Abstract
Free full text

The transcriptional cycle of HIV-1 in real-time and live cells
Abstract
RNA polymerase II (RNAPII) is a fundamental enzyme, but few studies have analyzed its activity in living cells. Using human immunodeficiency virus (HIV) type 1 reporters, we study real-time messenger RNA (mRNA) biogenesis by photobleaching nascent RNAs and RNAPII at specific transcription sites. Through modeling, the use of mutant polymerases, drugs, and quantitative in situ hybridization, we investigate the kinetics of the HIV-1 transcription cycle. Initiation appears efficient because most polymerases demonstrate stable gene association. We calculate an elongation rate of approximately 1.9 kb/min, and, surprisingly, polymerases remain at transcription sites 2.5 min longer than nascent RNAs. With a total polymerase residency time estimated at 333 s, 114 are assigned to elongation, and 63 are assigned to 3′-end processing and/or transcript release. However, mRNAs were released seconds after polyadenylation onset, and analysis of polymerase density by chromatin immunoprecipitation suggests that they pause or lose processivity after passing the polyA site. The strengths and limitations of this kinetic approach to analyze mRNA biogenesis in living cells are discussed.
Introduction
The human immunodeficiency virus (HIV) type 1 genome consists of a single transcription unit that is integrated into cellular chromatin. HIV-1 is highly dependent on transcriptional regulation: acutely infected cells synthesize high levels of virus, whereas latently infected cells transcribe little or no viral RNAs. This tight regulation is a critical feature of viral pathogenicity because it allows the virus to remain silent in the organism and prevents clearance by the current antiretroviral regimens (for review see Greene and Peterlin, 2002; Marcello, 2006).
The HIV-1 promoter is located in the U3 region of the 5′ long terminal repeat (LTR). Its transcription is performed by the cellular machinery, but it is boosted by the viral protein Tat (for reviews see Jeang et al., 1999; Marcello et al., 2001). In latently infected cells that do not produce Tat, the polymerases initiating at the HIV-1 promoter are unprocessive and unable to transcribe the entire viral genome. Lymphocyte-activating stimuli induce the HIV-1 promoter to produce small amounts of Tat, which starts a positive feedback loop by stimulating viral transcription (Weinberger et al., 2005). Tat recruits to the HIV-1 promoter the active form of the positive transcription elongation factor P-TEFb (Wei et al., 1998), which consists of a complex between cyclin T1 and Cdk9 (for review see Peterlin and Price, 2006). Tat binds to both cyclin T1 and the trans-activation–responsive region, an RNA element present at the 5′ end of all viral transcripts. This induces the formation of a tertiary complex on nascent RNAs, which brings Cdk9 into position to phosphorylate several components of the transcription machinery, including the C-terminal domain (CTD) of the large subunit of RNA polymerase II (RNAPII), and elongation factors DSIF and negative elongation factor (NELF; for review see Peterlin and Price, 2006). This converts RNAPII into a highly processive enzyme, which can transcribe the entire viral genome.
Production of HIV-1 mRNAs is not only regulated at the level of transcription but also at the level of splicing and polyadenylation (Schwartz et al., 1990; Ashe et al., 1997). HIV-1 possesses two polyadenylation sites, one in each LTR. Polyadenylation at the first site is suppressed by binding of the small nuclear RNP U1 to the major splice donor (SD1; Ashe et al., 1997), whereas polyadenylation at the second site is activated by upstream sequences present in the U3 region of the 3′ LTR (Ashe et al., 1995; Gilmartin et al., 1995).
Many of the regulatory events that determine the fate of HIV-1 RNAs occur at the transcription site. This includes the decision to initiate transcription, to elongate, to splice, and to process the RNA at the 3′ end. Most importantly, the relative kinetic rate of each process appears to be critical for their final outcome. For instance, the rate of cleavage at the first polyA site and the rate of U1 binding at SD1 determine the amount of read-through at this polyA site (Ashe et al., 1997). Similarly, the rate of splicing versus 3′-end formation at the second polyA site determines the amount of unspliced RNA available for Rev-mediated export (Malim et al., 1989). In these two cases, the kinetic competition is influenced by the rate of transcription elongation. For instance, cleavage at the first polyA site cannot be suppressed until the polymerase reaches SD1, and splicing cannot be competed out by 3′-end formation until the polymerase arrives at the second polyA site. Importantly, the rate of transcription elongation can be regulated, and this can control gene expression (de la Mata et al., 2003; Batsche et al., 2006).
Like HIV-1, many cellular genes are regulated at the level of transcription, and transcriptional regulation has been the subject of a large number of studies. However, although transcription by RNAPII is a fundamental process, we still lack a precise view of how transcription occurs in vivo. In particular, we lack detailed kinetic models describing mRNA synthesis and the cycle of RNAPII. In this study, we used FRAP to study these processes directly in living cells.
Results
Real-time analysis of mRNA biogenesis by photobleaching nascent RNAs
Previous studies have shown that tagging RNA with 24 binding sites for the coat protein of phage MS2 allows its detection in living cells with excellent sensitivity (Fusco et al., 2003; Shav-Tal et al., 2004). To analyze the biogenesis of HIV-1 mRNA, we inserted 24 MS2-binding sites in the 3′ untranslated region of an HIV vector that carried the elements required for RNA production (Fig. 1 A): the 5′ LTR, the major splice donor (SD1), the packaging signal Ψ, the Rev-responsive element, the splice acceptor A7 flanked by its regulatory sequences (exonic splicing enhancer and ESS3), and the 3′ LTR that drives 3′-end formation. Stable arrays of this reporter construct (pExo-MS2×24) were integrated into U2OS cells. Two clones (Exo1 and Exo2) showed robust trans-activation by Tat and other stimuli known to induce transcription of integrated HIV-1 promoters (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200706018/DC1). When expressed, the RNAs were distributed homogenously in the cytoplasm and concentrated in a bright spot in the nucleoplasm. This spot corresponded to the transcription site as it colocalized with RNAPII and was labeled with probes directed against the nontranscribed strand of the vector (Fig. 1 B). Several active genes localize near speckles (Smith et al., 1999; Moen et al., 2004). However, the HIV-1 transcription site rarely colocalizes with speckles labeled by the marker protein SC35 (Fig. 1 B).

Characterization of HIV-1 transcription sites. (A) Schematics of the HIV-1 reporters. (B) HIV-1 transcription sites contain a focal accumulation of mRNAs. Exo1 cells were transfected with a Tat expression vector and fixed 24 h later. (top) HIV-1 mRNAs and integrated DNA were detected by in situ hybridization. (middle) RNAPII was detected by immunofluorescence. (bottom) SC35 does not accumulate at HIV-1 transcription sites. Each field is 22 × 22 μm; insets are magnifications of the transcription sites (2.5 × 2.5 μm). Proteins were visualized by immunofluorescence. Bar, 8.8 μm.
To visualize nascent HIV-1 RNAs, a nuclear MS2-GFP fusion was expressed in Exo1 or Exo2 cells. In live cells, MS2-GFP was diffused in the nucleoplasm and concentrated in a spot at the transcription site (Fig. 2). FRAP is a powerful technique to study the dynamic properties of a fluorescent molecule, and we used it to study mRNA synthesis by photobleaching the nascent RNAs labeled with MS2-GFP. Indeed, when a transcription site is bleached, incoming polymerases will synthesize new MS2 sites, and this will result in recovery of the fluorescent signal. In this system, RNAs are visualized indirectly through MS2-GFP, and this may complicate the FRAP analysis (Braga et al., 2007). First, a slow diffusion rate of MS2-GFP may mask the neosynthesis of nascent RNAs. Second, a rapid dissociation of the RNA-bound MS2-GFP may lead to rapid recovery rates unrelated to the synthesis of new RNAs.

Analysis of HIV-1 mRNA biogenesis by FRAP of nascent RNAs. Exo1 (A–G) or Exo2 (E, H, and I) cells were transfected with vectors expressing Tat and MS2-GFP and analyzed by FRAP. (A and B) FRAP of MS2-GFP analyzed at a high frame rate (160 ms) and on a a short time scale (5 s) with a confocal microscope. (A, left) Image of a transfected Exo1 cell (25 × 20 μm). (right) Recovery curves of MS2-GFP in the nucleoplasm of U2OS cells (pink curve) or at the HIV-1 transcription sites (blue curve). (B) Snapshot of the FRAP experiment (5 × 2 μm). Left, prebleach; middle, after bleach; right, 5-s time point. The bleached area is indicated by a circle (1.5-μm diameter). (C–I) FRAP of MS2-GFP analyzed by tracking transcription sites in 3D for 6 min using a wide-field microscope. (C) Image sequence from a FRAP experiment; each field is 30 × 25 μm. The time after bleach is indicated together with the bleached area (2.5-μm diameter). (D) Recovery curves of MS2-GFP in the nucleoplasm of U2OS cells (pink curve) and at the HIV-1 transcription site (blue curve). (E) Comparison of clones Exo1 and Exo2. The postbleach values were set to zero. (F) Schematic of the mRNA biogenesis pathway used to model the FRAP curves. (G and H) Fits of Exo1 (G) and Exo2 (H) FRAP curves with single exponential (red curve) and a two-step model that comprises a linear increase followed by an exponential (pink and green, respectively). (I) Comparison of experimental (red and blue) and computer-simulated (green) FRAP curves. Error bars represent SD. Bar, 7.2 μm.
