to the sequence of the C-terminal domain of Tup1. Residue numbers are with reference to the full-length Tup1 protein and secondary structural elements, which were assigned by PROCHECK (Laskowski et al., 1993), are depicted with a cylinder for a 310 helix and ‘→’ for a β strand. The positions where mutations affect the Tup1–Matα2 interaction (Komachi and Johnson, 1997) are shown in blue; the positions of the structural mutations (Carrico and Zitomer, 1998) are shown in orange; and the residues with poor density are shown in green. The double underline is the site of the three residue insertion linking amino acids 388 and 432. (D) Alignment of the seven WD40 repeats of Gβ indicating residues that contact phosducin (Gaudet et al., 1996) (in blue), Gα (Sondek et al., 1996) (in red), or both phosducin and Gα (orange).
Table I.
| Native | EMTS | KAu(CN)2 | PIP | |
|---|---|---|---|---|
| Data collection (~23°C) | ||||
| resolution limit (Å) | 2.30 (2.38–2.30) | 2.65 (2.74–2.65) | 3.30 (3.41–3.30) | 3.00 (3.11–3.00) |
 completeness (%) (outer shell) completeness (%) (outer shell) | 98.8 (95.9) | 75.4 (58.3) | 67.1 (38.2) | 68.1 (62.9) |
 redundancy redundancy | 2.9 | 2.0 | 1.1 | 1.1 |
 <I/σ(I)> <I/σ(I)> | 9.8 | 8.6 | 5.3 | 5.5 |
 Rsym (%)a (outer shell) Rsym (%)a (outer shell) | 8.3 (30.8) | 6.6 (30.4) | 9.0 (19.0) | 6.7 (26.1) |
| MIR phasing (15–2.65 Å) | ||||
 Rderiv (%)b Rderiv (%)b | 23.5 | 15.0 | 17.7 | |
 resolution limit for phasing (Å) resolution limit for phasing (Å) | 2.65 | 4.00 | 4.50 | |
 Rcullis (%)c Rcullis (%)c | 0.61 | 0.83 | 0.89 | |
 phasing powerd phasing powerd | 2.07 | 0.97 | 0.89 | |
 mean figure of merite mean figure of merite | 0.39 | 3-fold NCS averaging and solvent flattening | ||
| Refinement | ||||
 CNSSolve, MLF target CNSSolve, MLF target | ||||
 bulk-solvent correction, individual B factors, tight NCS restraints bulk-solvent correction, individual B factors, tight NCS restraints | ||||
 resolution range (Å) resolution range (Å) | 30–2.3 | |||
 Rcryst/Rfree (%)f Rcryst/Rfree (%)f | 22.8/26.6 (all F) | |||
 r.m.s.d.g r.m.s.d.g | bonds 0.009 Å | angles 1.62° | ||
 average B (Å2) average B (Å2) | 30 (molecule A, 34; molecule B, 30; molecule C, 26) | |||
 r.m.s.d. for Cα superposition of NCS molecules r.m.s.d. for Cα superposition of NCS molecules | 0.11 Å | |||
 53 788 reflections 53 788 reflections | 51 067 working set | 2721 test set | ||
 7982 protein atoms 7982 protein atoms | 265 water molecules | |||
aRsym = 100 × ∑|I – <I>| / ∑I, where I is the integrated intensity of a given reflection.
bRderiv = 100 × ∑|Fderiv – Fnat| / ∑Fnat, where Fderiv and Fnat are the derivative and native structure factor amplitudes, respectively.
cRcullis = ∑||FPH(obs) ± FP(obs)| – FH(calc)| / ∑|FPH(obs) ± FP(obs)|, where FPH and FP are the observed derivative and native structure factor amplitudes, respectively, and FH is the calculated heavy-atom structure factor amplitude.
dPhasing power = [∑|FH|2/∑(|FPH(obs)| – |FPH(calc)|)2]1/2, where FH is the calculated heavy-atom structure factor amplitude and FPH(obs) and FPH(calc) are the observed and calculated derivative structure factor amplitudes, respectively.
eFigure of merit = <∑P(α)exp(iα)/∑P(α)>, where P(α) is the probability distribution for the phase α and i is the unit imaginary number.
fR factor = 100 × ∑|Fobs – Fcalc|/∑Fobs, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rcryst is the crystallographic R factor calculated with all of the data used in refinement (95%) and Rfree is the free R factor calculated with 5% of the data that is chosen randomly and not used in the refinement.
gr.m.s.d. is the root mean square deviation from ideal geometry.
Description of the fold
The C-terminal domain of Tup1 folds into a seven-bladed β propeller (residues 332–710) preceded by a 50 amino acid N-terminal subdomain (residues 283–331) that packs against the side of the propeller (Figure 1A and B). The propeller fold is characterized by seven blades that are pseudosymmetrically arranged around a central axis. Each blade of the propeller is a β sheet composed of four antiparallel β strands. Following the convention for WD40 propellers (Wall et al., 1995; Lambright et al., 1996), the blades are numbered 1–7 and, within each blade, the strands are labeled A, B, C and D from the inside to the outside of the propeller. The loops connecting the strands are labeled according to the two strands that they connect. The ‘top’ face of the propeller is defined as the face that contains the DA loops connecting consecutive blades whereas the ‘bottom’ face is the opposite surface. Six copies of the WD40 repeat were originally identified in the Tup1 sequence (Williams and Trumbly, 1990); however, an additional repeat, repeat 1, which is separated from the N-terminus of repeat 2 by an ~50 amino acid linker has also been noted (Komachi et al., 1994). The crystal structure confirms that Tup1 has seven copies of the WD40 sequence motif. Although the sequences of the WD40 regions of Tup1 and Gβ are only 23% identical, the propeller of Tup1 overlays remarkably well with that of Gβ, with an r.m.s.d. of 1.3 Å over 282 Cα atoms (Figure 3B). Furthermore, the global properties of the seven-bladed, WD40-repeat propeller, which were described for Gβ (Wall et al., 1995; Sondek et al., 1996), are also present in the Tup1 propeller structure. These similarities include the fact that the WD40 sequence repeat originally identified does not correspond to an individual blade. Beginning at the N-terminus of the first WD40 sequence repeat, the first strand is the outer strand (D) of blade 7 and then continues into strands A, B and C of the next blade, blade 1 (Figure 1A and C). The outer strand (D) of blade 1 comes from the next WD40 repeat and the pattern continues, with a single sequence repeat encompassing parts of two consecutive blades. The propeller ends with strand C of blade 7 from the final WD40 repeat. Thus, the propeller is closed by contacts in blade 7 between strand 7D from the first repeat and strand 7C from the seventh repeat, as is observed in Gβ (Wall et al., 1995; Sondek et al., 1996).

