Abstract
Free full text

RNA sequence analysis defines Dicer's role in mouse embryonic stem cells
Associated Data
Abstract
Short RNA expression was analyzed from Dicer-positive and Dicer-knockout mouse embroyonic stem (ES) cells, using high-throughput pyrosequencing. A correlation of miRNA quantification with sequencing frequency estimates that there are 110,000 miRNAs per ES cell, the majority of which can be accounted for by six distinct miRNA loci. Four of these miRNA loci or their human homologues have demonstrated roles in cell cycle regulation or oncogenesis, suggesting that a major function of the miRNA pathway in ES cells may be to shape their distinct cell cycle. Forty-six previously uncharacterized miRNAs were identified, most of which are expressed at low levels and are less conserved than the set of known miRNAs. Low-abundance short RNAs matching all classes of repetitive elements were present in cells lacking Dicer, although the production of some SINE- and simple repeat-associated short RNAs appeared to be Dicer-dependent. These and other Dicer-dependent sequences resembled miRNAs. At a depth of sequencing that approaches the total number of 5′ phosphorylated short RNAs per cell, miRNAs appeared to be Dicer's only substrate. The results presented suggest a model in which repeat-associated miRNAs serve as host defenses against repetitive elements, a function canonically ascribed to other classes of short RNA.
RNAi is a conserved set of gene regulatory mechanisms in which short RNA molecules guide protein complexes to suppress expression of complementary nucleic acid targets. Different classes of short RNAs, complexed with specific Argonaute protein family members, induce the degradation, prevent the translation, or prevent the transcription of their target RNA species (1).
In mammals, Argonaute proteins are thought to associate predominantly with a class of noncoding RNA genes termed microRNAs (miRNAs). miRNAs are essential regulators of diverse biological processes, including cell division, apoptosis, and metabolism (2). miRNA precursors are processed sequentially by the enzymes Drosha and Dicer to yield mature ≈22-nt long single-stranded miRNAs (3). miRNAs are thought to primarily influence gene expression by preventing productive translation of target mRNAs, although recent studies suggest that they may have other mechanisms of action (4, 5).
Other classes of short RNAs mediate different types of RNAi-based silencing. In Arabidopsis and Schizosaccharomyces pombe, Argonaute-associated short-interfering RNAs (siRNAs) cleave repetitive transcripts and nucleate heterochromatin at genomic repeats (6). These siRNAs require Dicer and an RNA-dependent RNA polymerase for biogenesis. Potentially analogous siRNA species were identified in mouse oocytes, although it is not clear whether these oocyte siRNAs nucleate heterochromatin (7). In animal germ cells, the Argonaute subfamily of Piwi proteins associate with Dicer-independent short RNAs, termed piRNAs. Like Arabidopsis and S. pombe siRNAs, piRNAs are thought to silence repetitive sequences at the level of transcription (8). Finally, in Caenorhabditis elegans, endogenous siRNAs exist that are thought to silence protein-coding genes at the posttranscriptional level (9). These siRNAs also require Dicer and RNA-dependent RNA polymerases for biogenesis and likely have 5′ di- or tri-phosphates instead of 5′ mono-phosphates (10, 11).
ES cells are derived from the inner cell mass of the blastocyst during the stage of development where epigenetic patterns of gene regulation are reestablished in preparation for implantation (12). ES cells can be propagated in vitro without the loss of pluripotency and induced to differentiate into specialized cell types when given appropriate cues, making them potential sources of tissue in regenerative therapies (13). Many cancers also have stem cell-like characteristics, underscoring the clinical relevance of ES cell biology (14).
Despite recent fundamental advances in the understanding of global ES cell chromatin architecture, much remains to be learned about the mechanisms by which ES cells maintain the pluripotent state (15). Specifically, ES cells lacking Dicer are viable, but are incapable of differentiation and display severe growth defects, indicating that the RNAi pathway is required for pluripotency and aspects of ES cell division (16, 17). Presumably, these defects are due to loss of miRNA biogenesis and not other types of short RNAs, because previous sequencing of short cDNA libraries revealed miRNAs to be the predominant class of short RNA in mouse ES cells (18, 19). However, Dicer is critical to the biogenesis of almost all classes of short RNAs described, with the potential exception of piRNAs; thus, it is possible that other previously unidentified RNAs contribute to the Dicer null ES cell mutant phenotype.