To obtain an estimate for the rate of exchange of the MS2-GFP protein on its target site, we used an abundant noncoding RNA, U3, which was modified to incorporate a single MS2-binding site. U3 is synthesized in the nucleoplasm and accumulates in nucleoli, where it plays essential functions during ribosomal RNA biogenesis (Kass et al., 1990). Previous work has shown that a fraction of the proteins associated with U3 exchange slowly between nucleoli and the nucleoplasm (Phair and Misteli, 2000), and it was therefore suspected that a fraction of U3 would also be stably associated with nucleoli. When nucleoli of cells expressing only MS2-GFP were bleached, fluorescence recovery was nearly complete within seconds (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200706018/DC1). In striking contrast, when nucleoli of cells expressing both U3-MS2 and MS2-GFP were bleached, only a fraction of the fluorescence was recovered, even after 10 min. This indicated that a part of U3-MS2 was immobile and that bleached MS2-GFP molecules stayed stably bound to the RNA during the course of the experiment. Furthermore, diffusion of MS2-GFP was rapid (15 μm2/s) and much faster than recovery at transcription sites (Fig. 2, A and B). Thus, the diffusion or dissociation of MS2-GFP was neglected in the analysis of FRAP experiments.
A substantial fraction of nascent RNAs are elongating
Recovery of Exo1 transcription sites showed that the synthesis of new RNAs occurred in <3 min and that virtually no RNAs were retained at the transcription site for a long time (immobile fraction of 3%; Fig. 2 D). A second clone expressing the same MS2-tagged RNA yielded similar kinetics (Exo2; Fig. 2 E), indicating that this was principally a property of the reporter.
RNA biogenesis occurs in a series of steps: transcription, 3′-end formation (cleavage and polyadenylation), and release from the transcription site (Fig. 2 F). To test whether elongation was rate limiting, we used a slow version of RNAPII, the hC4 mutant (de la Mata et al., 2003). This α-amanitin–resistant mutant was transfected in Exo1 cells, endogenous enzymes were inactivated with α-amanitin, and remaining transcription sites were analyzed by FRAP. As a control, we used another α-amanitin–resistant version of RNAPII that has no elongation defect (wild type [WTres]; de la Mata et al., 2003). FRAP curves obtained with WTres or the endogenous polymerase were nearly identical (Fig. 3). In contrast, recoveries were markedly slower with the hC4 mutant: 129 s for half-recovery versus 75 s for the WTres enzyme. To confirm that transcriptional elongation took a substantial part of the time that nascent RNAs spent at their transcription sites, we used camptothecin. This drug targets topoisomerase I, and it transiently cross-links it to DNA. A previous study has shown that this creates steric road blocks for polymerases, which result in increased pausing and a net slowdown of transcriptional elongation (Capranico et al., 2007). Indeed, when cells were treated with this drug, the recovery of nascent RNAs was slower: 170 s for half-recovery in Exo1 cells compared with 65 s in the case of untreated cells (Fig. 4 A).
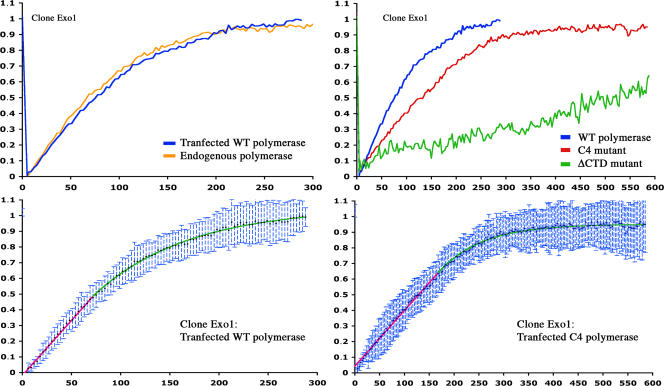
Effect of mutants of RNAPII on the synthesis of nascent RNAs. Exo1 cells were transfected with vectors expressing mutant forms of RNAPII, Tat, and MS2-GFP and were treated with α-amanitin before FRAP. (top left) Recovery curves of the endogenous and transfected WTRES RNAPII (orange and blue curves, respectively). (top right) Comparison between the WTRES (blue), hC4 (red), and ΔCTD (green) RNAPII. (bottom) Fit of the two-step model (straight line followed an exponential) with WTRES (left) and hC4 curves (right). Error bars represent SD.

Elongation is a limiting step in the recovery of nascent RNAs. (A) Recoveries of MS2-GFP in Exo1 cells treated with camptothecin. Cells were transfected with vectors expressing Tat and MS2-GFP, treated or untreated with camptothecin, and analyzed by FRAP. (left) Comparison of treated (green) with untreated (blue) cells. (right) Treated cells were fitted with the two-step model. Pink, straight line; green, exponential component. (B) Comparison of the MS2-GFP recovery curves for the short and long HIV-1 reporters. ExoLong cells were transfected with vectors expressing Tat and MS2-GFP and were analyzed by FRAP. (top left) Comparison of the recovery curves of Exo1 cells (blue), Exo2 (red), and ExoLong (green). (top right) Fit of the long reporter with the two-step model. (bottom left) Comparison of Exo1 and ExoLong recovery curves after normalization by the initial slope (initiation rate). (bottom right) Comparison of experimental (blue) and computer-simulated (green) FRAP curves for the long reporter. (C) Recoveries of MS2-GFP in HIV-1 reporters lacking U3. pTRIP_1_13 cells expressing the ΔU3 reporter (pTRIP) were transfected as described in A and analyzed by FRAP. Error bars represent SD.
To further establish that elongation was rate limiting, we developed a cell line (ExoLong) that integrated a second reporter, pExo-MS2 × 24-Long, which differed by 2.4 kb in the length of the sequence separating the MS2 repeat and the end of the gene. If elongation is rate limiting, the recovery should be slower for the long reporter, and the difference should represent the time taken to transcribe the additional sequences. As expected, transcription sites of ExoLong recovered more slowly: 126 s for half-recovery versus 65 s for the short reporter (Fig. 4 B). Altogether, these observations demonstrated that elongation took a substantial fraction of the time dedicated to mRNA production.
A two-step model comprising elongation and 3′-end processing can describe recoveries of nascent RNAs
In kinetic terms, transcription elongation is the repetition of an elementary step: the addition of one nucleotide. The stochastic basis of the process implies that the time taken to add a single nucleotide is variable and follows an exponential described by the rate constant kel. In contrast, when polymerases transcribe a large number of nucleotides, the repetition of the elementary step creates a statistical averaging such that the time taken to synthesize n nucleotides will be virtually constant for all polymerases and equal to n/kel. This feature of transcription elongation is distinctive, and it predicts a linear increase in signal during FRAP recovery, whereas single-step processes should result in exponentials (Fig. S3 B, available at http://www.jcb.org/cgi/content/full/jcb.200706018/DC1).
Elongation is the first step of mRNA biogenesis that can be observed in our FRAP experiments. Indeed, once the polymerase reaches the polyA site, the pre-mRNA is cleaved, polyadenylated, and released. These processes occur as a series of single steps and, thus, may be modeled as exponentials. We attempted to fit the FRAP curve with two components: a straight line for elongation followed by a single exponential for 3′-end formation and release. This two-step model fitted experimental data substantially better than a single exponential (Figs. 2, G and H; and 3). The short reporter gave 65 to 73 s for elongation and 44 to 31 s for the half-time of the exponential in Exo1 and Exo2 cells, respectively (Table I). The long reporter yielded 148 s for elongation and 65 s for the exponential. This translated into similar elongation rates of 1.79–2.03 kb/min. Cells treated with camptothecin or transfected with the slow mutant of RNAPII could also be fitted to this two-step model (Figs. 3 and 4 A). For the hC4 mutant, it yielded an elongation rate more than twofold slower than with WT RNAPII (0.8 kb/min), which is in agreement with in vitro data (Coulter and Greenleaf, 1985).
Table I.