Fig. 3. (A) Superposition of all seven blades of Tup1. The Cα backbone for each of the seven blades was aligned with respect to strands 3B and 3C. The four conserved side chains involved in the intrablade hydrogen-bonding network, referred to as the structural tetrad, are shown for blade 3 as ball-and-stick models colored according to atom type and hydrogen bonds are indicated with dashed lines. A representative blade from Gβ, blade 4, is shown for reference. Blade 1, light green; blade 2, red; blade 3, green; blade 4, purple; blade 5, pink; blade 6, cyan; blade 7, orange; Gβ, brown. (B) Superposition of the WD40 propellers. The Cα backbones for Gβ (blue; 1TBG) and Tup1 (yellow), which align with an r.m.s.d. of 1.3 Å, are shown in the same view as Figure 1A and labeled. Least-squares alignments for (A) and (B) were done in O (Jones et al., 1991) and figures generated with RIBBONS (Carson, 1997).
The structure of a single blade is highly conserved both within the Tup1 structure and between Tup1 blades and individual blades in Gβ. For example, blade 3 of Tup1 superimposes with other Tup1 blades with Cα–Cα r.m.s.ds of 0.21–0.88 Å, and with a typical blade of Gβ, blade 4, with an r.m.s.d. of 0.69 Å (Figure 3A). Therefore, the blades of Tup1 are as structurally similar to Gβ as they are to each other. A hallmark of blades in WD40-repeat propellers is the hydrogen-bonding network, or ‘structural tetrad’, observed between some of the most conserved residues in the WD40 repeat (Wall et al., 1995; Sondek et al., 1996). This tetrad is formed between Trp in strand C, Ser/Thr in strand B, His in the DA loop, and the nearly invariant Asp in the tight turn between strands B and C (Figures (Figures1C1C and and3A).3A). The interface between neighboring blades within the propeller domain is primarily hydrophobic, with residues on the adjacent faces of the β sheets in van der Waals contact. The overall packing of the blades within the propeller leads to the formation of a narrow channel along the ~7-fold symmetry axis of the propeller. The channel is ~12 Å in diameter (Cα to Cα) and is filled with solvent molecules that are bound by the carbonyl oxygens and amide groups from the inner strands (A) of each blade. Side chains from small residues, such as serine, cysteine and alanine, also line the channel, and a few larger side chains even extend into the interior of the channel, most notably Lys351 and Asp492. Side chain atoms from Asn354, Arg586 and Asn682, which are located on the bottom surface of the propeller and line the opening to the channel, form a cluster of hydrogen-bond acceptors and donors.
The greatest differences between the Tup1 and Gβ propellers occur in the loop regions. In most WD40 repeats, the length of the β strands and loops connecting the β strands are relatively constant, resulting in a typical length for a WD40 repeat of 40 amino acids. In general, variation in the length of a WD40 repeat occurs because of insertions in the loops. In Gβ, the loops within the WD40 repeats generally differ by no more than three residues. In contrast, the lengths of the loops in Tup1 are more variable, with some significantly longer than those found in Gβ. The lengths of the WD40 repeats of Tup1 vary with insertions of 6–60 amino acids in each of four loops (Figure 1C and D). In the Tup1 crystal structure, the density for each of these loops was poor and was often interpretable in only one of the three molecules of the asymmetric unit (see Materials and methods); thus, these loops are likely to be flexible. Within each blade, the AB and CD loops are the most flexible whereas the BC loops are fixed in length and less conformationally variable (Figure 3). An exception to the conformational invariance of the BC loop occurs in blade 1. This BC loop deviates from those in all of the other blades of both Tup1 and Gβ because it is shorter by one residue, and an Asn replaces the nearly invariant Asp, which is the most conserved residue in all WD40 repeats. This Asp is present in the BC loops of blades 2–7, where it participates in the structural tetrad, as described above, and forms a hydrogen bond with the backbone to stabilize the tight turn. Although the substitution of Asn for Asp is conservative, the shortened BC loop of blade 1 alters the position of Asn364 such that it can not make contacts analogous to the usual Asp (Figure 3B). Instead, in blade 1, Asn364 stabilizes the DA loop connecting blades 1 and 2 as it leads into strand 2A through hydrogen bonds with Leu444 N and Asp443 OD1. One additional consequence of the shortened BC loop is the unusual backbone conformation of the next residue, Lys365, which is classified as a Ramachandran outlier.
The N-terminal 50 amino acids do not form an independent globular domain, but rather form a subdomain that is joined to the propeller by β sheet interactions that extend one blade into a six-stranded sheet. Starting at the N-terminus of the model, the protein forms an extended strand (residues 283–286) followed by one turn of a 310 helix and then another extended strand (residues 296–298) followed by a second turn of a 310 helix. These two extended strands are antiparallel to each other and perpendicular to the axis of the propeller (Figure 1A and B). The second 310 helix is preceded by a loop that leads into the final two β strands of the N-terminal subdomain, n1 and n2. Strands n1 (residues 312–316) and n2 (residues 322–325) are antiparallel and interact using the typical hydrogen bonds between backbone atoms that are found in β sheets. In addition, strand n2 forms backbone hydrogen-bond contacts with strand 6D, extending blade 6 into a six-stranded antiparallel β sheet (Figure 2A). Interaction between the N-terminal and propeller subdomains is primarily mediated by hydrophobic residues from the N-terminal subdomain contacting side chains located near the interfaces of neighboring blades (Figure 2B). For example, residues from strand n2 of the blade 6 extension, Tyr321, Ile323 and Tyr325, are in van der Waals contact with side chains that line the faces of the sheets between neighboring blades 6 and 7. Phe301 makes non-polar contacts with Pro664, a side chain on the face of blade 6 that is adjacent to blade 5. The largest hydrophobic pocket is between blades 4 and 5, which is lined by residues Lys576, Asp597, Ser599, Lys601, Thr625 and Ile627, and accommodates His294, which is a side chain from the second extended strand in the N-terminal subdomain. These extensive non-polar contacts are further buttressed by polar contacts scattered across the interface. The interface between the N-terminal subdomain and the propeller buries 2395 Å2 of surface area. In contrast to the close association of the N-terminal subdomain of Tup1 with the propeller, the residues in Gβ that precede the propeller form a helical subdomain that makes few contacts with the propeller domain (Wall et al., 1995; Lambright et al., 1996). Instead, the N-terminal helix of Gβ is tightly associated with an accessory protein, Gγ, which interacts extensively with the propeller.