To further our understanding of Dicer function and the mechanisms by which short RNAs mediate gene regulation in ES cells, short RNA expression was profiled in four independently derived ES cell cDNA libraries, including a library made from Dicer null ES cells. From quantification of miRNA levels, we estimate that there are 130,000 5′ phosphorylated short RNAs per ES cell. Fifteen percent of these RNAs are generated independently of Dicer, and consist of: short noncoding RNA (ncRNA) fragments, promoter proximal RNAs (unpublished data), presumed breakdown products of mRNAs, and low-abundance, highly repetitive sequences. The remaining 85% of 5′ phosphorylated ES cell short RNAs consist of miRNAs or miRNA-like species that depend on Dicer for biogenesis. The majority of ES cell miRNAs appear to be generated by six distinct loci, four of which have been implicated in cell cycle control or oncogenesis. Notably, poorly conserved ES cell miRNA hairpins tend to overlap annotated repetitive elements, potentially connecting the miRNA pathway to host defense against accumulated repeats.
Results
Global Statistics of Short cDNA Libraries.
Four separate short cDNA libraries made from mouse ES cells were sequenced with high-throughput pyrosequencing (20). To determine whether classes of short RNAs other than miRNAs depend on Dicer for biogenesis, short cDNA libraries were made from a floxed Dicer ES cell line before and several months after deletion of the floxed region containing the key catalytic residues of Dicer's second RNase III domain (referred to as libraries Dicer+/+ and Dicer−/−, respectively). This Dicer deletion cell line has been used in previous studies (18, 21) and largely recapitulates the phenotypic defects observed from earlier studies of Dicer loss in mouse ES cells [supporting information (SI) Fig. 4] (16, 17). Additionally, to determine whether changes in DNA methylation correlate with expression of novel classes of mammalian short RNAs, libraries were sequenced from J1 ES cells before and five days after treatment with the DNA methyltransferase inhibitor 5-aza-deoxycytidine (referred to as libraries “J1” and “J1aza,” respectively; SI Fig. 5). The rationale for this experiment was based on observations made in Arabidopsis, where production of short RNAs by the RNAi-pathway stimulates DNA methylation at certain classes of repetitive elements (6). Subsequent sequencing and analysis indicated few significant differences between the J1 and J1aza cDNA libraries (data not shown), and, for the purpose of this study, they were treated primarily as expression replicates for Dicer-containing ES cell libraries. Because of strain and sex chromosome differences between J1 and Dicer+/+ ES cells, reads have only been compared between the Dicer+/+ and Dicer−/− libraries when considering the consequences of Dicer loss.
In total, the four libraries contained 418,093 reads representing 79,265 distinct sequences (Table 1). We focused our analysis on the 298,039 reads representing 29,016 distinct sequences that matched the mouse genome with 100% identity over their entire length. On average, 82% of all reads from the Dicer-positive libraries matched annotated miRNA hairpins, whereas 11% of reads matched other known ncRNAs (rRNAs, tRNAs, and small nuclear RNAs, etc.), and 7% of reads were previously uncharacterized short RNAs (referred to as “novel” sequences; Table 1). As expected, the Dicer−/− library was nearly devoid of miRNAs, and instead composed of other known ncRNAs (69%) and novel sequences (31%; Table 1).
Table 1.
Composition of cDNA libraries analyzed
| Feature | J1 | J1aza | Dicer+/+ | Dicer−/− |
|---|---|---|---|---|
| miRNA, % | 86.2 | 81.6 | 78.0 | 0.5 |
| rRNA, % | 4.8 | 4.2 | 9.3 | 43.8 |
| ncRNA, % | 2.4 | 4.1 | 1.0 | 7.9 |
| tRNA, % | 1.6 | 2.1 | 3.0 | 16.9 |
| Novel reads, % | 5.0 | 8.0 | 8.7 | 30.9 |
| Match mm7 | 104,220 | 115,304 | 45,320 | 33,195 |
| All reads | 149,986 | 155,934 | 57,834 | 54,339 |
Percentages represent the total number of reads matching the August 2005 build of the mouse genome (match mm7). Also shown is the total number of reads sequenced in each library (all reads). ncRNA, non-miRNA/non-rRNA/non-tRNA noncoding RNA. Novel reads, previously uncharacterized short RNAs.
Expression and Analysis of Known miRNAs.
