Abstract
Free full text

Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli.
Abstract
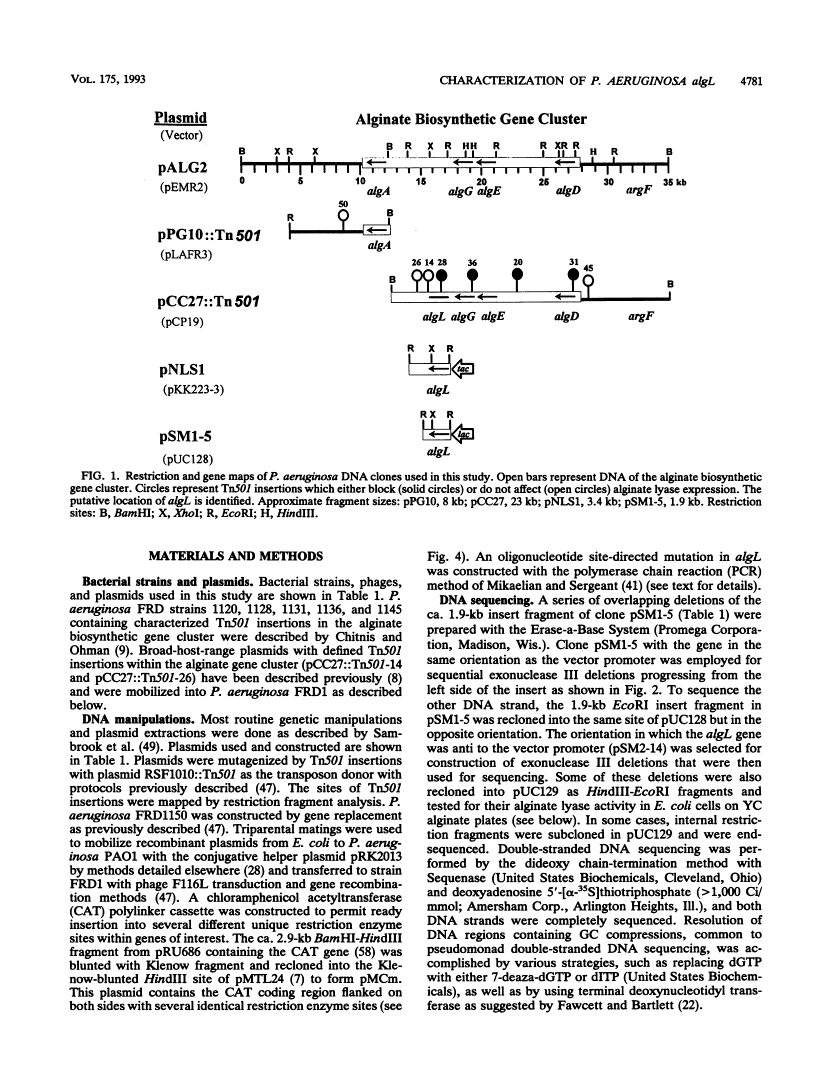
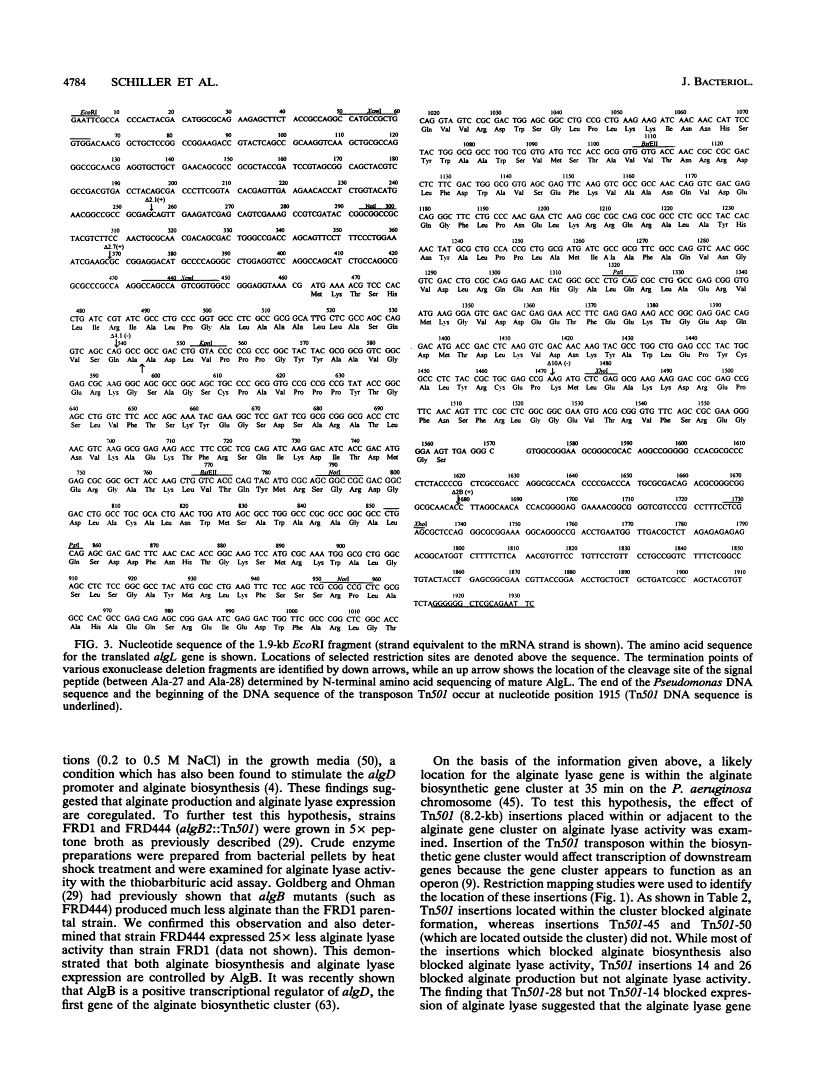
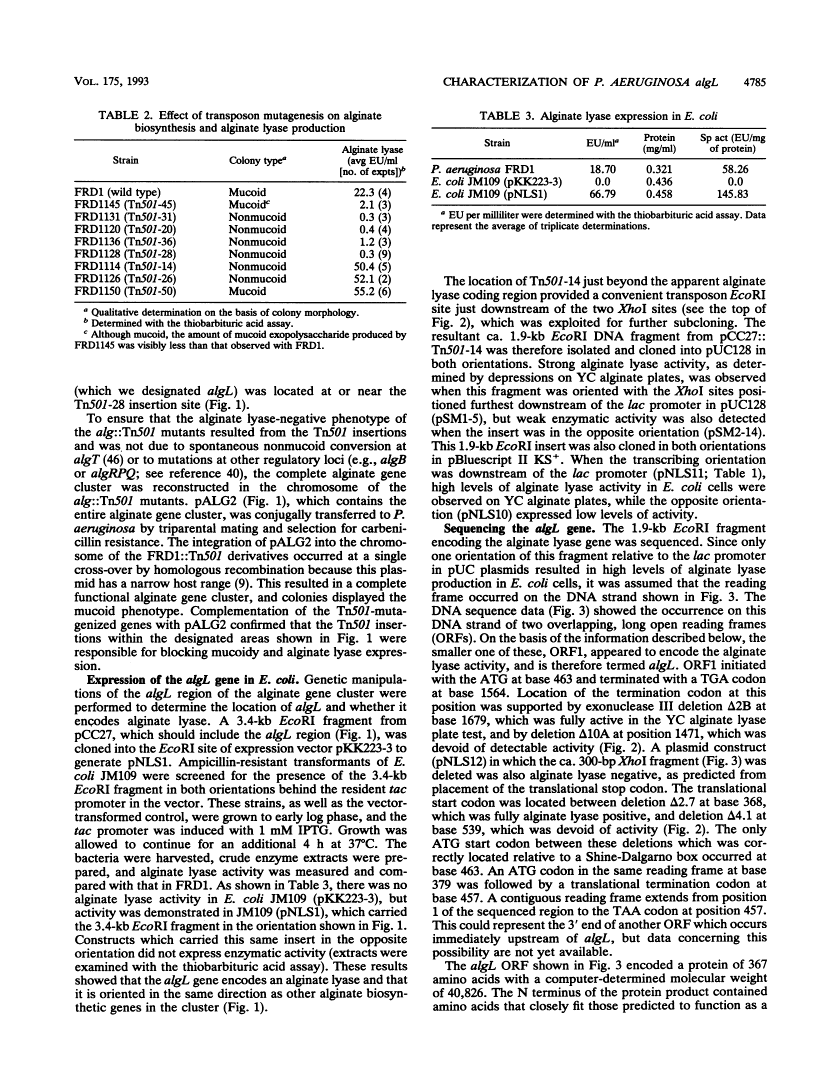
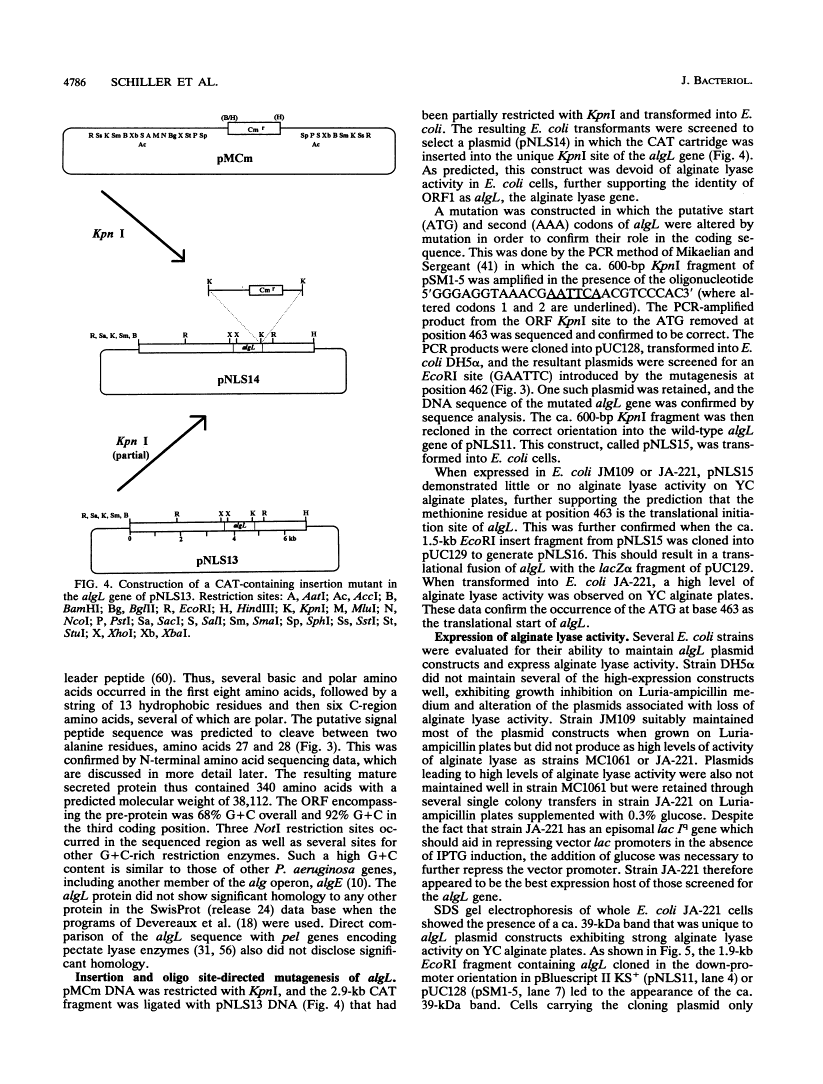
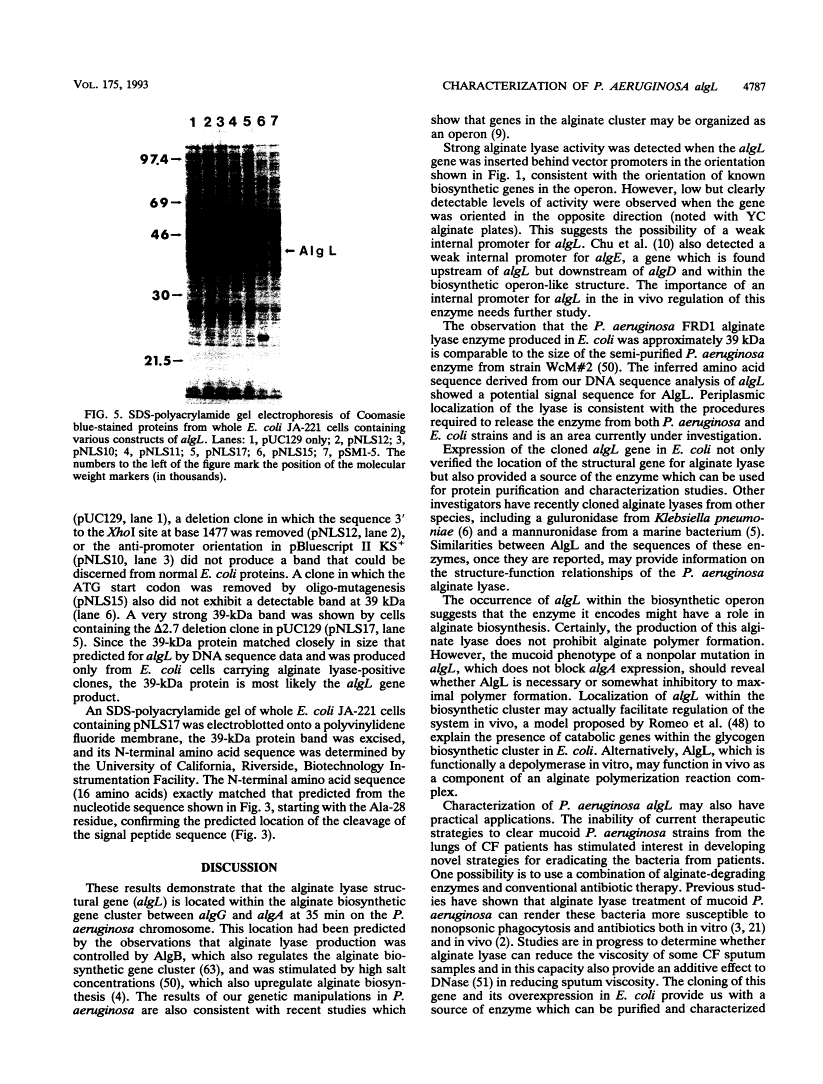
Mucoid strains of Pseudomonas aeruginosa produce a viscous exopolysaccharide called alginate and also express alginate lyase activity which can degrade this polymer. By transposon mutagenesis and gene replacement techniques, the algL gene encoding a P. aeruginosa alginate lyase enzyme was found to reside between algG and algA within the alginate biosynthetic gene cluster at 35 min on the P. aeruginosa chromosome. DNA sequencing data for algL predicted a protein product of ca. 41 kDa, including a 27-amino-acid signal sequence, which would be consistent with its possible localization in the periplasmic space. Expression of the algL gene in Escherichia coli cells resulted in the expression of alginate lyase activity and the appearance of a new protein of ca. 39 kDa detected on sodium dodecyl sulfate-polyacrylamide gels. In mucoid P. aeruginosa strains, expression of algL was regulated by AlgB, which also controls expression of other genes within the alginate gene cluster. Since alginate lyase activity is associated with the ability to produce and secrete alginate polymers, alginate lyase may play a role in alginate production.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (2.0M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartell PF, Orr TE, Lam GK. Polysaccharide depolymerase associated with bacteriophage infection. J Bacteriol. 1966 Jul;92(1):56–62. [Europe PMC free article] [Abstract] [Google Scholar]
- Bayer AS, Park S, Ramos MC, Nast CC, Eftekhar F, Schiller NL. Effects of alginase on the natural history and antibiotic therapy of experimental endocarditis caused by mucoid Pseudomonas aeruginosa. Infect Immun. 1992 Oct;60(10):3979–3985. [Europe PMC free article] [Abstract] [Google Scholar]