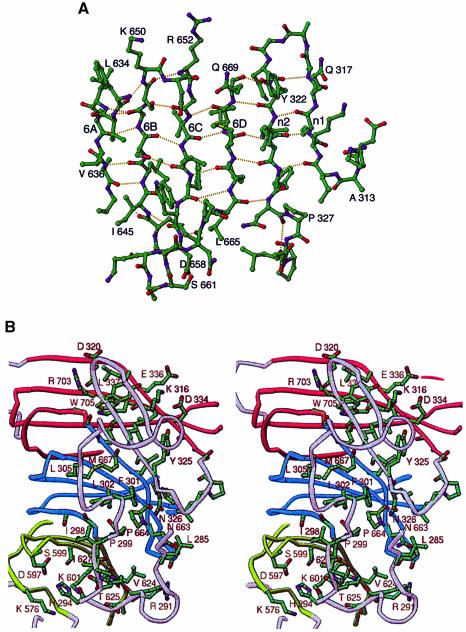
Fig. 2. Packing of the N-terminal fragment against the propeller. (A) Extended β sheet formed by blade 6 and strands n1 and n2 from the N-terminal fragment. Hydrogen bonds are indicated with dashed lines. (B) Stereoview of contacts between the N-terminal fragment (283–331) and the propeller. Side chain and backbone atoms are superimposed on the Cα trace. Some side chain labels were omitted for clarity. The approximate boundaries of each blade are shown in different colors (blade 5, light green; blade 6, blue; blade 7, red; N-terminal fragment, gray). This figure was generated with RIBBONS (Carson, 1997).
As discussed above, the protein used in the structure determination contains a deletion that replaces residues 389–431 with the three amino acid linker, KDP, which joins residues 282–388 to residues 432–713. In a comparison of seven Tup1 sequences from different species of fungi, only the S.cerevisiae and K.lactis Tup1 proteins contain an extended linker between blades 1 and 2. The linkers from these two Tup1 proteins bear little sequence similarity to one another, other than an abundance of polar residues. This linker is unlikely to play a functional role because the C.albicans Tup1 protein, which lacks this linker region, represses a genomic a-specific reporter gene in S.cerevisiae (Braun and Johnson, 1997). Moreover, in yeast cells lacking endogenous Tup1, a full-length Tup1 protein with the deletion of residues 389–431 complements for several and perhaps all of the Tup1 functions (K.Komachi and A.D.Johnson, personal communication), further suggesting that this region is not required for the known functions of the Tup1 protein. The linker in S.cerevisiae Tup1 is therefore likely to be a flexible loop that does not significantly contribute to the repressor properties of Tup1, particularly repression of the mating-type genes. In the crystal structure reported here, poor density is observed for the residues adjacent to the deleted linker, whereas the ordered region of the loop extends away from the top surface of the propeller and adopts a conformation that is distinct from all of the other DA loops between adjacent blades (Figure 3A). Although the conformation of the linker in the intact protein has not been determined, the linker is likely to be a flexible loop that does not form significant contacts with the rest of the C-terminal domain.
Although the C-terminal domain of Tup1 crystallizes with three molecules in the asymmetric unit, there is no evidence that the contacts observed between NCS-related molecules reflect formation of a possible biologically relevant multimer of the C-terminal domain. Little surface area is buried at the interface between NCS-related molecules, ranging from 192 to 407 Å2, indicating that the interaction energy is very weak (Janin and Rodier, 1995). In solution, both the linker-deleted fragment used in this study and a fragment of Tup1 containing residues 253–713 are monomeric, as determined by sedimentation equilibrium analytical ultracentrifugation (data not shown). Although the full-length Tup1 protein is a tetramer whose oligomerization is mediated by the N-terminal 91 residues (Varanasi et al., 1996), overexpression of the monomeric C-terminal domain in yeast partially suppresses the mating-type defects in cells whose TUP1 and SSN6 genes have been disrupted (Komachi et al., 1994). One interpretation of this result is that Tup1 can function, albeit weakly, as a monomer.
Mapping of Tup1 mutations
Studies of protein fragments and mutant proteins have shown definitively that the C-terminal domain of Tup1 interacts with at least one promoter-specific DNA-binding protein, Matα2 (Komachi et al., 1994; Komachi and Johnson, 1997). Eleven point mutations in Tup1 that specifically affect its interaction with Matα2 have been isolated (Komachi and Johnson, 1997). All of these mutations are located on the top face of the propeller, as shown in Figure 1A–C. Mutations found in the DA loops include C348R, Y445C, N673S and S674P; mutations found in the BC turns include E463N and K650N; and mutations found in strand A, near the top face, include S448P, Y489H, Y580H, L634S, I676T and I676V. With the exception of blade 4, mutations affecting interactions with Matα2 have been identified in every blade. Because of the repetitive and symmetric nature of the propeller fold, all of the DA loops and BC turns are found on the top surface of the propeller, whereas all of the AB and CD loops are found on the bottom surface of the propeller. All of these critical side chains cluster on the top surface of the propeller, near the center, and are accessible to solvent. Model building confirms that the side chain substitutions that disrupt α2-mediated repression are not structural mutations, because the mutant side chains can be modeled as one of the common side chain rotamers without steric clashes. Surface localization of these eleven mutations strongly supports the model that the Matα2 protein interacts with Tup1 via the top surface of the propeller (Komachi and Johnson, 1997).
A striking similarity in protein–protein interaction interfaces is seen when the putative Tup1–α2 interface is compared with both the Gβγ–Gα and Gβγ–phosducin complexes. In each of the complexes with Gβγ, the interaction between Gβγ and the partner protein is mediated by two surfaces on Gβγ: a side surface, which is unique for each complex, and a top surface, which is similar for both complexes (Wall et al., 1995; Lambright et al., 1996). The residues in Gβ that mediate these contacts with each of the partner proteins are highlighted in Figure 1D. Because the Tup1 mutations discussed above do not affect the general repression function of Tup1 (Komachi and Johnson, 1997), Tup1 most likely interacts with downstream proteins in the repression pathway, such as components of the transcription machinery, using a different subset of residues, perhaps a different face or side of the propeller. The location of the S448P mutation in strand A, pointing into the solvent channel, and the positioning of four more critical residues in strand A, around the top entrance into the channel, raises the possibility that Tup1 may also use at least a portion of the channel to interact with Matα2. The channel could accommodate an extended strand without major conformational adjustments, similar to the β hairpin that projects into the central channel in galactose oxidase, a seven-bladed propeller that does not contain WD40 repeats (Ito et al., 1994).
Point mutations in the C-terminal domain of Tup1 that have differential effects on the expression of four Ssn6–Tup1-repressed reporter genes have also been reported (Carrico and Zitomer, 1998) (Figure 1C). These mutations derepress the a-mating type and hypoxic reporter genes while having little effect on the expression of genes that regulate flocculence and glucose response. Four of the five mutations replace the conserved Ser/Thr that is located in strand B in the core of each blade with a proline (T460P, S647P, S593P and T695P). This Ser/Thr is at the center of the structural tetrad, described above, making hydrogen bonds with the conserved Trp in strand C of the same blade and with the conserved His in the DA loop leading into strand A of the same blade (Figure 3A). The fifth mutation substitutes proline for Ser595, which is within hydrogen-bonding distance of the backbone carbonyl oxygens of residues 577 and 578. The wild-type side chain at each of the five positions is involved in polar intramolecular contacts, either directly or through a bound water that is conserved in each of the three molecules in the asymmetric unit. Model building shows that the substitution of proline for each of the five Ser/Thr residues disrupts the hydrogen bonds detailed above and causes steric clashes with nearby backbone and side chain atoms. The conclusion from this analysis is that these five mutations are likely to perturb the overall structure of the propeller and therefore do not identify specific residues in Tup1 that interact with promoter-specific DNA-binding proteins or with other general repression targets. The differential effects on repression observed for these mutations probably arise because the various promoters have different requirements for the WD40 domain of Tup1.