To validate that the cDNA libraries accurately recapitulated short RNA expression in ES cells, the absolute numbers of seven known miRNAs were determined in J1 and Dicer+/+ ES cells, using the Direct miRNA assay (Table 2) (22). The Pearson correlation coefficients between the miRNA quantification and sequencing frequencies in the J1 and Dicer+/+ libraries were 0.62 and 0.95, respectively. Correlating miRNA quantification to sequencing frequency, we conclude that a single ES cell contains ≈110,000 miRNAs from a total pool of 130,000 5′ phosphorylated short RNAs. The calculated number of miRNAs per femtogram of total ES cell RNA is 5.4 ± 1.
Table 2.
Direct quantification of specific miRNAs per ES cell
| miRNA | J1 (quant) | J1 (reads) | Dicer+/+ (quant) | Dicer+/+ (reads) |
|---|---|---|---|---|
| miR15a | 290 ± 50 | 175 | 280 ± 20 | 293 |
| miR 15b | 950 ± 20 | 2,301 | 970 ± 40 | 1,621 |
| miR 16 | 1,130 ± 140 | 2,037 | 1090 ± 120 | 1,199 |
| miR 17–5p | 1,510 ± 110 | 795 | 1440 ± 170 | 1,509 |
| miR 19b | 2,140 ± 490 | 14,777 | 2340 ± 550 | 3,918 |
| miR 21 | 2,750 ± 410 | 6,172 | 1340 ± 450 | 2,272 |
| miR 30c | 250 ± 20 | 2,946 | 220 ± 40 | 379 |
The measured miRNA copy number is compared with the sequencing frequency per 130,000 reads in the J1 and Dicer+/+ libraries. Error is the SEM from 2 to 21 triplicate measurements. quant, quantified.
The number of reads obtained for each library approaches the total number of 5′ phosphorylated short RNAs per ES cell; thus, each cDNA library can be considered an accurate sampling of the spectrum of 5′ phosphorylated short RNAs in a single ES cell. With this in mind, the Dicer+/+ and J1 libraries were used to determine the most abundantly expressed ES cell miRNAs. Averaging values from the Dicer+/+ and J1 libraries estimates that 27 ES cell miRNAs are expressed at >1,000 molecules per cell, with the most abundant present at ≈5,000 molecules per cell (SI Table 4). When considering the 126 miRNAs that are expressed at least 50 molecules per cell, the average and median miRNA expression per cell is 713 and 231 molecules, respectively (SI Table 4).
The majority of miRNAs in both ES cell lines could be accounted for by six genomic loci, representing 76% and 69% of Dicer+/+ and J1 miRNAs, respectively (Table 3 and SI Tables 4 and 5). These include the miR15b/miR16 cluster, the miR17–92 cluster, miR21, the miR290–295 cluster, a repetitive miRNA cluster on chromosome 2 (SI Table 7), and an imprinted miRNA cluster on chromosome 12 (SI Table 7) (23). Certain of these miRNAs, specifically miR16 and several in the miR17–92 cluster, have multiple genomic locations that may contribute to expression. There were significant differences in expression of two of these miRNA clusters between J1 and Dicer+/+ ES cells, possibly due to differences in strain or sex. J1 ES cells appear to express the chromosome 12 cluster in higher abundance than Dicer+/+ ES cells, whereas Dicer+/+ ES cells appear to express the chromosome 2 cluster in higher abundance than J1 ES cells. The other four miRNA loci appeared quite similar in expression between the two cell types.
Table 3.
The major miRNAs expressed in ES cells
| miRNA cluster | J1 miRNAs, % | Dicer+/+ miRNAs, % |
|---|---|---|
| 290–295 | 23 | 29 |
| 17–92 | 17 | 11 |
| chr2 | 6 | 27 |
| chr12 | 14 | 4 |
| 21 | 6 | 2 |
| 15b/16 | 4 | 3 |
The genomic location and miRNAs contained in the chr2 and chr12 clusters are described in SI Table 7.
Validation of Known miRNAs.
Comparison of the Dicer+/+ and Dicer−/− libraries allowed for the genetic validation of miRNAs expressed in ES cells, because true miRNAs should be absent in the Dicer−/− library. Six annotated miRNA hairpins expressed in the Dicer−/− library had exact matches to ribosomal or small-nuclear ncRNAs and are thus probably incorrectly designated as miRNAs (denoted as “ncRNA” in SI Table 5). There were 2.5 times as many reads matching these six miRNA hairpins in the Dicer−/− library than the Dicer+/+ library, consistent with their being generated from Dicer-independent processing of abundant ncRNA transcripts and not miRNA hairpins. Excluding these six hairpins, the overall ratio of Dicer+/+ to Dicer−/− reads was 213:1 for 240 miRNA hairpins present in the Dicer+/+ and Dicer−/− libraries. This clear Dicer dependence of miRNA expression indicates that the previous annotation of mammalian miRNAs has been an accurate process.
