Abstract
Free full text

Regulation and initiation of cenB transcripts of Cellulomonas fimi.
Abstract
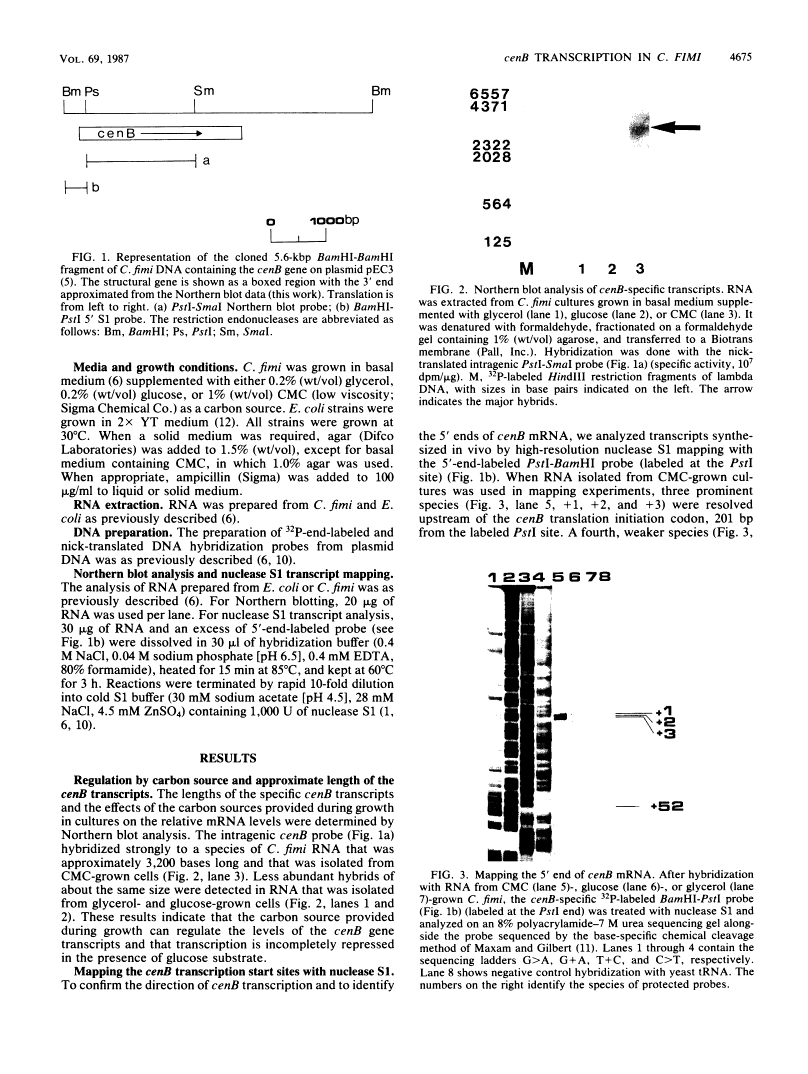
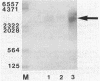
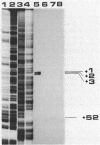
We characterized the in vivo transcription of the Cellulomonas fimi cenB gene, which encodes an extracellular endo-beta-1,4-glucanase (EC 3.2.1.4). By Northern blot (RNA blot) analysis, cenB mRNA was detected in C. fimi RNA preparations from glycerol-, glucose-, and carboxymethyl cellulose (CMC)-grown cells. The relative abundance of the specific mRNAs in these preparations appeared to depend on the carbon source provided, with the preparations from CMC-grown cells having the greatest amount of cenB transcripts, followed by glycerol- and glucose-grown cells. Therefore, the transcription of this gene could be regulated by the carbon source provided to C. fimi. High-resolution nuclease S1 protection studies were used to map cenB mRNA 5' termini with a unique 5'-labeled DNA probe and C. fimi RNA isolated in vivo. With this procedure, three 5' termini were found in abundance upstream of the translational initiation ATG codon in RNA preparations from C. fimi grown on CMC, while less-abundant 5' termini were found 52 bases closer to the ATG codon in RNA prepared from C. fimi grown on any one of the three substrates. These results are indicative of a tandem promoter arrangement, with the ATG-proximal promoter directing constitutive low-level cenB transcription and the more distal promoter directing higher levels of transcription under the inducing effects of the cellulosic substrate. The corresponding transcripts were not detected in S1 mapping experiments with RNA isolated in vivo from Escherichia coli clones harboring recombinant plasmids carrying C. fimi genomic inserts. Comparative analysis of the 5' -flanking DNA sequences of the cenB gene and the cenA and cex genes of C. fimi (N. M. Greenberg, R. A. J. Warren, D. G. Kilburn, and R. C. Miller, Jr., J. Bacteriol. 169:646-653, 1987) revealed a region of 50 bases in which these sequences displayed at least 64% homology.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (987K), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk AJ, Sharp PA. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. [Abstract] [Google Scholar]
- Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [Abstract] [Google Scholar]
- Fornwald JA, Schmidt FJ, Adams CW, Rosenberg M, Brawner ME. Two promoters, one inducible and one constitutive, control transcription of the Streptomyces lividans galactose operon. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2130–2134. [Europe PMC free article] [Abstract] [Google Scholar]
- Greenberg NM, Warren RA, Kilburn DG, Miller RC., Jr Regulation, initiation, and termination of the cenA and cex transcripts of Cellulomonas fimi. J Bacteriol. 1987 Feb;169(2):646–653. [Europe PMC free article] [Abstract] [Google Scholar]