- Bayer AS, Speert DP, Park S, Tu J, Witt M, Nast CC, Norman DC. Functional role of mucoid exopolysaccharide (alginate) in antibiotic-induced and polymorphonuclear leukocyte-mediated killing of Pseudomonas aeruginosa. Infect Immun. 1991 Jan;59(1):302–308. [Europe PMC free article] [Abstract] [Google Scholar]
- Berry A, DeVault JD, Chakrabarty AM. High osmolarity is a signal for enhanced algD transcription in mucoid and nonmucoid Pseudomonas aeruginosa strains. J Bacteriol. 1989 May;171(5):2312–2317. [Europe PMC free article] [Abstract] [Google Scholar]
- Brown BJ, Preston JF, Ingram LO. Cloning of alginate lyase gene (alxM) and expression in Escherichia coli. Appl Environ Microbiol. 1991 Jun;57(6):1870–1872. [Europe PMC free article] [Abstract] [Google Scholar]
- Caswell RC, Gacesa P, Lutrell KE, Weightman AJ. Molecular cloning and heterologous expression of a Klebsiella pneumoniae gene encoding alginate lyase. Gene. 1989 Jan 30;75(1):127–134. [Abstract] [Google Scholar]
- Chambers SP, Prior SE, Barstow DA, Minton NP. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988 Aug 15;68(1):139–149. [Abstract] [Google Scholar]
- Chitnis CE, Ohman DE. Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J Bacteriol. 1990 Jun;172(6):2894–2900. [Europe PMC free article] [Abstract] [Google Scholar]
- Chitnis CE, Ohman DE. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol. 1993 May;8(3):583–593. [Abstract] [Google Scholar]
- Chu L, May TB, Chakrabarty AM, Misra TK. Nucleotide sequence and expression of the algE gene involved in alginate biosynthesis by Pseudomonas aeruginosa. Gene. 1991 Oct 30;107(1):1–10. [Abstract] [Google Scholar]
- Cross A, Allen JR, Burke J, Ducel G, Harris A, John J, Johnson D, Lew M, MacMillan B, Meers P, et al. Nosocomial infections due to Pseudomonas aeruginosa: review of recent trends. Rev Infect Dis. 1983 Nov-Dec;5 (Suppl 5):S837–S845. [Abstract] [Google Scholar]
- Dahler GS, Barras F, Keen NT. Cloning of genes encoding extracellular metalloproteases from Erwinia chrysanthemi EC16. J Bacteriol. 1990 Oct;172(10):5803–5815. [Europe PMC free article] [Abstract] [Google Scholar]