Hypothesizing that a low level of Dicer-independent cleavage of pre-miRNA hairpins generated the few miRNA-matching reads in the Dicer−/− library, we further examined the sequence characteristics of the Dicer−/− miRNAs. Consistent with this hypothesis, the lengths of the Dicer−/− miRNA reads were more broadly distributed compared with the lengths of the Dicer+/+ miRNA reads (SI Fig. 6A). Fifty-eight percent of the Dicer−/− miRNA reads were 21–23 nt long, compared with 91% of the Dicer+/+ miRNA reads (P = 7 × 10−14). This difference was striking considering the similarity of the size distributions for all other known ncRNAs between the Dicer+/+ and Dicer−/− libraries (SI Fig. 6A).
Next, we examined the extent of miRNA processing variability in each library, defined here as the proportion of miRNA-matching reads that do not match the annotated 5′ and 3′ ends of mature miRNA sequences. Drosha defines the 5′ ends of mature miRNAs from the 5′ arm of pre-miRNA hairpins and the 3′ ends of mature miRNAs from the 3′ of pre-miRNA hairpins; Dicer defines the 5′ ends of mature miRNAs from the 3′ arm of pre-miRNA hairpins and the 3′ ends of mature miRNAs from the 5′ arm of pre-miRNA hairpins (SI Fig. 6B). If Dicer−/− miRNA reads were excised from pre-miRNA hairpins by a Dicer-independent mechanism, more miRNA processing variability might be expected in the Dicer−/− compared with the Dicer+/+ library. Also, the ends of Dicer−/− miRNAs that would normally be defined by Dicer might show greater processing variability compared with those defined by Drosha. Supporting these ideas, miRNA reads exhibited more processing variability in the Dicer−/− compared with the Dicer+/+ library (SI Fig. 6 C and D), and, although Dicer-processed miRNA ends showed more variability compared with Drosha-processed miRNA ends in all four libraries, this difference was greatest in the Dicer−/− library (SI Fig. 6 C and D). Although we cannot formally exclude the possibility that some miRNAs in the Dicer−/− library could be due to cross-contamination from Dicer-positive libraries, these clear differences in expression characteristics suggest that many of the miRNAs in the Dicer−/− library were generated by inefficient Dicer-independent processing of pre-miRNA hairpins.
Annotation of Novel miRNAs.
Using guidelines for miRNA annotation established by Ambros et al. (24) and incorporating rules for Drosha processing of primary miRNA transcripts (25, 26), 46 previously uncharacterized miRNAs were identified in the Dicer-positive libraries (see SI Table 6 and SI Dataset 1). These 46 miRNA hairpins generate miRNAs with 42 distinct seeds, defined as bases 2–7 from the 5′ end of the miRNA (27). Forty of these 42 seeds are previously uncharacterized. As a group, the miRNAs are expressed at low levels in ES cells and less conserved than the set of known miRNAs (Fig. 1A). Despite their low expression levels, most of the miRNAs were consistently present in each Dicer-containing ES cell library. Thirty-six of the 46 miRNAs were sequenced in at least two of the three libraries made from ES cells with functional Dicer, with 21 of these being present in all three Dicer-containing libraries. Twenty of the miRNAs mapped into large clusters of previously identified miRNAs on chromosomes 2, 12, and X (SI Table 7). Of the remaining 26 miRNA hairpins, only 2 were located within 5 kb of a known miRNA.
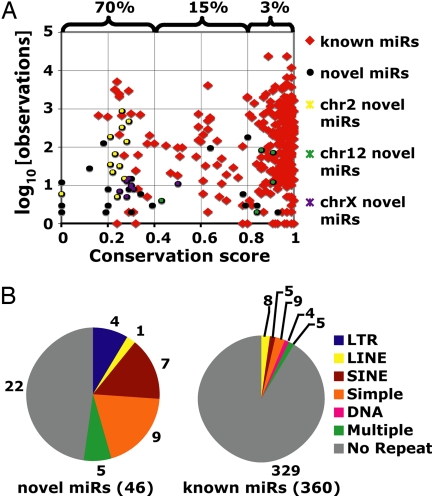
Conservation, expression, and repeat overlap of known and novel miRNA hairpins. (A) Conservation and ES cell expression of known and novel miRNA hairpins. The percentage of miRNA hairpins overlapping repeats is bracketed for three bins of conservation. The genomic locations of the chr2, 12, and X clusters are described SI Table 7. (B) Repeatmasker overlap of known and novel miRNA hairpins. Numbers refer to the total number of miRNA hairpins in each category. “Multiple” refers to those hairpins overlapping more than one class of repeat.