Analysis of conserved residues in seven Tup1 sequences
The sequences of all seven known fungal Tup1 homologs were compared in order to identify conserved residues that may be required for the in vivo function of Tup1. The sequence identity for each of the six homologs of S.cerevisiae Tup1 (residues 283–710) ranges from 50% for Dictyostelium discoideum (slime mold) Tup1 to 76% for K.lactis Tup1. The sequence identity over the 43 kDa C-terminal domain of Tup1 is extensive, with clusters of both high and low identity. A molecular surface representation of S.cerevisiae Tup1 that is colored by sequence identity highlights four regions of high sequence conservation (Figure 4). The largest region of sequence conservation is on the top surface centered around the channel and encompasses all of the DA and BC loops, in addition to all of the residues that were previously described as required for the interaction with Matα2 (Figure 4A). This area of high sequence conservation extends into the channel of propeller (Figure 4A). In contrast to the top surface, the bottom surface has only a small patch of sequence conservation that includes the AB loop of blade 2 (Figure 4B). The bottom surface also differs from the top surface because of a large region of negative electrostatic potential distributed over the entire bottom surface. The side surface of the propeller exhibits the lowest sequence conservation, with the exception of the outer strand (D) of blade 4 (Figure 4). Stretches of conserved buried residues are found in several of the blades in strand B; however, they presumably help to stabilize the overall protein fold rather than mediate protein–protein interactions (Figure 4). Most of the conserved buried residues, such as the Ser/Thr and Trp that are part of the conserved structural tetrad mentioned above, are characteristic of the WD40 repeat family (Neer et al., 1994).
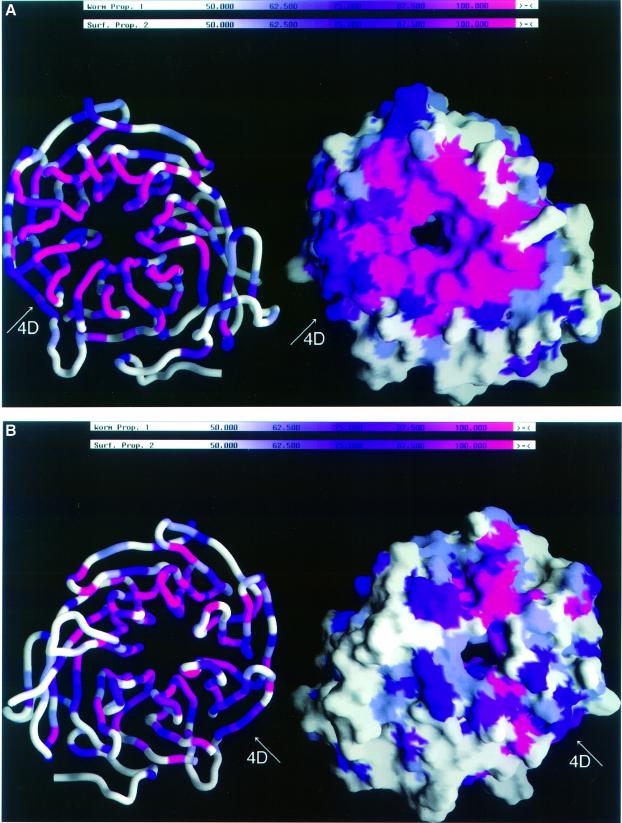
Fig. 4. Sequence identity of seven fungal Tup1 proteins mapped onto the molecular surface and Cα backbone of the C-terminal domain of the Tup1 structure. (A) View looking onto the top surface of the molecule, similar to the view in Figures Figures1A1A and and3B.3B. (B) View rotated 180° about the vertical axis in (A), looking onto the bottom surface of the molecule. The color gradient is a continuum from white to purple to pink, representing residues with <50% identity, residues with 75% identity and residues with 100% identity, respectively. Strand 4D displays significant sequence identity when compared with all of the other outer strands of the propeller. Tup1 sequences (P16649, P56094, P56093, O14432, Q09715, P78706 and O76734) were aligned using ClustalW 1.7 and displayed in GRASP (Nicholls et al., 1991).
The presence of four discrete regions of high sequence conservation suggests that all seven Tup1 homologs interact with one or more common proteins using these conserved surfaces. One possibility suggested by our structure and by the mutation data presented above is that the C-terminal domain of Tup1 uses residues centered around the channel on the top surface of the propeller to interact with the Matα2 protein. It is possible that this surface is conserved in all seven species where Tup1 homologs are found because they all have a Matα2-like protein. Matα2 homologs have been found in K.lactis (M.Redd and A.D.Johnson, unpublished data) and in C.albicans (Hull and Johnson, 1999); however, they have yet to be identified in other species of fungi. An intriguing possibility is that Tup1 uses one or more of the conserved surfaces to interact with proteins in the general repression pathway, such as components of the transcription machinery or chromatin.
Implications for Tup1 function
A universal feature of Tup1–Ssn6-mediated repression is that sequence-specific DNA-binding proteins must bind upstream of the promoters of the genes that are regulated in order to recruit Tup1–Ssn6 to the repression complex, thereby causing transcriptional repression. The best characterized of the many protein–protein interactions necessary for Tup1–Ssn6-mediated repression are between the Tup1–Ssn6 complex and Matα2. Biochemical studies have demonstrated direct interactions between the N-terminal domain of Matα2 and the WD40 domain of Tup1 (Komachi et al., 1994; Komachi and Johnson, 1997), as well as between the C-terminal DNA-binding domain of Matα2 and the TPR motifs of Ssn6 (Smith et al., 1995). Three of the four residues in Matα2 whose substitutions disrupt the interaction with Tup1 are located in the extreme N-terminus of Matα2 (Ile4, Leu9 and Leu10), referred to as the terminal peptide; the fourth residue is Gly71 (Komachi et al., 1994). Extending the analogy of the protein–protein interactions between Gα–Gβγ and phosducin–Gβγ discussed above, Matα2 may contact the top surface of Tup1 with one section of the protein while contacting another surface, such as the conserved side surface on the outer strand (D) of blade 4, with the N-terminal peptide. Many other types of protein–protein contacts involving one or more interacting interfaces between Matα2 and the conserved surfaces of Tup1 can be envisioned. For example, the location of one of the mutations in Tup1 that affects its interaction with Matα2 in the central channel suggests that Matα2 may sit on the top surface of Tup1 with the terminal peptide protruding into the central channel. Side chains that line the opening to the channel on the opposite surface could position the peptide through hydrogen-bond interactions. Other DNA-binding proteins that regulate transcription through the Tup1–Ssn6 pathway, such as Mig1, Nrg1, Rox1 and Crt1, may bind either Tup1 or Ssn6, or even both proteins (Treitel and Carlson, 1995; Tzamarias and Struhl, 1995; Huang et al., 1998; Ostling et al., 1998; Park et al., 1999). Each of these proteins binds DNA via a different class of DNA-binding domain, and none shares a global sequence similarity with another. Some of the properties of the interaction between the DNA-binding proteins and the Tup1–Ssn6 complex, such as the use of the conserved Tup1 surfaces, may be shared among different protein–protein combinations.