Kinetic parameters derived from FRAP analysis of nascent HIV-1 RNAs
| Clone | Exo1 | Exo1 Camptoth | Exo2 | ExoLong | Exo1 Pol-WTRES | Exo1 Pol-hC4 |
|---|---|---|---|---|---|---|
| Length to 3′ end (nt) | 2,187 | 2,187 | 2,187 | 4,580 | 2,187 | 2,187 |
| Immobile fraction (%) | 3 | 10 | 4 | 5.5 | 0 | 5 |
| t 1/2 (s)a | 65 | 170 | 63 | 126 | 75 | 129 |
| t 1/2 monoexponential (s)b | 60.2 | ND | 53.3 | 95.0 | ND | ND |
| Linear phase (s)c | 64.6 | 68 | 73.3 | 148.1 | 70.7 | 165.1 |
| t mean exponential (s)c | 63.5 | 189 | 44.9 | 93.6 | 95.9 | 87.1 |
| t 1/2 exponential (s)c | 44 | 131 | 31.1 | 64.9 | 66.5 | 60.4 |
| Elongation rate (kb/min)c | 2.03 | 1.52 | 1.79 | 1.85 | 1.86 | 0.79 |
The proportion of time dedicated to elongation versus 3′-end processing can also be estimated by direct comparison of the short and long reporters. Indeed, if initiation rates are equal, the intensity of the MS2 signal at various transcription sites should be proportional to the time that the RNA spends there. When the curves of the short and long reporters were normalized to their initiation rate, that is, to their initial slopes, the long reporter accumulated 1.5 times more RNAs at the transcription site (Fig. 4 B). If one assumes that the time required for elongation is proportional to the RNA length, whereas other steps are identical for the two reporters, this value corresponds to half of the time spent on elongation for the short reporter and 71% for the long one (see Materials and methods). This was in agreement with the values obtained by fitting the FRAP curves with the two-step model (Table I).
Estimation of the relative rates of elongation and 3′-end formation by quantitative in situ hybridization
Next, we performed quantitative in situ hybridization with oligonucleotide probes that hybridized along the gene. Incompletely transcribed RNAs should yield more signals with probes hybridizing at their 5′ end, whereas full-length RNAs should yield equal signals with 5′ and 3′ probes (Femino et al., 1998). Because incompletely transcribed RNAs correspond to elongating molecules, whereas full-length RNAs are at the stage of 3′-end processing or transcript release, the ratio of 5′ to 3′ probes can be used to estimate the relative time taken by elongation versus 3′-end processing and release. Four sets of Cy3-labeled probes were used (Fig. 5): the first hybridized in exon 1 (E1), the second in the intron (I), the third at the splice acceptor site (I-E2), and the last in exon 2 (E2), immediately before the active polyA site. When signals were normalized with a Cy5-labeled probe against the MS2 repeat, 5′ probes gave 2.2-fold more signal than 3′ probes (Fig. 5). To confirm this, we hybridized E1-Cy5 and E2-Cy3 probes simultaneously, and signals at the transcription sites were normalized to signals in the cytoplasm. Again, we found twofold more E1 probe at transcription sites (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200706018/DC1). From these values, we could estimate that half of the polymerases that have reached the MS2 repeat are elongating toward the polyA site, whereas the other half are at the stage of 3′-end formation (see Materials and methods), which is in agreement with FRAP experiments.

Measurements of nascent RNA species by quantitative in situ hybridization. (A) Schematic of the short reporter with the position of the probes indicated. The relative amount of probe corresponds to the Cy3/Cy5 (MS2) ratio measured at the transcription site of Exo1 cells. The predicted ratios were estimated from an elongation rate of 2.03 kb/min and 63.5 s for 3′-end formation (see Materials and methods). (B) Polyadenylated HIV-1 mRNAs do not accumulate at the transcription site of Exo1 cells. (top) Cells were hybridized with a mixture of pA+-Cy3 and MS2-Cy5 probes. (bottom) Cells expressing PABN1-GFP were hybridized with an MS2-Cy3 probe. Insets are magnifications of the transcription sites (2.5 × 2.5 μm). (C) Images of single cells with the various HIV-1 probes. Exo1 cells were transfected with Tat and hybridized with a mixture of Cy3-/MS2-Cy5 probes. (B and C) Field size is 20 × 20 μm. Bar, 6.7 μm.
3′-end formation occurs at various rates, whereas release of HIV-1 mRNAs occurs seconds after the onset of polyadenylation
To further characterize RNA species present at transcription sites, we used a probe specific for polyadenylated HIV-1 mRNAs. This probe yielded robust signals in the cytoplasm but only faint signals at transcription sites (11% of E2 signal; Fig. 5 B). This was unlikely the result of a failure of the probe to hybridize to its target because the nuclear polyA-binding protein PABN1 was also not detected there (Fig. 5 B). This indicated that RNAs were released rapidly once polyadenylation started. Indeed, with a total 3′-end processing time of 63.5 s (Table I), we calculated that cleavage/polyadenylation took 54.7 s and mRNA release took 8.8 s (see Materials and methods).
The CTD of the large subunit of RNAPII has been proposed to connect the polymerase with the 3′-end processing machinery (for review see Meinhart et al., 2005). In addition, RNAPII lacking the CTD have defects in transcription initiation and in elongating through nucleosomal templates (Meininghaus et al., 2000). Remarkably, transcription sites generated by this RNAPII mutant recovered very slowly after photobleaching (half-recovery of 550 s vs. 75 s for WTres; Fig. 3). A defect in initiation should result in lower amounts of fluorescent signal at the transcription site, but it should not affect the time that polymerases spend on the gene. Thus, the defects we observed arose from a reduced ability to synthesize or process nascent RNAs, which is in agreement with biochemical data.
To confirm that alteration of 3′-end processing could slow down the release of HIV-1 RNA, we made a reporter containing mutations near the polyA site. Besides the canonical AAUAAA and G/U-rich element, HIV-1 contains upstream elements that are required for efficient 3′-end formation. These elements are present in the U3 region of the LTR, partly explaining why the upstream polyA site in the 5′ LTR is not used (Ashe et al., 1995; Gilmartin et al., 1995). The removal of these activating sequences does not prevent 3′-end formation but renders the process inefficient (Gilmartin et al., 1995). For instance, polyA sites inserted downstream of the HIV-1 3′ LTR are normally not used, but they become active when the U3 region of the 3′ LTR is removed (Ashe et al., 1995). Thus, we created cell lines expressing HIV-1 reporters lacking the U3 region in the 3′ LTR (clone pTRIP_1_13). Remarkably, when MS2-GFP was bleached at these transcription sites, the signal recovered much more slowly than for WT HIV-1 reporters: half-recovery took 400 s instead of 65 s (Fig. 4 C). Thus, slowing down the rate of 3′-end formation correspondingly increased the time that nascent RNAs remained at their transcription site.
Computer-simulated mRNA synthesis
To better understand HIV-1 RNA biogenesis, we simulated the behavior of individual polymerases in silico. For each time interval, each polymerase could initiate, elongate by one nucleotide, cleave and polyadenylate, and eventually release their associated RNA. The probabilities to perform these steps were the means of the measured rate constants (see Materials and methods). Predicted FRAP curves approximated those obtained experimentally with both the short (Fig. 2 I) and long reporters (Fig. 4 B). Simulated SDs were also in range with experimental ones. Close examination of simulations indicated that a large part of the SD was caused by stochastic variation in the total number of polymerases on the gene, which affected the prebleach values and the extent of recovery.
Interestingly, the parameters measured with the short reporter were slightly too rapid to accurately predict the recovery curve of the long one (Table I and Fig. 4 B). An explanation for this observation might reside in the fact that RNAPII can pause because pausing is expected to be more frequent with longer sequences. Another interesting possibility would be that the site of integration of the reporter in the genome generates some variation in the kinetics of mRNA synthesis and processing.
FRAP analysis of RNAPII
Although the MS2-GFP FRAP assay provides a direct measurement of RNAPII activity, it provides little information on the events that occur either before the polymerase reaches the MS2 repeat or after it releases its mRNA. To gain a complete view of the transcription cycle, we repeated the FRAP assay with fluorescent subunits of RNAPII. HIV-1 transcription sites were identified with a red variant of MS2 (MS2-mCherry), and GFP-tagged subunit C of RNAPII was bleached in the nucleoplasm or at HIV-1 transcription sites. Recoveries at the HIV-1 transcription site were much slower than in the nucleoplasm, suggesting that most polymerases present at the HIV-1 gene array were engaged in transcription (Fig. 6). Interestingly, recovery of the polymerase was substantially slower than recovery of nascent RNAs: in Exo1 cells, half-recovery took 200 s for the polymerase but only 66 s for nascent RNAs.