Consistent with the novel miRNAs being less conserved than the set of known miRNAs, 24 of the 46 miRNA hairpins overlapped at least partially with annotated repetitive elements. By comparison, only 31 known miRNA hairpins overlap repeats in the set of 360 mouse miRNAs that map to the mm7 build of the mouse genome (Fig. 1B). As expected, the proportion of miRNA hairpins overlapping repeats decreases as miRNA conservation increases (Fig. 1A).
Analysis of Repeat-Overlapping Novel Reads.
A small number of short RNAs overlapping highly repetitive sequences existed in each of the four libraries, defined as those sequences with at least 20 exact matches to the genome (SI Table 8; see SI Text for further analysis). The 1,211 unique sequences in this group were represented by 1,991 reads and had 3,935,923 total hits to the genome covering ≈48 Mb of DNA. Based on correlations of miRNA quantification with sequencing frequency (Table 2), as a class these repetitive RNAs are present at ≈225–750 copies per ES cell. There were no strong biases in the first nucleotide or length of these highly repetitive short RNAs, although there were slightly more sequences beginning with U compared with the set of novel sequences with less than 20 matches to the genome (Fig. 2A). Examining the length distribution of repetitive sequences, we observed a peak above background at 22 nt (Fig. 2B). This peak is due solely to a Dicer-independent short RNA that is antisense to the primer-binding site of the early transposon repeat, an endogenous retrovirus abundantly expressed in the early mouse embryo and ES cells (28).
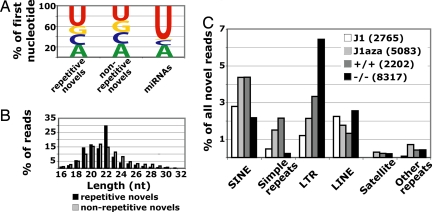
Analysis of highly repetitive novel sequences. (A) First nucleotide distribution of highly repetitive novel sequences (≥20 hits to the genome) compared with nonrepetitive novel sequences (<20 hits to the genome) and known miRNAs. (B) Length distribution of highly repetitive novel sequences compared with all nonrepetitive novel sequences. (C) Repeatmasker classification of highly repetitive novel sequences, represented as proportions of novel reads per library. The number of novel reads per library is in parentheses.
The proportions of repetitive sequences overlapping SINE and simple repeats were significantly lower in the Dicer−/− compared with the Dicer+/+ library (Fig. 2C). This suggests either that certain SINE- and simple repeat-associated RNAs are processed by Dicer from precursor double-stranded RNA, or that a transcriptional difference between Dicer+/+ and Dicer−/− cells results in differential expression of these short RNAs. Northern blots showed no significant difference in full-length SINE B1 RNA levels between Dicer+/+ and Dicer−/− ES cells (SI Fig. 7), arguing against the latter hypothesis.
In contrast, short RNAs overlapping centromeric satellite repeats, long interspersed nuclear elements, and LTR elements were clearly not dependent on Dicer for biogenesis (Fig. 2C). This was surprising, because previous studies have suggested that Dicer-dependent siRNAs processed from long double-stranded RNA precursors are important for silencing of these elements (7, 16, 29). The Dicer−/− ES cells analyzed here maintain genomic DNA methylation at satellite repeats and long interspersed nuclear elements (SI Fig. 4E), demonstrating that RNAi is not required for maintenance of global repeat methylation and suggesting that loss of centromeric silencing in certain Dicer null ES cells lines may be an indirect effect of Dicer loss (16).
Few Non-miRNA Dicer-Dependent Sequences Are Expressed in ES Cells.