- JACOB F, MONOD J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961 Jun;3:318–356. [Abstract] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. [Abstract] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. [Abstract] [Google Scholar]
- O'Neill G, Goh SH, Warren RA, Kilburn DG, Miller RC., Jr Structure of the gene encoding the exoglucanase of Cellulomonas fimi. Gene. 1986;44(2-3):325–330. [Abstract] [Google Scholar]
- Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. [Abstract] [Google Scholar]
- Wong WK, Gerhard B, Guo ZM, Kilburn DG, Warren AJ, Miller RC., Jr Characterization and structure of an endoglucanase gene cenA of Cellulomonas fimi. Gene. 1986;44(2-3):315–324. [Abstract] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.169.10.4674-4677.1987
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/169/10/4674.full.pdf
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/169/10/4674
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/169/10/4674
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jb.169.10.4674-4677.1987
Article citations
Regulation of cellulase synthesis in batch and continuous cultures of Clostridium thermocellum.
J Bacteriol, 187(1):99-106, 01 Jan 2005
Cited by: 69 articles | PMID: 15601693 | PMCID: PMC538832
Regulation of expression of cellulosomes and noncellulosomal (hemi)cellulolytic enzymes in Clostridium cellulovorans during growth on different carbon sources.
J Bacteriol, 186(13):4218-4227, 01 Jul 2004
Cited by: 31 articles | PMID: 15205424 | PMCID: PMC421611
Quantitative determination of cellulase concentration as distinct from cell concentration in studies of microbial cellulose utilization: analytical framework and methodological approach.
Biotechnol Bioeng, 77(4):467-475, 01 Feb 2002
Cited by: 16 articles | PMID: 11787020
The glucanases of Cellulomonas.
Biotechnol Adv, 15(2):315-331, 01 Jan 1997
Cited by: 7 articles | PMID: 14538714
The biological degradation of cellulose.
FEMS Microbiol Rev, 13(1):25-58, 01 Jan 1994
Cited by: 422 articles | PMID: 8117466
Review
Go to all (11) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Regulation, initiation, and termination of the cenA and cex transcripts of Cellulomonas fimi.
J Bacteriol, 169(2):646-653, 01 Feb 1987
Cited by: 19 articles | PMID: 3804971 | PMCID: PMC211827
Unusual sequence organization in CenB, an inverting endoglucanase from Cellulomonas fimi.
J Bacteriol, 173(1):308-314, 01 Jan 1991
Cited by: 55 articles | PMID: 1987122 | PMCID: PMC207188
The tertiary structure of endo-beta-1,4-glucanase B (CenB), a multidomain cellulase from the bacterium Cellulomonas fimi.
Glycobiology, 2(4):321-326, 01 Aug 1992
Cited by: 7 articles | PMID: 1421753
Cellulose-binding polypeptides from Cellulomonas fimi: endoglucanase D (CenD), a family A beta-1,4-glucanase.
J Bacteriol, 175(7):1910-1918, 01 Apr 1993
Cited by: 32 articles | PMID: 8458833 | PMCID: PMC204259