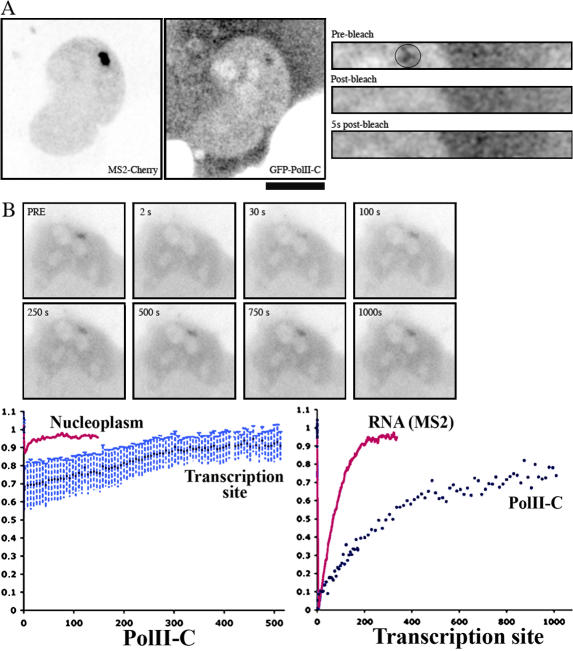
Dynamic of RNAPII at HIV-1 transcription sites. Exo1 cells were transfected with vectors expressing GFP-PolII-C and MS2-Cherry and were analyzed on a confocal (A) or wide-field microscope (B). (A) FRAP of RNAPII analyzed at a high frame rate (160 ms) and on a short time scale (5 s). (left) Image of a transfected cell (25 × 25 μm). (right) Snapshots from a FRAP experiment (8 × 2 μm). The circle indicates the area of bleach. (B) FRAP recovery of GFP-PolII-C analyzed by tracking transcription sites in 3D for 15 min with a wide-field microscope. (top) Image sequence from a FRAP experiment; each field is 25 × 25 μm. (left graph) Recovery curves in the nucleoplasm of transfected U2OS cells (pink) or at the HIV-1 transcription site (blue). Data were normalized to prebleach values. (right graph) Comparison of the recoveries of nascent RNAs (pink) and the polymerase (blue). Data were normalized between pre- and postbleach values. Bar, 9.2 μm.
To extract more information from the recovery curves, they were fitted with the diffusion/reaction model developed by Sprague et al. (2004). This model assumes that the bleached spot contains identical binding sites that are equally distributed in space, and it allows one to derive diffusion coefficients, binding time (tb), and the delay between two binding events (td). The recovery curves obtained with the subunit C of RNAPII were fitted to this model (see Materials and methods). In Exo1 cells, we found that the polymerase resided for 333 s at the HIV-1 transcription site and diffused for 10 min before engaging a second transcription cycle.
When the residency time of the polymerase was compared with that of nascent RNAs, it was obvious that the polymerase remained on the gene longer than expected. Indeed, elongating through the reporter should take 114 s, and 3′-end formation should take 63 s. Thus, 156 s were missing to match the 333 s of the residency time of the polymerase. One possibility could be that RNAPII proceeds after the 3′ LTR before terminating transcription. To test this possibility, we performed chromatin immunoprecipitation experiments. As expected, RNAPII was enriched within the HIV gene after Tat induction (primer sets A and B; Fig. 7 A). Surprisingly however, a PCR fragment located 220–420 bases downstream of the polyA site did not show a comparable enrichment (primer set C), indicating that RNAPII stops rapidly after the end of the gene and is released without proceeding much through neighboring sequences.

Density of RNAPII along the HIV-1 reporter. (A) RNAPII chromatin immunoprecipitation analysis of the HIV-1 reporter. Exo1 cells were incubated with GST-Tat or a mock control and subjected to chromatin immunoprecipitation with anti-RNAPII and the indicated primer sets (A, B, and C). Primer set C is located 220–420 nucleotides after the polyA site. B13 corresponds to a control region in the genome. (middle) PCR reactions on the input and immunoprecipitated chromatin. The size of the DNA fragments is indicated above the panels. (bottom) Quantification of the PCR reaction (mean of two experiments). (B) Schematic of the HIV-1 transcription cycle. The promoter, transcribed, and polyA regions of the HIV-1 reporter are schematized. The proposed residency times of the polymerases (gray disks) in various areas of the gene are indicated.
Discussion
MS2-GFP as a tool to analyze the dynamics of specific RNAs
FRAP is a powerful tool to analyze the dynamics of tagged proteins. In this study, we photobleached MS2-GFP to analyze the turnover of nascent mRNAs tagged with the MS2 repeat. This supposes that there is little dissociation of the MS2-GFP–RNA complex during the course of the experiment. The MS2 protein variant we use has a mutation that increases its affinity by 7.5-fold (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200706018/DC1; Lim and Peabody, 1994). In addition, the RNA-binding site also contains a mutation that decreases the off rate of the protein by 100-fold such that the complex has a half-life of 7 h at 4°C (Table S1; Lowary and Uhlenbeck, 1987). To directly analyze the stability of the complex in vivo, we tagged an RNA stably associated with nucleoli (U3) and found by FRAP analysis that little dissociation occurred within the first 10 min of the experiment. Although it could be argued that the complex might be less stable in the nucleoplasm than in the dense nucleoli, analysis of mutant mRNA (clone pTRIP_1_13) or polymerase (ΔCTD) set a minimal value of 10 min for the half-life of the complex in the nucleoplasm. MS2-GFP has also been previously used to analyze the diffusion of MS2-tagged mRNA in the nucleoplasm (Shav-Tal et al., 2004). Interestingly, similar values (although not identical) were obtained when diffusion was measured by single-particle tracking or photoactivation, which is also in agreement with a slow dissociation rate of MS2-GFP in vivo. Thus, the contribution of the dissociation of MS2-GFP in the FRAP curves modeled in this study is expected to be small and was neglected in the analyses.
To analyze the biogenesis of mRNA in live cells and real time, we engineered cell lines that contained many copies of the reporter integrated in a single place within the chromatin (75 copies for Exo1 cells). This amplifies the signal such that many events become visible by microscopy analysis. For instance, this system can be used to analyze the dynamic of the Tat–pTEFb complex on nascent HIV-1 RNAs (Molle et al., 2007). However, the repeated structure of the transgene might affect mRNA biogenesis in unknown ways. Clearly, the next frontier will be to perform similar analysis on single-copy genes.
The use of live cell imaging technologies allows the analysis of mRNA biogenesis in intact cells and with an unprecedented temporal resolution. However, this process is highly complex and is composed of a myriad of successive and interconnected steps. The drawback of these analyses is therefore that it is difficult to separate each step for detailed analysis. In addition, complete modeling of mRNA biogenesis yields a very large number of variables that cannot be determined and measured in a simple FRAP experiment. In this study, we have attempted to alleviate these limitations by the use of simplified models, which can thus be constrained by experimental data. The reliability of the models was then assessed through the use of mutant RNA, mutant polymerases, drugs, and quantitative in situ hybridization.
Kinetics of mRNA biogenesis and the transcription cycle of RNAPII
In this study, we were able to visualize mRNA transcription and processing in real time and single cells by fluorescence tagging of HIV-1 RNAs. By performing FRAP analysis on RNAPII and nascent RNAs through MS2-GFP, we could obtain a view of the entire HIV-1 transcription cycle. For this, the polymerase recovery curves were fitted with a diffusion- binding model and compared with that of nascent RNAs. This indicated that during the 333 s that the polymerase resided at the HIV-1 transcription site, 114 s could be attributed to elongation, and 63 s could be attributed to 3′-end processing and transcript release. The remaining 156 s could be the result of initiation or termination. In an attempt to discriminate these possibilities, we investigated the localization of Xrn2, a 5′ → 3′ exonuclease involved in transcription termination (West et al., 2004). Xrn2 is loaded cotranscriptionally at the end of genes (Luo et al., 2006), and it degrades the 3′ cleavage product generated by the 3′-end maturation of mRNAs (West et al., 2004). Interestingly, Xrn2 was present only in a minute amount at the HIV-1 transcription site (unpublished data), suggesting that termination and polymerase release may be rapid. Thus, a large fraction of the missing 156 s could be the result of initiation. In this view, as many as 47% of the polymerases could be initiating transcription at the promoter, 34% would be undergoing processive elongation, and 19% would be processing the RNA at the 3′-end (Fig. 7 B). However, it is equally possible that the steps between pre-mRNA cleavage and the entry of Xrn2 account for some of the missing 156 s. Likewise, an alternative hypothesis to explain the long residency time of the polymerase might be that a fraction of the polymerase would reengage transcription on the same gene by a looping mechanism, as described in yeast (O'Sullivan et al., 2004; Ansari and Hampsey, 2005). Finally, we also cannot exclude that some polymerases present at the HIV-1 transcription sites are stalled or are involved in as yet uncharacterized but slow processes.
Transcription initiation
We found that although the residency time of the polymerase at the HIV-1 gene was 333 s, the diffusion time between two binding events was 10 min. Previous FRAP experiments of the large subunit of RNAPII at random nucleoplasmic sites have indicated that for cellular genes, polymerases should spend only a third of their time engaged in transcription (Kimura et al., 2002). Because the half-life of transcription was estimated as 20 min, it was deduced that polymerases spend as much as 90 min diffusing between two transcriptional cycles (Kimura et al., 2002). Thus, initiation at the HIV-1 promoter is nearly 10 times more frequent than at cellular genes. This could be caused by a high efficiency of transcription initiation at the HIV-1 site or, alternatively, by a much higher density of active genes at the HIV-1 transcription site. Interestingly, we did not detect a rapid component in the recovery curves of RNAPII (Fig. 6 A), which corresponds to polymerases undergoing rapid binding and dissociation from the promoter (Darzacq et al., 2007). In contrast, the vast majority of polymerases appeared transiently immobilized on the HIV-1 gene and engaged in productive transcription. This suggested that the initiation of HIV-1 transcription was indeed very efficient. A high efficiency of initiation would be consistent with biochemical studies that have shown that the HIV-1 promoter is constantly occupied by the polymerase (Jeang et al., 1999; Marcello et al., 2001). In addition, it is well established that a major mode of trans-activating the HIV-1 promoter is at the level of elongation through the binding of Tat to nascent RNAs, and this requires that the promoter can efficiently initiate transcription (for reviews see Jeang et al., 1999; Marcello et al., 2001).