- Darzins A, Wang SK, Vanags RI, Chakrabarty AM. Clustering of mutations affecting alginic acid biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1985 Nov;164(2):516–524. [Europe PMC free article] [Abstract] [Google Scholar]
- Davidson IW, Sutherland IW, Lawson CJ. Purification and properties of an alginate lyase from a marine bacterium. Biochem J. 1976 Dec 1;159(3):707–713. [Europe PMC free article] [Abstract] [Google Scholar]
- Deretic V, Dikshit R, Konyecsni WM, Chakrabarty AM, Misra TK. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989 Mar;171(3):1278–1283. [Europe PMC free article] [Abstract] [Google Scholar]
- Deretic V, Gill JF, Chakrabarty AM. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987 Jan;169(1):351–358. [Europe PMC free article] [Abstract] [Google Scholar]
- Deretic V, Mohr CD, Martin DW. Mucoid Pseudomonas aeruginosa in cystic fibrosis: signal transduction and histone-like elements in the regulation of bacterial virulence. Mol Microbiol. 1991 Jul;5(7):1577–1583. [Abstract] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. [Europe PMC free article] [Abstract] [Google Scholar]
- Doubet RS, Quatrano RS. Isolation of marine bacteria capable of producing specific lyases for alginate degradation. Appl Environ Microbiol. 1982 Sep;44(3):754–756. [Europe PMC free article] [Abstract] [Google Scholar]
- Dunne WM, Jr, Buckmire FL. Partial purification and characterization of a polymannuronic acid depolymerase produced by a mucoid strain of Pseudomonas aeruginosa isolated from a patient with cystic fibrosis. Appl Environ Microbiol. 1985 Sep;50(3):562–567. [Europe PMC free article] [Abstract] [Google Scholar]
- Eftekhar F, Speert DP. Alginase treatment of mucoid Pseudomonas aeruginosa enhances phagocytosis by human monocyte-derived macrophages. Infect Immun. 1988 Nov;56(11):2788–2793. [Europe PMC free article] [Abstract] [Google Scholar]
- Fawcett TW, Bartlett G. An effective method for eliminating "artifact banding" when sequencing double-stranded DNA templates. Biotechniques. 1990 Jul;9(1):46–48. [Abstract] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. [Europe PMC free article] [Abstract] [Google Scholar]