Another transcriptional corepressor that is also a member of the WD40-repeat family of proteins is the Drospholia Groucho protein, which regulates developmental gene transcription (Fisher and Caudy, 1998). The sequence similarity between Groucho and Tup1 is limited, although both proteins share common features such as N-terminal-mediated tetramerization, direct interaction with DNA-binding proteins and seven WD40 repeats (~20% identity with the WD40 repeats in Tup1) (Varanasi et al., 1996; Chen et al., 1998; Fisher and Caudy, 1998). It is unclear whether or not any mechanistic details of repression are shared between these two proteins. For example, Groucho is recruited to specific promoters through direct interaction with a tetrapeptide at the C-terminus of partner DNA-binding proteins (Fisher and Caudy, 1998) whereas there is no general signature for the analogous recruitment of the Tup1–Ssn6 complex. Tup1-mediated repression may involve interactions with the general transcription machinery (Wahi et al., 1998) whereas Groucho appears to act by recruiting a histone deacetylase (Chen et al., 1999). One common feature is that both Tup1 and Groucho have been shown to interact directly with the N-terminal tails of histones (Edmondson et al., 1996; Palaparti et al., 1997). An indication that Tup1 and Groucho may have some functional similarity is that two of the conserved surfaces of Tup1, which were described above and are shown in Figure 4, are also conserved in Groucho and its mammalian homologs, the transducin-like Enhancer of split (TLE) proteins. The conserved region on the top surface of the propeller is also present in Groucho/TLE, but somewhat smaller. In addition, the small region of sequence conservation on the bottom surface, which is in the AB loop of blade 2, is also found in Groucho proteins. The observation that these regions of identity are conserved across the Tup1 and Groucho/TLE proteins raises the exciting possibility that they may share a common binding partner, linking the mechanisms of transcription in WD40-repeat transcriptional corepressors. Additional structural and biochemical studies on Tup1 and Groucho repression complexes will enhance our understanding of how corepressors regulate gene expression.
Materials and methods
Preparation of the C-terminal domain of Tup1
The Tup1 protein fragment used in the structural study, Tup1cΔ, was expressed in Escherichia coli from plasmid pER7, which encodes a 43 kDa C-terminal fragment of S.cerevisiae Tup1 containing amino acids 282–388 and amino acids 432–713 joined by a three amino acid linker, KDP. pER7 was constructed by PCR amplification of the appropriate cDNA from pKK719 (a gift from K.Komachi), which encodes a full-length version of the Tup1 protein containing the deletion described above. The amplified cDNA was subcloned into the NdeI and XhoI sites in the polylinker of the T7 expression vector, pHB40P.
BL21(DE3) cells transformed with pER7 were grown at 37°C in Luria–Bertani medium with carbenicillin (100 µg/ml) to an optical density of 0.9 at 600 nm. Cell cultures were equilibrated at 30°C prior to induction with 0.4 mM isopropyl-β-d-thiogalactopyranoside. Cultures were harvested by centrifugation 3 h later, and the pellets were stored at –80°C. Thawed cells were resuspended in lysis buffer (2 M urea, 100 mM Tris pH 8, 500 mM NaCl, 1 mM EDTA, 10 mM β-ME, 0.1% (v/v) NP-40 and 0.1 mg/ml AEBSF) and lysed by passage through a microfluidizer (Microfluidics) at 100 psi. After centrifuging the lysate for 30 min at 8000 r.p.m. in a GSA rotor, polyethyleneimine (PEI) was added to the lysis supernatant to a final concentration of 0.5% (v/v) and stirred for 60 min at 4°C. The PEI precipitate was pelleted by centrifuging for 20 min at 12 000 r.p.m. in a GSA rotor and discarded. Solid ammonium sulfate was added to the PEI supernatant until 30% saturation was reached. After stirring at 4°C for 40 min, insoluble proteins were pelleted by centrifugation for 20 min at 8000 r.p.m. in a GSA rotor. The pellet was discarded, and solid ammonium sulfate was slowly added to the supernatant until 65% saturation was reached. After stirring at 4°C for 40 min, insoluble proteins were pelleted by centrifuging for 20 min at 8000 r.p.m. in a GSA rotor. The 65% ammonium sulfate pellet, which contained Tup1cΔ, was resuspended in 1 M urea, 50 mM Tris pH 8, 250 mM NaCl, 1 mM EDTA and 10 mM β-ME and dialyzed overnight against 20 mM bis-Tris propane pH 9.5, 20 mM NaCl, 1 mM EDTA, 10 mM β-ME and 0.4 mM AEBSF. The soluble protein after dialysis was clarified by centrifuging for 30 min at 18 000 r.p.m. in an SS34 rotor. Tup1cΔ was further purified on a Q Sepharose FF 26/10 column (Amersham Pharmacia Biotech) at pH 9.5 with a gradient from 2 to 250 mM NaCl and a phenyl Sepharose HP 16/10 column (Amersham Pharmacia Biotech) at pH 7.6 with a gradient from 1 to 0 M ammonium sulfate. The final purification step was carried out on a HiLoad S200 Superdex 26/60 gel filtration column (Amersham Pharmacia Biotech) equilibrated with 20 mM Tris pH 8, 150 mM NaCl, 1 mM EDTA and 0.5 mM bis(2-mercaptoethyl)sulfone (BMS) (Calbiochem). Peak fractions were pooled, dialyzed against storage buffer (10 mM Tris pH 8, 50 mM NaCl and 1 mM DTT or 0.5 mM BMS), and concentrated in a stirred-cell concentrator (Amicon) to 20 mg/ml. The protein was stored in aliquots at –80°C until needed.
Tup1c was expressed in BL21(DE3) cells transformed with pMR44, a plasmid that encodes S.cerevisiae Tup1 amino acids 253–713. Expression and purification of Tup1c was similar to that described above for Tup1cΔ.