Transcription elongation
By analyzing the turnover of nascent RNAs, we found that nascent transcripts were elongated at a rate of 1.89 kb/min. Four important controls support these numbers. First, increasing the length of the mRNA increased the total synthesis time but yielded a similar elongation rate. Second, quantitative in situ hybridization indicated that a substantial fraction of nascent RNAs were at the level of 3′-end processing or transcript release, and the fraction of incompletely transcribed RNAs was consistent with the calculated elongation rate. Third, a slow mutant of RNAPII induced a reduced transcription rate. Fourth, treating cells with camptothecin, an inhibitor of elongation, also reduced the rate of RNA synthesis.
RNAPII is a fundamental enzyme in the cell. However, few studies were able to directly measure its activity in vivo. In vitro kinetic analyses of human RNAPII have yielded elongation rates between 0.9 and 1.8 kb/min and have suggested that the chemical step is rate limiting (Burton et al., 2005). Our in vivo data are in the upper range of these values, indicating that RNAPII may work close to its maximal speed. The giant gene of human dystrophin and a long yeast gene have also been used to evaluate elongation rates (Tennyson et al., 1995; Mason and Struhl, 2005). These studies yielded values of 2.4 and 2 kb/min, which is in agreement with our measures. The range of values obtained in different biological systems have observed elongation rates ranging from 1.5 kb/min for living bacteria to 5.7 kb/min for eukaryotic ribosomal RNA genes (Dundr et al., 2002; Golding et al., 2005). Our model system provides a tool of choice for the quantitative analysis of transcription in real time and living cells.
In vitro, single-molecule analyses of bacterial RNA polymerases have shown that the time taken to transcribe a DNA segment can vary substantially between individual molecules, and at least part of this variability can be attributed to polymerase pausing (Herbert et al., 2006). In higher eukaryotes, pausing is also a well-known phenomenon, and there is some evidence that the elongation rate can regulate gene expression (Batsche et al., 2006). We describe elongation as the repetition of an elementary step and model it as a straight line in the recovery curves. This does not take into account the polymerase heterogeneity that can be caused by pausing and may directly contribute to the exponential component of the curve. Although determining polymerase pausing time may not be easy in many cases, it seems that pausing occurs when cells were treated with camptothecin. Indeed, in this case, the MS2-GFP FRAP assay indicates a large increase in the exponential component of the recovery curve. Because camptothecin is believed to physically arrest the polymerase, it is tempting to speculate that this increase represents pausing.
3′-end processing and termination
3′-end formation is a critical step during mRNA biogenesis, yet the rates of this reaction are not known. Our study indicates a rapid release of mRNA once polyadenylation is initiated (9 s). In contrast, the preceding steps were relatively slow (55 s). These steps probably correspond to the cleavage of the pre-mRNA, as indicated by the much longer residency time of a mutant RNA known to have a slow rate of 3′-end cleavage (pTRIP_1_13). However, we cannot rule out other possibilities. For instance, cleavage could be rapid, but a quality control step could occur before polyadenylation and could prevent mRNA release. Indeed, yeast data indicate that a quality control step occurs at the level of transcript release (Jensen at al., 2001), and detailed localization studies in mammalian cells have shown that some transcript can accumulate at the transcription site after detaching from the gene (Smith et al., 1999; Johnson et al., 2000). It is also possible that the cleaved RNA could be released before being polyadenylated.
However, with the most likely possibility being that cleavage is slow and rapidly followed by polyadenylation, we estimated that cleavage and release occurred in 55 s and 9 s, respectively. Remarkably, when the U3 sequences of the 3′ LTR were removed, the rate of 3′-end formation was dramatically reduced. The half-life of these RNAs at the transcription site was 400 s, against 65 s for WT RNAs. This indicates that the cleavage reaction may occur as much as 10 times more slowly for the mutant RNAs. This was consistent with previous biochemical analyses that have shown that U3 contains sequences that can markedly stimulate 3′-end formation by enhancing the recruitment of cleavage and polyadenylation specificity factor (Gilmartin et al., 1995). Thus, we expect that the rates of 3′-end formation and polymerase read-through may differ markedly from gene to gene depending on the strength of the polyA site. In particular, situations in which alternative splicing regulates polyA site usage implies that 3′-end formation at the first site is slow enough to let the polymerase reach the splice junctions.
Remarkably, ChIP analysis of the distribution of RNAPII along the HIV-1 gene indicated that the density of polymerases dropped sharply a few hundred bases after the polyA site, similar to what has been observed recently with an HSP70 gene of Drosophila melanogaster (Zhang and Gilmour, 2006). Because cleavage is estimated at 55 s and elongation is estimated at 2 kb/min, this suggests that polymerases pause and/or loose their processivity after passing the polyA site. This would be consistent with several previous studies. First, run-on assays and in vitro transcription reactions have suggested that the polymerase looses its processivity as it passes the polyA site (Nag et al., 2006). Second, pause sites have been found adjacent to polyA sites (Gromak et al., 2006). Third, it has recently been shown that the 3′-end processing factor Pcf11 and the exonuclease Xrn2 have a role in promoting transcription termination at the end of cellular genes (West et al., 2004; Zhang and Gilmour, 2006) by dismantling paused elongation complexes (Gromak et al., 2006; Zhang and Gilmour, 2006). Altogether, our data are in agreement with previously proposed models in which the 3′-end processing machinery would assemble on nascent RNAs, whereas the polymerase would pause after passing the polyA site. Completion of assembly and pre-mRNA cleavage would occur in about one minute, after which the mRNA would be rapidly released and the polymerase would be dismantled.
Materials and methods
Cells and plasmids
U2OS cells were cultivated at 37°C in DME containing 10% FCS. For live cell imaging, cells were maintained in the same medium, except it did not contain riboflavin and phenol red (Fusco et al., 2003). Stable transformants were obtained with the calcium-phosphate procedure by cotransfecting a 20-fold excess of the vector of interest with Ptk-Hygro and selecting cells with 132 μg/ml hygromycin. Individual clones were expanded, and their gene copy number was measured by quantitative PCR using DNA from U1 cells as reference (two copies per genome). Exo1 and ExoLong contained 75 and 70 copies, respectively.
For live cell experiments, cells were plated on glass, transiently transfected with LipofectAMINE with vectors expressing Tat and MS2-GFP, and analyzed 24 h later at 37°C in a nonfluorescent media (Fusco et al., 2003). For polymerase replacement, cells were treated for 2.5 h with 100 μg/ml α-amanitin before FRAP. Control experiments showed that this was sufficient to induce the disappearance of transcription sites in cells that did not express α-amanitin–resistant forms of the polymerase. Treatment of cells for 2 h with 10 μg/ml actinomycin D resulted in the disappearance of the spots in all cases.
Plasmids expressing MS2-GFP and the hC4 and WTRES mutant of RNAPII have been described previously (de la Mata et al., 2003; Fusco et al., 2003). Plasmid expressing PABPN1-GFP was a gift from M. Carmo-Fonseca (Institute of Molecular Medicine, University of Lisbon, Portugal). MS2-Cherry and GFP-PolII-C were created with the Gateway system (Invitrogen). U3-MS2 was created by inserting a single MS2 site in the apical loop of the rat U3B.7 gene. pExo-MS2×24 plasmid was derived from the plasmid pEV731 (Jordan et al., 2001) by cloning 24×MS2 repeats into the ClaI–XhoI sites. pExo-MS2×24-Long was constructed by inserting a cassette coding for CFP with the peroxisome localization signal SKL, the internal ribosome entry site from encephalomyocarditis virus, and the thymidine kinase from HSV-1 into the unique XhoI site (Marcello and Giaretta, 1998). The clone pTRIP that lacked U3 sequences in the 3′ LTR originated from a similar vector.