- Flynn JL, Ohman DE. Use of a gene replacement cosmid vector for cloning alginate conversion genes from mucoid and nonmucoid Pseudomonas aeruginosa strains: algS controls expression of algT. J Bacteriol. 1988 Jul;170(7):3228–3236. [Europe PMC free article] [Abstract] [Google Scholar]
- Friedman AM, Long SR, Brown SE, Buikema WJ, Ausubel FM. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. [Abstract] [Google Scholar]
- Goldberg JB, Ohman DE. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984 Jun;158(3):1115–1121. [Europe PMC free article] [Abstract] [Google Scholar]
- Goldberg JB, Ohman DE. Construction and characterization of Pseudomonas aeruginosa algB mutants: role of algB in high-level production of alginate. J Bacteriol. 1987 Apr;169(4):1593–1602. [Europe PMC free article] [Abstract] [Google Scholar]
- Hansen JB, Doubet RS, Ram J. Alginase enzyme production by Bacillus circulans. Appl Environ Microbiol. 1984 Apr;47(4):704–709. [Europe PMC free article] [Abstract] [Google Scholar]
- Hinton JC, Sidebotham JM, Gill DR, Salmond GP. Extracellular and periplasmic isoenzymes of pectate lyase from Erwinia carotovora subspecies carotovora belong to different gene families. Mol Microbiol. 1989 Dec;3(12):1785–1795. [Abstract] [Google Scholar]
- Holloway BW, Krishnapillai V, Morgan AF. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979 Mar;43(1):73–102. [Europe PMC free article] [Abstract] [Google Scholar]
- Hoshino T, Kageyama M. Purification and properties of a binding protein for branched-chain amino acids in Pseudomonas aeruginosa. J Bacteriol. 1980 Mar;141(3):1055–1063. [Europe PMC free article] [Abstract] [Google Scholar]
- Keen NT, Dahlbeck D, Staskawicz B, Belser W. Molecular cloning of pectate lyase genes from Erwinia chrysanthemi and their expression in Escherichia coli. J Bacteriol. 1984 Sep;159(3):825–831. [Europe PMC free article] [Abstract] [Google Scholar]
- Keen NT, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988 Oct 15;70(1):191–197. [Abstract] [Google Scholar]