Because Dicer is involved in the production of short RNAs other than miRNAs in several organisms, we next sought to determine what non-miRNA short RNAs might be Dicer-dependent in ES cells. Sequences present at least three times in the Dicer+/+ library and absent in the Dicer−/− library were flagged as potentially dependent on Dicer for biogenesis (referred to as “Dicer-dependent” below) and subjected to further analysis. There were 50 distinct sequences, represented by 233 reads in the Dicer+/+ and 139 reads in the J1 and J1aza libraries, which matched these criteria and were not annotated above as novel miRNAs. Consistent with their being Dicer products, the length distribution of these sequences peaked more sharply at ≈21 nt when compared with all other novel sequences in the Dicer+/+ library (Fig. 3A; P = 4.0 × 10−5). The Dicer-dependent short RNAs are biased toward sequences that begin with A compared with the set of all novel reads, although this bias is not as strong as the U bias seen for known miRNAs (Fig. 3B). As expected from the analysis of highly repetitive reads, these sequences were enriched in SINE and simple repeat elements compared with the set of novel sequences that did not meet the criteria for Dicer dependence (SI Fig. 8). Two groups of Dicer-dependent sequences, composed of 48 and 87 reads, were related in sequence (Fig. 3C). Both of these sequence groups appeared to be repeat-derived, with group 1 composed entirely of SINE B1 overlapping reads and group 2 displaying more heterogeneity with respect to its repeat overlap (Fig. 3C).

Description of Dicer-dependent novel sequences. (A) Length distribution of Dicer-dependent novel sequences compared with all other Dicer+/+ and Dicer−/− novel sequences. (B) First nucleotide distribution of Dicer-dependent novel sequences. (C) Two groups of Dicer-dependent sequences share sequence similarity. Shown are identified sequence motifs along with aligning sequences, total reads by library, number of genome matches, and overlapping repeats.
The possibility that Dicer-dependent sequences represent endogenous siRNAs processed by Dicer from long double-stranded RNA was examined. Endogenous siRNAs processed from a single precursor would be expected to cluster near other short RNA sequences. In contrast, Dicer-dependent novel sequences do not cluster with any greater frequency than novel sequences not defined as Dicer-dependent. Twenty-two percent of novel sequences both defined and not defined as Dicer-dependent fell within 500 bases of at least one other short RNA from the set of 25,040 nonrepetitive sequences present in all four libraries (10 of 45 Dicer-dependent sequences and 2,493 of 11,493 other novel sequences; nonrepetitive sequences were defined as having <20 matches to the genome). Moreover, of the 10 Dicer-dependent sequences that did cluster near other short RNA loci, eight overlapped protein-coding genes in the sense orientation, again not consistent with these sequences being canonical siRNAs involved in gene silencing processes.
Instead of representing a class of endogenous siRNAs, it seems likely that many of these Dicer-dependent sequences are miRNA-like reads whose surrounding genomic sequences did not form prototypical miRNA hairpins. The two groups of related Dicer-dependent sequences are in support of this hypothesis (Fig. 3C). The five SINE B1-associated sequences from group 1 aligned to hairpins, which were miRNA-like, but did not meet the minimum requirements for miRNA hairpin base-pairing used in this study (SI Dataset 1). The group 2 sequences are related to known miRNAs on chromosome 2 (SI Table 7), and two sequences from this group also aligned to miRNA-like hairpins with poorly defined secondary structure (SI Dataset 1). Again, these observations are consistent with group 2 sequences being miRNA-like and not siRNA-like in origin.
Sequences present less than three times in the Dicer+/+ library were not evaluated for Dicer dependence, because the transcriptional program of Dicer+/+ and Dicer−/− ES cells is likely quite different and minor differences in short RNA expression between the two cell types would be expected. There remained 1,096 novel sequences, each present and represented by less than three reads in the Dicer+/+ library, which were absent in the Dicer−/− library and potentially dependent on Dicer for biogenesis. Although some are expected to be Dicer products, as a class they clearly differed from the Dicer-dependent sequences described above; most notably, these sequences exhibited a broad length distribution uncharacteristic of Dicer products (SI Fig. 9). Thus, if non-miRNA Dicer-dependent short RNAs are expressed in ES cells, they are beyond the limits of detection in the cDNA libraries analyzed here.
Discussion
Of the estimated 130,000 5′ phosphorylated short RNAs in an ES cell, 85% are Dicer-dependent miRNAs or miRNA-like species and 15% are Dicer-independent short RNAs. These Dicer-independent RNAs consist primarily of short ncRNA species, promoter proximal RNAs that are likely the products of paused RNA polymerase II (unpublished data), presumed breakdown products of mRNAs, and highly repetitive short-RNA sequences.