Chromatin immunoprecipitation
Chromatin immunoprecipitation analysis was performed on Exo1 cells treated with GST-Tat essentially as previously described (Lusic et al., 2003; du Chene et al., 2007). Primer set A corresponds to primer sets for Nuc1 in Lusic et al. (2003) and map to the promoter-proximal region. Primer set B corresponds to a region of the vector proximal to the 3′ LTR (primer Bfw [5′-CATGGAGCAATCACAAGTAGC-3′] and primer Brv [5′-ATCTTGTCTTCGTTGGGAGTG-3′]). Primer set C maps 3′ of the 3′ LTR within the backbone of the vector (primer Cfw [5′-AGCATCTGGCTTACTGAAGCAG-3′] and primer Crv [5′-ATCGGTGATGTCGGCGATATAG-3′]). Primer set B13 corresponds to an unrelated genomic region, as described in Lusic et al. (2003). Quantification of immunoprecipitated material was performed by semiquantitative PCR and normalized for input DNA and for B13. The antibody against RNAPII was purchased from Santa Cruz Biotechnology, Inc. (N-20).
In situ hybridization
In situ hybridization was performed as previously described (Fusco et al., 2003). The formamide concentration was 50% in the hybridization and washing mixture except for the pA+ probe (hybridized and washed at 30% formamide) and the MS2 probe (10% formamide). The sequences of the probes were as follows (X stands for amino-allyl-T): HIV_I_E2 (5′-A X GGGTTGGGAGGTGGGTC X GAAACGATAATGGTGAAT X A); HIV_E1a (5′-A X GAGAGCTCCTCTGG X TTCCCTTTCGCTT X CAAGTCCCTGTTC X A); HIV_E1b (5′-A X TCTTGCCGTGCGCGCT X CAGCAAGCCGAGTCCTGCGT X A); HIV_intron1 (5′-A X TCTCGCACCCATC X CTCTCCTTCTAGCC X CCGCTAGTCAAAAT X A); HIV_intron2 (5′-A X AACTGCGAATCGT X CTAGCTCCCTGCT X GCCCATACTATATGTT X A); HIV_Exon2a (5′-A X GTGGCTAAGATC X ACAGCTGCCTTGTAAG X CATTGGTCTTAAA X A); HIV_Exon2b; (5′-A X ATCTTGTCTTCG X TGGGAGTGAATTAGCCC X TCCAGTCCCCC X A); HIV_pA+ (5′-A X TTTTTTTTTTTTTTTTTTTTTTTTTT X TGAAGCACTCAAGGCAAGC X A); MS2 (transcribed strand; 5′-A X GTCGACCTGCAGACA X GGGTGATCCTCA X GTTTTCTAGGCAAT X A); and MS2 (nontranscribed strand; 5′-A X AGTATTCCCGGG X TCATTAGATCC X AAGGTACCTAATTGC X A). The modified oligonucleotide probes for RNA FISH were synthesized by J-M. Escudier (Plateforme de synthèse d'Oligonucléotides modifiés de l'Interface Chimie Biologie de l'ITAV).
For quantitative measurements, 3D image stacks were collected and deconvolved with Hyugens (Bitplane AG). Background was removed, and the total light intensity at the transcription site was calculated and divided by the number of planes. The number of molecules was then computed from a calibration curve of the probes (Fusco et al., 2003), or, alternatively, transcription site signals were normalized to the ones of the cytoplasm. For each probe, 15–40 transcription sites were analyzed.
For Cy3/Cy5 quantification relative to the MS2-Cy5 probe (Fig. 4), cells were hybridized sequentially with the Cy3 and MS2-Cy5 probes (the MS2 probes hybridized at the lower stringency than the other probes). Images were taken in both channels, and the amount of light at the transcription site was calculated using the same mask for both colors. The Cy3/Cy5 ratios were then corrected for the specific activity of each probe by measuring the signals of an equimolar solution of the Cy3 and Cy5 probes under the microscope. The SDs were E1 (0.55), I (1.55), I-E2 (0.35), and E2 (0.16).
For the quantification of E1-Cy5 versus E2-Cy3 (Fig. S4), the two probes were hybridized simultaneously, and the signal of the two probes at the transcription site was normalized to the amount of signal in the cytoplasm. For this, each image stack was summed along the depth axis (z), and the projected signal was quantified along a line (six pixels wide) that passed across the transcription site. The two probes were normalized by first removing the background and then equalizing the cytoplasmic signals. The surface of the peak at the transcription sites was measured for both probes, and the values of exon 1 were divided by that of exon 2.
Immunofluorescence and image acquisition of fixed cells
Immunofluorescence was performed as previously described (Marcello et al., 2003). Anti–Pol II (all isoforms) was used at the following dilutions: 8WG16 at 1:100 and anti-SC35 (Sigma-Aldrich) at 1:100. Fluorescent images of fixed cells were captured on a 100× NA 1.4 wide-field microscope (DMRA; Leica) equipped with a camera (CoolSNAP HQ; Roper Scientific) and was controlled by MetaMorph software (Universal Imaging Corp.). Stacks of wide-field images were deconvolved with Huygens (Bitplane AG) and mounted with Photoshop (Adobe).
FRAP experiments
For live cell imaging, cells were maintained at 37°C in appropriate medium (Fusco et al., 2003). Two microscopic setting were used to perform FRAP. For analysis of rapid recoveries (Fig. 2, A and B), we used a confocal microscope (Meta LSM510; Carl Zeiss MicroImaging, Inc.) with a 100× NA 1.4 objective. MS2-GFP at transcription sites or in the nucleoplasm was bleached at 488 nm in a circle of 1.5-μm diameter at full laser power and for one passage (bleaching time of 100 ms). Recoveries were measured at a high frame rate (one image for ≤160 ms) and for a short time (up to 10 s) using ≤1% of the power of the 488-nm laser line. Images were analyzed as previously described (Phair and Misteli, 2000) by recording the fluorescence of the bleached region. Background was removed, intensities at each time point were corrected for bleaching by dividing them by the total cell fluorescence, and these values were finally normalized by dividing them with the fluorescent intensity before the bleach. In Figs. 2 (E–I), 3, and 6 B (right), postbleach values were additionally set to zero.
When recovery of transcription sites had to be recorded for periods exceeding 10 s, we used an adapted microscopic setting. A microscope (TE200; Nikon) equipped for both confocal and wide-field imaging was used with a 100× NA 1.45 objective. Transcription sites were bleached with the confocal port using a circular region of 2.5-μm diameter (bleaching time of 1 s). Recoveries were then recorded in the wide-field port using MetaMorph (Universal Imaging Corp.) with an excitatory light of low intensity. Images were recorded with an EM-CCD camera (Cascade 512K; Roper Scientific). Stacks of nine images 0.5 μm apart were collected every 3 s (one stack took 0.5–1 s). For image analysis, fluorescence intensities were measured in a small parallelepiped (1 × 1 × 1.5 μm) placed at the most intense area of the transcription site. This operation was performed automatically by a macro that was created in ImageJ software (National Institutes of Health). This automatic tracking of transcription sites in 3D allowed us to correct cell movements and to minimize signal from diffusing MS2-GFP. When a nucleoplasmic region devoid of transcription site was bleached, the cube was placed at the center of the bleached region. The values obtained were then treated and normalized as in the previous paragraph except that the postbleach value was taken at the 5-s time point. Indeed, at this time, the diffusing pool of MS2-GFP had come back very close to its equilibrium, allowing us to neglect the diffusion of MS2-GFP in the analysis (Fig. 2). In all figures except Fig. 2 C, the postbleach value was set to zero to facilitate comparison of the curves.
Diffusion coefficients
Diffusion coefficients were measured by FRAP on transfected HeLa cells. For 21 μm2/s of free GFP, we exploited the solution for free diffusion described by Soumpasis (1983). For 15 μm2/s MS2-GFPnls, we exploited the solution proposed for the reaction-diffusion model by Sprague et al. (2004).
Curve fitting
For the MS2-GFP FRAP experiment, the curve of 10–20 cells were averaged and fitted with a straight line followed by an exponential: f(t) = A + α × t, t ≤ telong; f(t) = A + α × telong + (B − A − α × telong) × (1 − exp[−α × (t − telong)/(B −A − α × telong)]), t > telong. telong is the time point at which the line converts into an exponential and corresponds to the length of the linear phase. A is the intensity at the zero time point, α is the slope of the initial linear part, and B is the immobile fraction. The minimization routine of the C++ GNU Scientific Library (http://www.gnu.org/software/gsl/) was used for finding the minimum of the chi square. Chi square values were 0.06, 0.06, 0.04, 0.01, and 0.04 for Exo1, Exo2, ExoLong, Wtres, and hC4, respectively.
Recovery curves of RNA polymerase at the transcription site were fitted with the binding-only simplification of the diffusion/reaction model developed by Sprague et al. (2004): f(t) = Ceq × exp[−Koff × t], where Koff is the binding off rate and Ceq is the ratio of on and off rates. This model was fitted with the R (http://www.r-project.org/) nonlinear least squares function. This yielded a residency time of 333 s and a delay between two binding events of 660 s.
Computer simulations of mRNA biogenesis
The software was written in Metal, a BASIC emulator for Mac computers (http://www.iit.edu/~sarimar/GDS/metal.html). It simulated a small population of RNA polymerase molecules using a stochastic model: during an elementary time period, polymerases in a given state had a certain probability to perform the next reaction in the pathway. We used the following circular scheme: inactive → first nucleotide transcribed → second nucleotide transcribed…last nucleotide transcribed → pre-mRNA is cleaved → mRNA is released and polymerase is inactive.