- Krishnapillai V. A novel transducing phage. Its role in recognition of a possible new host-controlled modification system in Pseudomonas aeruginosa. Mol Gen Genet. 1972;114(2):134–143. [Abstract] [Google Scholar]
- Lange B, Wingender J, Winkler UK. Isolation and characterization of an alginate lyase from Klebsiella aerogenes. Arch Microbiol. 1989;152(3):302–308. [Abstract] [Google Scholar]
- Linker A, Evans LR. Isolation and characterization of an alginase from mucoid strains of Pseudomonas aeruginosa. J Bacteriol. 1984 Sep;159(3):958–964. [Europe PMC free article] [Abstract] [Google Scholar]
- May TB, Shinabarger D, Maharaj R, Kato J, Chu L, DeVault JD, Roychoudhury S, Zielinski NA, Berry A, Rothmel RK, et al. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin Microbiol Rev. 1991 Apr;4(2):191–206. [Europe PMC free article] [Abstract] [Google Scholar]
- Mikaelian I, Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 1992 Jan 25;20(2):376–376. [Europe PMC free article] [Abstract] [Google Scholar]
- Miller JF, Mekalanos JJ, Falkow S. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science. 1989 Feb 17;243(4893):916–922. [Abstract] [Google Scholar]
- Ohman DE, Chakrabarty AM. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981 Jul;33(1):142–148. [Europe PMC free article] [Abstract] [Google Scholar]
- Ohman DE, West MA, Flynn JL, Goldberg JB. Method for gene replacement in Pseudomonas aeruginosa used in construction of recA mutants: recA-independent instability of alginate production. J Bacteriol. 1985 Jun;162(3):1068–1074. [Europe PMC free article] [Abstract] [Google Scholar]
- Romeo T, Kumar A, Preiss J. Analysis of the Escherichia coli glycogen gene cluster suggests that catabolic enzymes are encoded among the biosynthetic genes. Gene. 1988 Oct 30;70(2):363–376. [Abstract] [Google Scholar]
- Shak S, Capon DJ, Hellmiss R, Marsters SA, Baker CL. Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9188–9192. [Europe PMC free article] [Abstract] [Google Scholar]