At a depth of sequencing approaching the total number of 5′ phosphorylated short RNAs per ES cell, the miRNA was the only class of short RNA found to be Dicer-dependent. Other classes of Dicer-dependent short RNAs found in many nonmammalian organisms do not appear to be expressed in ES cells. Specifically not observed were the Dicer-dependent heterochromatic siRNAs, analogous to those seen in Arabidopsis and S. pombe, that have been proposed to guide the silencing of ES cell centromeric repeats (16). Whereas short RNAs corresponding to highly repetitive sequences were detected at low levels in the ES cells analyzed here, their biogenesis was Dicer-independent. Moreover, the potential mammalian counterparts to these siRNAs, piRNAs, were also not detected in the analyzed libraries, nor were C. elegans-like siRNAs that are anti-sense to mRNAs (see SI Text). Direct comparison of the Dicer+/+ and Dicer−/− libraries did detect a small number of sequences, representing 0.5% of all Dicer+/+ reads, which appeared Dicer-dependent and were not annotated as miRNAs; however, many of these sequences appeared miRNA-like. In summary, the presented data favor the hypothesis that Dicer's sole catalytic role in ES cells is to produce miRNAs, and that the phenotypic consequences of ES cell Dicer deletion are due solely to miRNA loss (16, 17).
In total, 323 distinct miRNA sequences were observed in the J1 and Dicer+/+ libraries. The most abundant of these have implied functions consistent with the severe growth defects of Dicer null ES cells; miR21, the miR17–92 cluster, the miR15b/16 cluster, and the miR290–295 cluster, or their human homologues, have demonstrated roles in cell-cycle regulation or oncogenesis (30–33). Almost half of the 110,000 ES cell miRNAs can be accounted for by these four loci, suggesting that a major function of the miRNA pathway in ES cells is to contribute to the control of cell division.
Close to two-thirds of the 323 ES cell miRNAs are expressed at <50 copies per cell. A subset of these lowly expressed miRNAs may play important roles in defining the ES cell state; however, many may have more critical roles in cell types other than ES cells, especially those that are the most conserved. Considering the latter possibility, their apparent ES cell expression could be due to the existence of a small number of differentiated cells within a larger population of undifferentiated ES cells. Alternatively, the diverse set of lowly expressed miRNAs might reflect the heterogeneity of regulatory systems inherent within a pluripotent ES cell population.
Many of the least-conserved ES cell miRNA hairpins overlap annotated repetitive elements, suggesting that particular miRNAs may partially function to silence complementary repeat-containing RNAs (34, 35). This repression could occur through a canonical miRNA-based targeting mechanism, resulting in the translational inhibition and targeting to cellular processing bodies of repeat-containing RNAs with seed complements to repeat-derived miRNAs. Alternatively, the most repetitive miRNA sequences have the potential to direct cleavage of transcripts with perfect or near perfect complementarity. Finally, in certain cases, it is possible that recognition of the miRNA hairpin itself may be the initiating signal for a silencing event in cis.
In mouse oocytes, repetitive sequences appear to be under Dicer-dependent repression. Certain repeat-containing mRNAs were found to be expressed at higher levels in Dicer−/− compared with Dicer+/+ oocytes (36). Further, expression of EGFP reporters with retrotransposon-derived 3′ UTRs was repressed in mouse oocytes (7). These repressive effects were conjectured to be due to endogenous siRNA species arising from genomic repeats (7, 36). Similarly, long interspersed nuclear element retrotransposition has been proposed to be repressed by Dicer-dependent siRNA species in human cells (29). The apparent absence of analogous siRNA species in mouse ES cells, coupled with the observed relationship between miRNAs and repetitive elements, suggests that in certain contexts the miRNA pathway may perform functions canonically thought of as siRNA-specific. This hypothesis argues for the reevaluation of repressive effects associated with mammalian repetitive elements and potentially has important implications during early mouse development, where repetitive element expression is dynamic (37).
Methods
ES Cell Culture and Manipulation.