The kinetic rates of each step were determined from the experimental data: elongation by one nucleotide, 31.5 s−1 (1.89 kb/min); cleavage, 0.018 s−1 (54.7 s); and mRNA release, 0.11 s−1 (8.8 s). To estimate initiation rates, we measured the number of RNA molecules by quantitative in situ hybridization using the MS2 probe. We found that a mean of 105 RNA molecules was present at the transcription site of Exo1 cells (±50). Because FRAP experiments estimated that the MS2-tagged RNAs stayed a mean of 128 s at this site, the initiation rate was deduced to 0.99 s−1 for the entire array. The total number of polymerases simulated was calculated such that 105 molecules of MS2-tagged RNA were present at the transcription site at equilibrium. At the start of the simulation, the population of polymerase was distributed according to the equilibrium. Then, at each time point of the simulation and for each polymerase, a random draw determined whether the next step of the pathway occurred or not. The probabilities of success were calculated to match the rate constants, and the time resolution was small enough to ensure that a single event per polymerase could occur. A variable fluorescent value was attributed to each polymerase, which corresponded to the number of nucleotides transcribed in the MS2 repeat. For the simulation of FRAP, all fluorescence was set to zero, and recovery was plotted as a function of time. The values obtained were treated as experimental data (i.e., normalized between the pre- and postbleach values).
Estimation of transcription elongation versus processing/release time by direct comparison of the FRAP curves of the short and long reporter
Curves normalized for their initiation rate (the initial slope of the curve) are shown in Fig. 4 B and indicate that 1.5 times more RNAs accumulated at the transcription site of the long reporter than for the short one. If initiation rates are constant, the amount (i) of fluorescent RNA at the transcription site is proportional to the time interval (t) between the moment polymerase reaches the MS2 site and the moment RNA leaves the transcription site. Thus, t = k × i, where k is a constant. However, t can be decomposed in two components: a variable one as a result of elongation (t1), which is proportional the length (l) of RNA that remains to be transcribed, and a constant time (t2), representing the time required for 3′-end processing and release. Thus, k × i = a × l + t2, where a is a constant. Because l is 2,180 for the short reporter and 4,580 for the long one, it follows that t1 is 1.2 times t2 for the short reporter and 2.5 times t2 for the long one.
Estimation of elongation versus processing time by quantitative in situ hybridization in Exo1 cells
The ratio between 5′ and 3′ probes depends on the relative amount of time that the polymerase spends on the transcription site once it reaches these hybridization sites. The same hypothesis was made as above by assuming that the time that a nascent RNA remains at the transcription site is decomposed into a variable part related to elongation and a constant part related to all other processes. In this case, the intensity (i) at a given hybridization site can be written as k × i = a × l + t2, where l is the length separating the hybridization site from the polyA site and k and a are constants. Assuming an elongation rate of 2.03 kb/min and a 3′-end processing time of 63.5 s (in Exo1 cells), one can calculate the values given in Fig. 5.
Estimation of cleavage and release rates in Exo1 cells
The FRAP curves yield a total time for 3′-end formation and release of 67.3 s on average. 3′-end formation can be decomposed in cleavage, polyadenylation, and transcript release. This latter rate can be estimated from the amount of polyadenylated mRNA present at the transcription site of Exo1 cells. The polyadenylated species represents 11% of the exon 2 signal, meaning that when a polymerase reaches exon 2, the nascent RNA then spends 89% of its time completing transcription and 3′-end processing and 11% as a polyadenylated species. Because the exon 2 probe is 560 nucleotides away from the polyA site, it should take polymerases 16.6 s to go from there to the polyA site (at 2.03 kb/min; value from Exo1 cells) and then a further 63.5 s to process the RNA (value from Exo1 cells). Thus, polyadenylated RNAs should remain at transcription sites 11% of 80.1 s (i.e., 8.8 s, yielding a rate of 0.11 s−1). Because a total time of 63.5 s is required for 3′ processing, the time required for cleavage/polyadenylation was then estimated at 54.7 s (0.018 s−1).
Online supplemental material
Fig. S1 shows that transcription of HIV-1 mRNA is induced by Tat and PMA/ionomycin in Exo1 cells. Fig. S2 shows that MS2-GFP is stably bound to its target RNA in vivo. Fig. S3 shows that elongation can be modeled with a straight line. Fig. S4 shows quantification of exon 1 versus exon 2 at the HIV-1 transcription site. Table S1 provides RNA binding properties of the coat protein of phage MS2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200706018/DC1.
Acknowledgments
We thank R. Bordonné, G. Pegoraro, and T. Vasselon for critical readings of the manuscript. We are particularly indebted to X. Darzacq and R.H. Singer for their help and advice as well as for the scientific exchanges that occurred during the course of this project.
This work was supported by the Association pour la recherche sur le cancer (grant 3109), the European Community Systems Biology of RNA Metabolism in Yeast project (grant LSHG-CT-2005-518280), and the Network of Excellence European Alternative Splicing Network. A. Marcello was supported by the European Community Specific Targeted Research Projects consortium (grant 012182), a Human Frontier Science Program Young Investigators grant, and the AIDS project of the Istituto Superiore di Sanità of Italy. A. Kornblihtt is a Howard Hughes Medical Institute International Research Scholar and a career investigator of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). M. de la Mata is a recipient of a CONICET fellowship. E. Basyuk was supported by a fellowship from Agence nationale de recherche sur le sida.
Notes
S. Boireau, P. Maiuri, and E. Basyuk contributed equally to this paper.
Abbreviations used in this paper: CTD, C-terminal domain; HIV, human immunodeficiency virus; LTR, long terminal repeat; RNAPII, RNA polymerase II; WT, wild type.
References
- Ansari, A., and M. Hampsey. 2005. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 19:2969–2978. [Europe PMC free article] [Abstract] [Google Scholar]
- Ashe, M., P. Griffin, W. James, and N. Proudfoot. 1995. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 9:3008–3025. [Abstract] [Google Scholar]
- Ashe, M.P., L.H. Pearson, and N.J. Proudfoot. 1997. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 16:5752–5763. [Europe PMC free article] [Abstract] [Google Scholar]
- Batsche, E., M. Yaniv, and C. Muchardt. 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 13:22–29. [Abstract] [Google Scholar]
- Braga, J., J.G. McNally, and M. Carmo-Fonseca. 2007. A reaction-diffusion model to study RNA motion by quantitative fluorescence recovery after photobleaching. Biophys. J. 92:2694–2703. [Europe PMC free article] [Abstract] [Google Scholar]
- Burton, Z.F., M. Feig, X.Q. Gong, C. Zhang, Y. Nedialkov, and Y. Xiong. 2005. NTP-driven translocation and regulation of downstream template opening by multi-subunit RNA polymerases. Biochem. Cell Biol. 83:486–496. [Abstract] [Google Scholar]
- Capranico, G., F. Ferri, M. Fogli, A. Russo, L. Lotito, and L. Baranello. 2007. The effects of camptothecin on RNA polymerase II transcription: roles of DNA topoisomerase I. Biochimie. 89:482–489. [Abstract] [Google Scholar]
- Coulter, D., and A.L. Greenleaf. 1985. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J. Biol. Chem. 260:13190–13196. [Abstract] [Google Scholar]
- Darzacq, X., Y. Shav-Tal, V. de Turris, Y. Brody, S.M. Shenoy, R.D. Phair, and R.H. Singer. 2007. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14:796–806. [Europe PMC free article] [Abstract] [Google Scholar]
- de la Mata, M., C.R. Alonso, S. Kadener, J.P. Fededa, M. Blaustein, F. Pelisch, P. Cramer, D. Bentley, and A.R. Kornblihtt. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 12:525–532. [Abstract] [Google Scholar]
- du Chene, I., E. Basyuk, Y. Lin, R. Triboulet, A. Knezevich, C. Chable-Bessia, C. Mettling, V. Baillat, J. Reynes, P. Corbeau, et al. 2007. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26:424–435. [Europe PMC free article] [Abstract] [Google Scholar]
- Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L. Rothblum, R. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 298:1623–1626. [Abstract] [Google Scholar]
- Femino, A.M., F.S. Fay, K. Fogarty, and R.H. Singer. 1998. Visualization of single RNA transcripts in situ. Science. 280:585–590. [Abstract] [Google Scholar]