- Shinabarger D, Berry A, May TB, Rothmel R, Fialho A, Chakrabarty AM. Purification and characterization of phosphomannose isomerase-guanosine diphospho-D-mannose pyrophosphorylase. A bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J Biol Chem. 1991 Feb 5;266(4):2080–2088. [Abstract] [Google Scholar]
- Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. [Europe PMC free article] [Abstract] [Google Scholar]
- Stevens RA, Levin RE. Purification and characteristics of an alginase from Alginovibrio aquatilis. Appl Environ Microbiol. 1977 May;33(5):1156–1161. [Europe PMC free article] [Abstract] [Google Scholar]
- Tamaki SJ, Gold S, Robeson M, Manulis S, Keen NT. Structure and organization of the pel genes from Erwinia chrysanthemi EC16. J Bacteriol. 1988 Aug;170(8):3468–3478. [Europe PMC free article] [Abstract] [Google Scholar]
- Thomassen MJ, Demko CA, Doershuk CF. Cystic fibrosis: a review of pulmonary infections and interventions. Pediatr Pulmonol. 1987 Sep-Oct;3(5):334–351. [Abstract] [Google Scholar]
- Ubben D, Schmitt R. A transposable promoter and transposable promoter probes derived from Tn1721. Gene. 1987;53(1):127–134. [Abstract] [Google Scholar]
- VOGEL HJ, BONNER DM. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [Abstract] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. [Abstract] [Google Scholar]
- von Riesen VL. Digestion of algin by Pseudomonas maltophilia and Pseudomonas putida. Appl Environ Microbiol. 1980 Jan;39(1):92–96. [Europe PMC free article] [Abstract] [Google Scholar]
- WEISSBACH A, HURWITZ J. The formation of 2-keto-3-deoxyheptonic acid in extracts of Escherichia coli B. I. Identification. J Biol Chem. 1959 Apr;234(4):705–709. [Abstract] [Google Scholar]
- Wozniak DJ, Ohman DE. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J Bacteriol. 1991 Feb;173(4):1406–1413. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.175.15.4780-4789.1993
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc204930?pdf=render
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/175/15/4780
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/175/15/4780