Crystallization and structure determination
Tup1cΔ crystals were grown by hanging-drop vapor diffusion by mixing an equal volume of protein and reservoir solution containing 50–100 mM bis-Tris propane pH 9, 0–50 mM NaCl, 23–26% (w/v) polyethylene glycol 6000 and 2 mM dithiothreitol or 1 mM BMS, and allowing the drop to equilibrate with the reservoir at 20°C. Crystals grew to an average size of 0.1 × 0.1 × 1 mm in ~1 week. Native crystals were transferred to a drop containing well solution immediately prior to data collection. Heavy-atom derivatives were prepared by soaking crystals in a solution containing 50 mM bis-Tris propane pH 9 and 24% (w/v) PEG 6000 with the heavy-atom reagent (0.5 mM EMTS for 12 h, 1 mM KAu(CN)2 for 22 h or 0.5 mM PIP for 12 h). Data were collected at room temperature on a RAXIS IIc detector equipped with a rotating-anode Rigaku RU-200 generator with CuKα radiation. Attempts to cryocool the crystals at –180°C were unsuccessful. All data were integrated and reduced using the DENZO/SCALEPACK program suite (Otwinowski and Minor, 1997), and subsequent manipulations were performed with the CCP4 program suite (CCP4, 1994). Intensity data were converted to amplitudes with TRUNCATE (CCP4, 1994), and data sets were scaled with SCALEIT (CCP4, 1994). Tup1cΔ crystals form in the space group P31 with unit cell dimensions of a = b = 119.28 Å, c = 77.07 Å, α = β = 90°, γ = 120° and contain three molecules in the asymmetric unit with a solvent content of 50%, as calculated from the Matthew’s coefficient (Matthews, 1968).
The structure of Tup1cΔ was solved by MIR. Heavy-atom positions were located using a combination of molecular replacement, difference Fourier and Patterson methods. Because mirror symmetry in the diffraction pattern suggested that the crystal contained apparent 2-fold symmetry axes perpendicular to the 3-fold screw axis and along the a and b crystallographic axes, as well as along the ab diagonal, the space group was originally assigned as P3121 or P3221. A molecular replacement search with AMoRe (Navaza, 1994) in space groups P3121 and P3221 was performed using a model constructed from the propeller of Gβ (1TBG; residues B47–B340) with all amino acids that were not identical between Tup1 and Gβ substituted with alanines. The best molecular replacement solution was obtained in the space group P3121 with one molecule in the asymmetric unit. Model phases were calculated with SFALL (CCP4, 1994) and used in a cross-difference Fourier map to locate the position of the first mercury atom. Self-difference Fourier maps were calculated to assign the remaining mercury positions, and phases calculated from the mercury derivative were used in cross-difference Fourier maps to locate all additional heavy-atom positions in the gold and platinum derivatives. All heavy-atom sites were verified by Patterson methods, and the six mercury sites correlated with the cysteine positions in Tup1, which are not conserved in Gβ, confirming the accuracy of the rotation of the 7-fold pseudosymmetric molecular replacement model. Heavy-atom refinement and phase calculation with MLPHARE (CCP4, 1994) resulted in an overall figure of merit of 0.51 (15–2.7 Å). The electron density map was improved by solvent flattening and histogram matching with DM (CCP4, 1994), and a model was built using O (Jones et al., 1991). No phase information from the molecular replacement solution was included in the calculation of electron density maps.
After tracing ~85% of the main chain atoms and 85% of the side chains for which there were main chain atoms, the model underwent several rounds of positional refinement and B-factor refinement using CNS (Brunger et al., 1998) with the MLF target (Adams et al., 1997). After each round, σA-weighted 3Fo – Fc, 2Fo – Fc and Fo – Fc maps were calculated, and the model was adjusted to agree with the density. Despite reasonable geometry and an excellent Ramachandran profile, refinement using all of the data from 30 to 2.3 Å stalled at Rfree/Rcryst = 42/38%, indicating a possible error in the space group assignment. The entire structure solution process was recalculated in the lower symmetry space group P31, and a molecular replacement search using the Tup1 model that was built in the original space group, P3121, was used to locate two molecules in the asymmetric unit. Subsequent analysis of the isomorphous difference Pattersons and cross-difference Fourier maps, however, suggested the presence of a third molecule. Heavy-atom site assignment and phase calculations proceeded as described above with a total of 13 mercury, six gold and five platinum sites resulting in an overall figure of merit of 0.39 (15–2.65 Å). The resulting electron density maps, which confirmed the presence of a third molecule, were improved by NCS averaging of the three molecules in the asymmetric unit using the RAVE program suite (Jones, 1992; Kleywegt and Jones, 1994). All subsequent model manipulations were done in O (Jones et al., 1991). The three molecules in the asymmetric unit are related by non-crystallographic symmetry as follows: molecules A and B are related by a 120° rotation about an axis parallel to the crystallographic 31 axis, molecules B and C are related by a non-crystallographic dyad parallel to the crystallographic b axis, and molecules C and A are related by a dyad parallel to the crystallographic [110] axis.
The model was refined with several rounds of positional refinement, restrained group and individual B-factor refinement, and simulated-annealing with torsion angle refinement followed by calculation of σA-weighted 3Fo – Fc, 2Fo – Fc and Fo – Fc maps and manual model rebuilding. Strict NCS was imposed in the initial rounds and then relaxed to a set of restraints that minimized the free R factor. For most of the model, with the exceptions detailed below, an effective force constant for non-crystallographic symmetry (NCS) positional restraints of 300 kcal/mol Å2 was applied. For the two loops for which there was defined density in all three monomers (residues 301–308 and 682–691), the force constant was relaxed to 75 kcal/mol Å2. Only backbone atoms were restrained for those residues whose sidechains were not ordered in all three molecules. In addition, residues involved in NCS contacts and loops not defined for all molecules were excluded. Waters were placed into Fo – Fc peaks of at least 3σ with the requirement that there be at least one reasonable hydrogen-bonding partner. The structure has been refined with tight NCS restraints in CNS (Brunger et al., 1998) using all data from 30 to 2.3 Å, to a crystallographic R value of 22.8% and a free R value of 26.6%. The final model is similar for each of the three molecules in the asymmetric unit and includes residues 283–383, 441–566, 573–605, 621–710 for molecule A (9.7% of side chains modeled as alanines); residues 283–383, 443–566, 573–605, 621–710 for molecule B (5.7% of side chains modeled as alanines); residues 283–384, 442–606, 621–710 for molecule C (8.4% of side chains modeled as alanines); and 265 water molecules. An analysis with PROCHECK (Laskowski et al., 1993) shows that the model has excellent stereochemistry, with 88.1% of the total non-glycine and non-proline residues lying in the most favorable regions of the Ramachandran plot and three residues (Lys365 in each of the three molecules) in the disallowed regions. The average temperature factor for each of the three protein molecules in the asymmetric unit is 34, 30 and 26 Å2 for molecules A, B and C, respectively. All superpositions and distances referred to in the text are with respect to molecule C, which is the most complete molecule and has the lowest overall average temperature factor. The buried surface area at the N-terminal subdomain–propeller interface was calculated using the method of Lee and Richards (1971) with a probe radius of 1.4 Å as implemented in CNS (Brunger et al., 1998).