- Fusco, D., N. Accornero, B. Lavoie, S. Shenoy, J. Blanchard, R. Singer, and E. Bertrand. 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13:161–167. [Europe PMC free article] [Abstract] [Google Scholar]
- Gilmartin, G.M., E.S. Fleming, J. Oetjen, and B. Graveley. 1995. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 9:72–83. [Abstract] [Google Scholar]
- Golding, I., J. Paulsson, S.M. Zawilski, and E.C. Cox. 2005. Real-time kinetics of gene activity in individual bacteria. Cell. 123:1025–1036. [Abstract] [Google Scholar]
- Greene, W.C., and B.M. Peterlin. 2002. Charting HIV's remarkable voyage through the cell: basic science as a passport to future therapy. Nat. Med. 8:673–680. [Abstract] [Google Scholar]
- Gromak, N., S. West, and N.J. Proudfoot. 2006. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell Biol. 26:3986–3996. [Europe PMC free article] [Abstract] [Google Scholar]
- Herbert, K., A. La Porta, B. Wong, R. Mooney, K. Neuman, R. Landick, and S. Block. 2006. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 125:1083–1094. [Europe PMC free article] [Abstract] [Google Scholar]
- Jeang, K.T., H. Xiao, and E.A. Rich. 1999. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 274:28837–28840. [Abstract] [Google Scholar]
- Jensen, T.H., K. Patricio, T. McCarthy, and M. Rosbash. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell. 7:887–898. [Abstract] [Google Scholar]
- Johnson, C., D. Primorac, M. McKinstry, J. McNeil, D. Rowe, and J.B. Lawrence. 2000. Tracking COL1A1 RNA in osteogenesis imperfecta. Splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J. Cell Biol. 150:417–432. [Europe PMC free article] [Abstract] [Google Scholar]
- Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726–1738. [Europe PMC free article] [Abstract] [Google Scholar]
- Kass, S., K. Tyc, J. Steitz, and B. Sollner-Webb. 1990. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 60:897–908. [Abstract] [Google Scholar]
- Kimura, H., K. Sugaya, and P.R. Cook. 2002. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159:777–782. [Europe PMC free article] [Abstract] [Google Scholar]
- Lim, F., and D.S. Peabody. 1994. Mutations that increase the affinity of a translational repressor for RNA. Nucleic Acids Res. 22:3748–3752. [Europe PMC free article] [Abstract] [Google Scholar]
- Lowary, P.T., and O. Uhlenbeck. 1987. An RNA mutation that increase the affinity of an RNA-protein interaction. Nucleic Acids Res. 15:10483–10493. [Europe PMC free article] [Abstract] [Google Scholar]
- Luo, W., A.W. Johnson, and D.L. Bentley. 2006. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 20:954–965. [Europe PMC free article] [Abstract] [Google Scholar]
- Lusic, M., A. Marcello, A. Cereseto, and M. Giacca. 2003. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 22:6550–6561. [Europe PMC free article] [Abstract] [Google Scholar]
- Malim, M.H., J. Hauber, S.Y. Le, J.V. Maizel, and B.R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 338:254–257. [Abstract] [Google Scholar]
- Marcello, A. 2006. Latency: the hidden HIV-1 challenge. Retrovirology. 3:7. [Europe PMC free article] [Abstract] [Google Scholar]
- Marcello, A., and I. Giaretta. 1998. Inducible expression of herpes simplex virus thymidine kinase from a bicistronic HIV1 vector. Res. Virol. 149:419–431. [Abstract] [Google Scholar]
- Marcello, A., M. Zoppe, and M. Giacca. 2001. Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life. 51:175–181. [Abstract] [Google Scholar]
- Marcello, A., A. Ferrari, V. Pellegrini, G. Pegoraro, M. Lusic, F. Beltram, and M. Giacca. 2003. Recruitment of human cyclin T1 to nuclear bodies through direct interaction with the PML protein. EMBO J. 22:2156–2166. [Europe PMC free article] [Abstract] [Google Scholar]
- Mason, P.B., and K. Struhl. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 17:831–840. [Abstract] [Google Scholar]
- Meinhart, A., T. Kamenski, S. Hoeppner, S. Baumli, and P. Cramer. 2005. A structural perspective of CTD function. Genes Dev. 19:1401–1415. [Abstract] [Google Scholar]
- Meininghaus, M., R.D. Chapman, M. Horndasch, and D. Eick. 2000. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J. Biol. Chem. 275:24375–24382. [Abstract] [Google Scholar]
- Moen, P.T.J., C. Johnson, M. Byron, L. Shopland, I. de la Serna, A. Imbalzano, and J. Lawrence. 2004. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol. Biol. Cell. 15:197–206. [Europe PMC free article] [Abstract] [Google Scholar]
- Molle, D., P. Maiuri, S. Boireau, E. Bertrand, A. Knezevich, A. Marcello, and E. Basyuk. 2007. A real-time view of the TAR:Tat:pTEFb complex at HIV-1 transcription sites. Retrovirology. 4:36. [Europe PMC free article] [Abstract] [Google Scholar]
- Nag, A., K. Narsinh, A. Kazerouninia, and H. Martinson. 2006. The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity. RNA. 12:1534–1544. [Europe PMC free article] [Abstract] [Google Scholar]
- O'Sullivan, J., S. Tan-Wong, A. Morillon, B. Lee, J. Coles, J. Mellor, and N. Proudfoot. 2004. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 36:1014–1018. [Abstract] [Google Scholar]
- Peterlin, B.M., and D.H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 23:297–305. [Abstract] [Google Scholar]
- Phair, R., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–609. [Abstract] [Google Scholar]
- Schwartz, S., B.K. Felber, D.M. Benko, E.M. Fenyo, and G.N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency. J. Virol. 64:2519–2529. [Europe PMC free article] [Abstract] [Google Scholar]
- Shav-Tal, Y., X. Darzacq, S. Shenoy, D. Fusco, S. Janicki, D. Spector, and R. Singer. 2004. Dynamics of single mRNPs in nuclei of living cells. Science. 304:1797–1800. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith, K., P. Moen, K. Wydner, J. Coleman, and J. Lawrence. 1999. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 144:617–629. [Europe PMC free article] [Abstract] [Google Scholar]
- Soumpasis, D.M. 1983. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41:95–97. [Europe PMC free article] [Abstract] [Google Scholar]
- Sprague, B.L., R.L. Pego, D.A. Stavreva, and J.G. McNally. 2004. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86:3473–3495. [Europe PMC free article] [Abstract] [Google Scholar]
- Tennyson, C.N., H.J. Klamut, and R.G. Worton. 1995. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat. Genet. 9:184–190. [Abstract] [Google Scholar]
- Wei, P., M.E. Garber, S.M. Fang, W.H. Fischer, and K.A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 92:451–462. [Abstract] [Google Scholar]
- Weinberger, L.S., J.C. Burnett, J.E. Toettcher, A.P. Arkin, and D.V. Schaffer. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 122:169–182. [Abstract] [Google Scholar]
- West, S., N. Gromak, and N.J. Proudfoot. 2004. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 432:552–525. [Abstract] [Google Scholar]
- Zhang, Z., and D. Gilmour. 2006. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol. Cell. 21:65–74. [Abstract] [Google Scholar]
Articles from The Journal of Cell Biology are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1083/jcb.200706018
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jcb/article-pdf/179/2/291/1333176/jcb_200706018.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Human RNA Polymerase II Segregates from Genes and Nascent RNA and Transcribes in the Presence of DNA-Bound dCas9.
Int J Mol Sci, 25(15):8411, 01 Aug 2024
Cited by: 0 articles | PMID: 39125980 | PMCID: PMC11312690
Kinetic pathway of HIV-1 TAR cotranscriptional folding.
Nucleic Acids Res, 52(10):6066-6078, 01 Jun 2024
Cited by: 0 articles | PMID: 38738640
Infectious Virus Tracking by Fluorescent Live Cell Imaging in Primary Cells.
Methods Mol Biol, 2807:3-14, 01 Jan 2024
Cited by: 0 articles | PMID: 38743217
Transcriptional Stochasticity as a Key Aspect of HIV-1 Latency.
Viruses, 15(9):1969, 21 Sep 2023
Cited by: 4 articles | PMID: 37766375 | PMCID: PMC10535884
Review Free full text in Europe PMC
The predictors of unsuppressed viremia among PLHIV: a cross-sectional study in Ghana.
BMC Public Health, 23(1):1113, 09 Jun 2023
Cited by: 1 article | PMID: 37296400 | PMCID: PMC10257285
Go to all (131) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Fast transcription rates of RNA polymerase II in human cells.
EMBO Rep, 12(12):1280-1285, 01 Dec 2011
Cited by: 60 articles | PMID: 22015688 | PMCID: PMC3245692
Measuring the kinetics of mRNA transcription in single living cells.
J Vis Exp, (54):e2898, 25 Aug 2011
Cited by: 3 articles | PMID: 21904295 | PMCID: PMC3253721
In vivo dynamics of RNA polymerase II transcription.
Nat Struct Mol Biol, 14(9):796-806, 05 Aug 2007
Cited by: 426 articles | PMID: 17676063 | PMCID: PMC4942130
Real-time imaging of the HIV-1 transcription cycle in single living cells.
Methods, 53(1):62-67, 25 Jun 2010
Cited by: 23 articles | PMID: 20600934
Review