Generation of Dicer+/+ and Dicer−/− ES cells and of J1aza RNA is described in SI Text. miRNA quantification was performed essentially as described in ref. 38. Briefly, trypsinized ES cells were counted and lysed directly in TRIzol. Single-stranded siRNA (1.5 or 3 pmol) was spiked into TRIzol solutions and quantified to normalize for short RNA recovery. From 15 preparations, the average total RNA per ES cell was 20 pg, and the average short RNA recovery was 76%. miRNA levels were quantified by using the Direct assay (22). miRNA molarity per sample was determined by comparison to standard curves of synthetic miRNAs and normalized for short RNA recovery. miRNA per cell values were obtained by dividing miRNA copy number per sample by the number of ES cell equivalents of RNA measured per assay. The number of 5′ phosphorylated short RNAs per ES cell, 130,000, was obtained by dividing the miRNA copy number per cell by the sequencing frequency of each quantified miRNA (SI Table 4) and taking the average for seven miRNAs quantified in J1 and Dicer+/+ ES cells. Mature miRNAs sequenced per library included those truncated on their 3′ end by one nucleotide and those extending beyond the annotated 3′ end.
Short cDNA Library Preparation and Read Processing.
Short cDNA libraries were made as described in ref. 38. Gel purifications of short RNA/DNA species extended from 16 to slightly past 30 nt. Downstream analysis was performed on sequences with perfect matches to either: the National Center for Biotechnology Information build 35 of the mouse genome (mm7), miRBase8.2 (39, 40), tRNA sequences (41), the noncode RNA database (42), ENSEMBL noncoding RNAs (43), or the complete rDNA repeating unit (44). Conservation and repeat information was obtained by using the University of California Santa Cruz table browser (45) (see SI Text for details).
Acknowledgments
We thank A. Young and J. Neilson for critical reading of this manuscript; G. Zheng, C. Whittaker, and G. Ruby for advice on bioinformatic analysis; M. Lindstrom for figure help; D. Livingston for Dicer antibodies; and the Broad Institute for pyrosequencing. This work was supported by National Institutes of Health Fellowship 5-F32-HD051190 (to A.C.S.), the Crick-Jacobs Center for Theoretical and Computational Biology (G.W.Y.), United States Public Health Service Grants RO1-GM34277 (from the National Institutes of Health), PO1-CA42063 from the National Cancer Institute (to P.A.S.) and partially by National Cancer Institute Grant P30-CA14051.
Abbreviation
| miRNA | microRNA. |
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE9306). Analyzed sequences are provided in SI Tables 9–13.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709193104/DC1.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0709193104
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2084302?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.0709193104
Article citations
Development of a Modular miRNA-Responsive Biosensor for Organ-Specific Evaluation of Liver Injury.
Biosensors (Basel), 14(9):450, 20 Sep 2024
Cited by: 0 articles | PMID: 39329825 | PMCID: PMC11430419
Circular RNA in cancer.
Nat Rev Cancer, 24(9):597-613, 29 Jul 2024
Cited by: 3 articles | PMID: 39075222
Review
Antiviral Defence Mechanisms during Early Mammalian Development.
Viruses, 16(2):173, 24 Jan 2024
Cited by: 1 article | PMID: 38399949 | PMCID: PMC10891733
Review Free full text in Europe PMC
IRE1-mediated degradation of pre-miR-301a promotes apoptosis through upregulation of GADD45A.
Cell Commun Signal, 21(1):322, 09 Nov 2023
Cited by: 2 articles | PMID: 37946177 | PMCID: PMC10634081
Pluripotency-Associated microRNAs in Early Vertebrate Embryos and Stem Cells.
Genes (Basel), 14(7):1434, 12 Jul 2023
Cited by: 1 article | PMID: 37510338 | PMCID: PMC10379376
Review Free full text in Europe PMC
Go to all (230) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE9306
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs.
Genes Dev, 22(20):2773-2785, 01 Oct 2008
Cited by: 561 articles | PMID: 18923076 | PMCID: PMC2569885
Structure of precursor microRNA's terminal loop regulates human Dicer's dicing activity by switching DExH/D domain.
Protein Cell, 6(3):185-193, 31 Dec 2014
Cited by: 14 articles | PMID: 25549615 | PMCID: PMC4348242
Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues.
Nucleic Acids Res, 41(5):3339-3351, 15 Jan 2013
Cited by: 67 articles | PMID: 23325850 | PMCID: PMC3597668
Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?
Curr Stem Cell Res Ther, 4(3):168-177, 01 Sep 2009
Cited by: 59 articles | PMID: 19492978
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: P01 CA042063
Grant ID: P01-CA42063
Grant ID: P30 CA014051
Grant ID: P30-CA14051
NICHD NIH HHS (2)
Grant ID: 5-F32-HD051190
Grant ID: F32 HD051190
NIGMS NIH HHS (2)
Grant ID: R01-GM34277
Grant ID: R01 GM034277