Acknowledgements
We thank N.LaRonde-LeBlanc, C.Garvie, A.VanDemark, J.Aishima and C.Foster for critical reading of the manuscript. E.Lattman, A.Gittis, A.Batchelor, D.Piper and R.Campbell provided crystallographic advice. K.Komachi provided plasmid pKK719. We also thank S.Soisson for help in displaying sequence identity using GRASP. This work was supported by a National Science Foundation Predoctoral Fellowship to E.R.S. and a grant from the National Science Foundation (MCB 98-08412) to C.W. Coordinates have been deposited in the Protein Data Bank (accession number 1ERJ).
References
- Adams P.D., Pannu,N.S., Read,R.J. and Brunger,A.T. (1997) Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl Acad. Sci. USA, 94, 5018–5023. [Europe PMC free article] [Abstract] [Google Scholar]
- Balasubramanian B., Lowry,C.V. and Zitomer,R.S. (1993) The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol. Cell. Biol., 13, 6071–6078. [Europe PMC free article] [Abstract] [Google Scholar]
- Braun B.R. and Johnson,A.D. (1997) Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science, 277, 105–109. [Abstract] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [Abstract] [Google Scholar]
- Carlson M. (1997) Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol., 13, 1–23. [Abstract] [Google Scholar]
- Carrico P.M. and Zitomer,R.S. (1998) Mutational analysis of the Tup1 general repressor of yeast. Genetics, 148, 637–644. [Europe PMC free article] [Abstract] [Google Scholar]
- Carson M. (1997) Ribbons. Methods Enzymol., 277, 493–505. [Abstract] [Google Scholar]
- Chen G., Nguyen,P.H. and Courey,A.J. (1998) A role for Groucho tetramerization in transcriptional repression. Mol. Cell. Biol., 18, 7259–7268. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev., 13, 2218–2230. [Europe PMC free article] [Abstract] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [Abstract] [Google Scholar]
- Cooper J.P., Roth,S.Y. and Simpson,R.T. (1994) The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev., 8, 1400–1410. [Abstract] [Google Scholar]
- Deckert J., Torres,A.M., Hwang,S.M., Kastaniotis,A.J. and Zitomer,R.S. (1998) The anatomy of a hypoxic operator in Saccharomyces cerevisiae. Genetics, 150, 1429–1441. [Europe PMC free article] [Abstract] [Google Scholar]
- Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [Abstract] [Google Scholar]
- Fisher A.L. and Caudy,M. (1998) Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev., 12, 1931–1940. [Abstract] [Google Scholar]
- Fong H.K., Hurley,J.B., Hopkins,R.S., Miake-Lye,R., Johnson,M.S., Doolittle,R.F. and Simon,M.I. (1986) Repetitive segmental structure of the transducin β subunit: homology with the CDC4 gene and identification of related mRNAs. Proc. Natl Acad. Sci. USA, 83, 2162–2166. [Europe PMC free article] [Abstract] [Google Scholar]
- Gaudet R., Bohm,A. and Sigler,P.B. (1996) Crystal structure at 2.4 Å resolution of the complex of transducin βγ and its regulator, phosducin. Cell, 87, 577–588. [Abstract] [Google Scholar]
- Hall M.N. and Johnson,A.D. (1987) Homeo domain of the yeast repressor α2 is a sequence-specific DNA-binding domain but is not sufficient for repression. Science, 237, 1007–1012. [Abstract] [Google Scholar]
- Huang L., Zhang,W. and Roth,S.Y. (1997) Amino termini of histones H3 and H4 are required for a1–α2 repression in yeast. Mol. Cell. Biol., 17, 6555–6562. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang M., Zhou,Z. and Elledge,S.J. (1998) The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell, 94, 595–605. [Abstract] [Google Scholar]
- Hull C.M. and Johnson,A.D. (1999) Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science, 285, 1271–1275. [Abstract] [Google Scholar]
- Ito N., Phillips,S.E., Yadav,K.D. and Knowles,P.F. (1994) Crystal structure of a free radical enzyme, galactose oxidase. J. Mol. Biol., 238, 794–814. [Abstract] [Google Scholar]
- Jabet C., Sprague,E.R., VanDemark,A.P. and Wolberger,C. (2000) Characterization of the N-terminal domain of the yeast transcriptional repressor Tup1/Ssn6. Proposal for an association model of the repressor complex Tup1 × Ssn6. J. Biol. Chem., 275, 9011–9018. [Abstract] [Google Scholar]
- Janin J. and Rodier,F. (1995) Protein–protein interaction at crystal contacts. Proteins, 23, 580–587. [Abstract] [Google Scholar]
- Jones T.A. (1992) A, yaap, asap, @#*? A set of averaging programs. In Dodson,E.J., Gover,S. and Wolf,W. (eds), Molecular Replacement. SERC Daresbury Laboratory, Warrington, UK, pp. 91–105. [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [Abstract] [Google Scholar]
- Keleher C.A., Redd,M.J., Schultz,J., Carlson,M. and Johnson,A.D. (1992) Ssn6–Tup1 is a general repressor of transcription in yeast. Cell, 68, 709–719. [Abstract] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) Halloween.. masks and bones. In Bailey,S., Hubbard,R. and Walle,D. (eds), From First Map to Final Model. SERC Daresbury Laboratory, Warrington, UK, pp. 59–66. [Google Scholar]
- Komachi K. and Johnson,A.D. (1997) Residues in the WD repeats of Tup1 required for interaction with α2. Mol. Cell. Biol., 17, 6023–6028. [Europe PMC free article] [Abstract] [Google Scholar]
- Komachi K., Redd,M.J. and Johnson,A.D. (1994) The WD repeats of Tup1 interact with the homeo domain protein α2. Genes Dev., 8, 2857–2867. [Abstract] [Google Scholar]
- Kuchin S. and Carlson,M. (1998) Functional relationships of Srb10–Srb11 kinase, carboxy-terminal domain kinase CTDK-I and transcriptional corepressor Ssn6–Tup1. Mol. Cell. Biol., 18, 1163–1171. [Europe PMC free article] [Abstract] [Google Scholar]
- Lambright D.G., Sondek,J., Bohm,A., Skiba,N.P., Hamm,H.E. and Sigler,P.B. (1996) The 2.0 Å crystal structure of a heterotrimeric G protein. Nature, 379, 311–319. [Abstract] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lee B. and Richards,F.M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol., 55, 379–400. [Abstract] [Google Scholar]
- Li Y., Bjorklund,S., Jiang,Y.W., Kim,Y.J., Lane,W.S., Stillman,D.J. and Kornberg,R.D. (1995) Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl Acad. Sci. USA, 92, 10864–10868. [Europe PMC free article] [Abstract] [Google Scholar]
- Liao S.M., Zhang,J., Jeffery,D.A., Koleske,A.J., Thompson,C.M., Chao,D.M., Viljoen,M., van Vuuren,H.J. and Young,R.A. (1995) A kinase–cyclin pair in the RNA polymerase II holoenzyme. Nature, 374, 193–196. [Abstract] [Google Scholar]