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Candidate genes involved in biosynthesis and degradation of the main extracellular matrix polysaccharides of brown algae and their probable evolutionary history.
BMC Genomics, 25(1):950, 10 Oct 2024
Cited by: 0 articles | PMID: 39390408 | PMCID: PMC11468063
Proteomic profiling spotlights the molecular targets and the impact of the natural antivirulent umbelliferone on stress response, virulence factors, and the quorum sensing network of Pseudomonas aeruginosa.
Front Cell Infect Microbiol, 12:998540, 30 Nov 2022
Cited by: 8 articles | PMID: 36530435 | PMCID: PMC9748083
Role of Exopolysaccharides of Pseudomonas in Heavy Metal Removal and Other Remediation Strategies.
Polymers (Basel), 14(20):4253, 11 Oct 2022
Cited by: 5 articles | PMID: 36297831 | PMCID: PMC9609410
Review Free full text in Europe PMC
Characterization of a Novel Polysaccharide Lyase Family 5 Alginate Lyase with PolyM Substrate Specificity.
Foods, 11(21):3527, 06 Nov 2022
Cited by: 0 articles | PMID: 36360141 | PMCID: PMC9655155
Cloning and Characterization of a Novel Endo-Type Metal-Independent Alginate Lyase from the Marine Bacteria Vibrio sp. Ni1.
Mar Drugs, 20(8):479, 26 Jul 2022
Cited by: 4 articles | PMID: 35892947 | PMCID: PMC9331746
Go to all (74) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sequence of the algL gene of Pseudomonas aeruginosa and purification of its alginate lyase product.
Gene, 131(1):1-8, 01 Sep 1993
Cited by: 50 articles | PMID: 8370530
Alginate synthesis in Pseudomonas aeruginosa: the role of AlgL (alginate lyase) and AlgX.
J Bacteriol, 178(3):625-632, 01 Feb 1996
Cited by: 47 articles | PMID: 8550492 | PMCID: PMC177704
Analysis and expression of algL, which encodes alginate lyase in Pseudomonas syringae Pv. syringae.
DNA Seq, 12(5-6):455-461, 01 Dec 2001
Cited by: 5 articles | PMID: 11913796
Molecular modeling and redesign of alginate lyase from Pseudomonas aeruginosa for accelerating CRPA biofilm degradation.
Proteins, 84(12):1875-1887, 25 Oct 2016
Cited by: 5 articles | PMID: 27676452