- Matthews B.W. (1968) Solvent content of protein crystals. J. Mol. Biol., 33, 491–497. [Abstract] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Neer E.J., Schmidt,C.J., Nambudripad,R. and Smith,T.F. (1994) The ancient regulatory-protein family of WD-repeat proteins [published erratum appears in Nature (1994) 371, 812]. Nature, 371, 297–300. [Abstract] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [Abstract] [Google Scholar]
- Nonet M.L. and Young,R.A. (1989) Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics, 123, 715–724. [Europe PMC free article] [Abstract] [Google Scholar]
- Ostling J., Cassart,J.P., Vandenhaute,J. and Ronne,H. (1998) Four hydrophobic amino acid residues in the C-terminal effector domain of the yeast Mig1p repressor are important for its in vivo activity. Mol. Gen. Genet., 260, 269–279. [Abstract] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [Abstract] [Google Scholar]
- Palaparti A., Baratz,A. and Stifani,S. (1997) The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem., 272, 26604–26610. [Abstract] [Google Scholar]
- Park S.H., Koh,S.S., Chun,J.H., Hwang,H.J. and Kang,H.S. (1999) Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 2044–2050. [Europe PMC free article] [Abstract] [Google Scholar]
- Redd M.J., Arnaud,M.B. and Johnson,A.D. (1997) A complex composed of tup1 and ssn6 represses transcription in vitro. J. Biol. Chem., 272, 11193–11197. [Abstract] [Google Scholar]
- Shimizu M., Roth,S.Y., Szent-Gyorgyi,C. and Simpson,R.T. (1991) Nucleosomes are positioned with base pair precision adjacent to the α2 operator in Saccharomyces cerevisiae. EMBO J., 10, 3033–3041. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith R.L., Redd,M.J. and Johnson,A.D. (1995) The tetratricopeptide repeats of Ssn6 interact with the homeo domain of α2. Genes Dev., 9, 2903–2910. [Abstract] [Google Scholar]
- Sondek J., Bohm,A., Lambright,D.G., Hamm,H.E. and Sigler,P.B. (1996) Crystal structure of a G-protein βγ dimer at 2.1Å resolution [published erratum appears in Nature (1996) 379, 847]. Nature, 379, 369–374. [Abstract] [Google Scholar]
- Thompson C.M., Koleske,A.J., Chao,D.M. and Young,R.A. (1993) A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell, 73, 1361–1375. [Abstract] [Google Scholar]
- Treitel M.A. and Carlson,M. (1995) Repression by SSN6–TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl Acad. Sci. USA, 92, 3132–3136. [Europe PMC free article] [Abstract] [Google Scholar]
- Tzamarias D. and Struhl,K. (1994) Functional dissection of the yeast Cyc8–Tup1 transcriptional co-repressor complex. Nature, 369, 758–761. [Abstract] [Google Scholar]
- Tzamarias D. and Struhl,K. (1995) Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8–Tup1 corepressor complex to differentially regulated promoters. Genes Dev., 9, 821–831. [Abstract] [Google Scholar]
- Varanasi U.S., Klis,M., Mikesell,P.B. and Trumbly,R.J. (1996) The Cyc8 (Ssn6)–Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol. Cell. Biol., 16, 6707–6714. [Europe PMC free article] [Abstract] [Google Scholar]
- Wahi M. and Johnson,A.D. (1995) Identification of genes required for α2 repression in Saccharomyces cerevisiae. Genetics, 140, 79–90. [Europe PMC free article] [Abstract] [Google Scholar]
- Wahi M., Komachi,K. and Johnson,A.D. (1998) Gene regulation by the yeast Ssn6–Tup1 corepressor. Cold Spring Harb. Symp. Quant. Biol., 63, 447–457. [Abstract] [Google Scholar]
- Wall M.A., Coleman,D.E., Lee,E., Iniguez-Lluhi,J.A., Posner,B.A., Gilman,A.G. and Sprang,S.R. (1995) The structure of the G protein heterotrimer Gi α1β1γ2. Cell, 83, 1047–1058. [Abstract] [Google Scholar]
- Williams F.E. and Trumbly,R.J. (1990) Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 6500–6511. [Europe PMC free article] [Abstract] [Google Scholar]
- Williams F.E., Varanasi,U. and Trumbly,R.J. (1991) The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol. Cell. Biol., 11, 3307–3316. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1093/emboj/19.12.3016
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc203344?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/emboj/19.12.3016
Article citations
Mechanism and evolutionary origins of alanine-tail C-degron recognition by E3 ligases Pirh2 and CRL2-KLHDC10.
Cell Rep, 42(9):113100, 06 Sep 2023
Cited by: 4 articles | PMID: 37676773 | PMCID: PMC10591846
Systematic analysis of tup1 and cyc8 mutants reveals distinct roles for TUP1 and CYC8 and offers new insight into the regulation of gene transcription by the yeast Tup1-Cyc8 complex.
PLoS Genet, 19(8):e1010876, 11 Aug 2023
Cited by: 2 articles | PMID: 37566621 | PMCID: PMC10446238
Functional characterization and comparative analysis of gene repression-mediating domains interacting with yeast pleiotropic corepressors Sin3, Cyc8 and Tup1.
Curr Genet, 69(2-3):127-139, 01 Mar 2023
Cited by: 1 article | PMID: 36854981 | PMCID: PMC10163088
PQBP5/NOL10 maintains and anchors the nucleolus under physiological and osmotic stress conditions.
Nat Commun, 14(1):9, 04 Jan 2023
Cited by: 3 articles | PMID: 36599853 | PMCID: PMC9813255
Tup1 is critical for transcriptional repression in Quiescence in S. cerevisiae.
PLoS Genet, 18(12):e1010559, 21 Dec 2022
Cited by: 4 articles | PMID: 36542663 | PMCID: PMC9815585
Go to all (88) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Genes & Proteins (Showing 7 of 7)
- (1 citation) UniProt - O76734
- (1 citation) UniProt - Q09715
- (1 citation) UniProt - O14432
- (1 citation) UniProt - P16649
- (1 citation) UniProt - P78706
- (1 citation) UniProt - P56094
- (1 citation) UniProt - P56093
Show less
Protein structures in PDBe
-
(2 citations)
PDBe - 1ERJView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Characterization of the N-terminal domain of the yeast transcriptional repressor Tup1. Proposal for an association model of the repressor complex Tup1 x Ssn6.
J Biol Chem, 275(12):9011-9018, 01 Mar 2000
Cited by: 28 articles | PMID: 10722750
Identification of residues in the TPR domain of Ssn6 responsible for interaction with the Tup1 protein.
FEBS Lett, 473(1):37-41, 01 May 2000
Cited by: 17 articles | PMID: 10802055
Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2.
J Mol Biol, 309(5):1007-1015, 01 Jun 2001
Cited by: 70 articles | PMID: 11399075
Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes.
Trends Biochem Sci, 25(7):325-330, 01 Jul 2000
Cited by: 216 articles | PMID: 10871883
Review





