Abstract
Free full text

Afadin: A Novel Actin Filament–binding Protein with One PDZ Domain Localized at Cadherin-based Cell-to-Cell Adherens Junction
Abstract
A novel actin filament (F-actin)–binding protein with a molecular mass of ~205 kD (p205), which was concentrated at cadherin-based cell-to-cell adherens junction (AJ), was isolated and characterized. p205 was purified from rat brain and its cDNA was cloned from a rat brain cDNA library. p205 was a protein of 1,829 amino acids (aa) with a calculated molecular mass of 207,667 kD. p205 had one F-actin–binding domain at 1,631–1,829 aa residues and one PDZ domain at 1,016– 1,100 aa residues, a domain known to interact with transmembrane proteins. p205 was copurified from rat brain with another protein with a molecular mass of 190 kD (p190). p190 was a protein of 1,663 aa with a calculated molecular mass of 188,971 kD. p190 was a splicing variant of p205 having one PDZ domain at 1,009–1,093 aa residues but lacking the F-actin–binding domain. Homology search analysis revealed that the aa sequence of p190 showed 90% identity over the entire sequence with the product of the AF-6 gene, which was found to be fused to the ALL-1 gene, known to be involved in acute leukemia. p190 is likely to be a rat counterpart of human AF-6 protein. p205 bound along the sides of F-actin but hardly showed the F-actin–cross-linking activity. Northern and Western blot analyses showed that p205 was ubiquitously expressed in all the rat tissues examined, whereas p190 was specifically expressed in brain. Immunofluorescence and immunoelectron microscopic studies revealed that p205 was concentrated at cadherin-based cell-to-cell AJ of various tissues. We named p205 l-afadin (a large splicing variant of AF-6 protein localized at adherens junction) and p190 s-afadin (a small splicing variant of l-afadin). These results suggest that l-afadin serves as a linker of the actin cytoskeleton to the plasma membrane at cell-to-cell AJ.
In various cellular events, such as cell adhesion, cell motility, and cell shape determination, specialized membrane domains are formed with transmembrane proteins, such as cell adhesion molecules, receptors, and channels, and these domains are often associated with the actin cytoskeleton (Geiger, 1983, 1989; Geiger and Ginsberg, 1991; Turner and Burridge, 1991; Luna and Hitt, 1992; Tsukita et al., 1992, 1997; Bretscher, 1993). The linkage between the actin cytoskeleton and the plasma membrane plays crucial roles in these cellular events, and proteins linking the actin cytoskeleton to the transmembrane proteins have been identified. However, the molecular basis of the linkage between the actin cytoskeleton and the plasma membrane is not fully understood.
To understand this molecular linkage, the cell adhesion sites have most extensively been studied (Geiger, 1983, 1989; Geiger and Ginsberg, 1991; Turner and Burridge, 1991; Luna and Hitt, 1992; Tsukita et al., 1992, 1997; Bretscher, 1993). The actin filament (F-actin)1–associated cell adhesion sites are subclassed into two types: cell-to-cell and cell-to-matrix adherens junctions (AJ). Many linker proteins have been identified at cell-to-cell AJ where cadherins interact with each other at the extracellular surface (Takeichi, 1988; Geiger and Ginsberg, 1991; Takeichi, 1991; Tsukita et al., 1992). The cytoplasmic domain of cadherin is associated with cytoplasmic proteins such as α-, β-, and γ-catenins (Ozawa et al., 1989; Nagafuchi et al., 1991; Takeichi, 1991; Tsukita et al., 1992). α-Catenin directly interacts with F-actin (Rimm et al., 1995). α-Catenin also interacts indirectly with F-actin through α-actinin and/or ZO-1 (Knudsen et al., 1995; Itoh et al., 1997). Vinculin, another F-actin–binding protein, is concentrated at cell-to-cell AJ, although its interacting molecule at cell-to-cell AJ has not yet been identified (Geiger and Ginsberg, 1991; Tsukita et al., 1992). At cell-to-matrix AJ where integrin interacts with matrix proteins at the extracellular surface, the cytoplasmic domain interacts directly or indirectly with F-actin–binding proteins, including α-actinin, vinculin, and talin (Jockusch et al., 1995). Thus, many F-actin– binding proteins appear to serve as linkers of the actin cytoskeleton to the plasma membrane cadherin and integrin.
The linkage between the actin cytoskeleton and the plasma membrane is also important for neuron-specific events, such as growth cone pathfinding and the subsequent formation and maintenance of synaptic junction (Mitchison and Kirschner, 1988; Smith, 1988; Bentley and O'Connor, 1994; Lin et al., 1994; Tanaka and Sabry, 1995). However, it remains to be clarified which molecules link the actin cytoskeleton to the plasma membrane in these neuron-specific events. To address this issue, we attempted to isolate novel F-actin–binding proteins from the rat brain by using a blot overlay method with 125I-labeled F-actin. We had isolated a neural tissue-specific F-actin–binding protein that is concentrated at synapse and named it neurabin (Nakanishi et al., 1997). Neurabin has one F-actin– binding domain and one PDZ domain. The PDZ domain has been found in a variety of proteins, some of which are localized at cell-to-cell junctions, such as PSD-95/SAP90 at synaptic junction (Cho et al., 1992; Kistner et al., 1993), Dlg at septate junction (Woods and Bryant, 1991), and ZO-1 and ZO-2 at tight junction (Stevenson et al., 1986; Gumbiner, 1991; Itoh et al., 1993; Tsukita et al., 1993; Willott et al., 1993; Jesaitis and Goodenough, 1994). Recent studies have revealed that the PDZ domain binds to unique COOH-terminal motifs of target proteins (Saras and Heldin, 1996), which are found in a large number of transmembrane proteins, such as N-methyl-d-aspartate receptors and Shaker-type K+ channel (Kim et al., 1995; Kornau et al., 1995; Niethammer et al., 1996). It is likely, therefore, that neurabin serves as a linker of the actin cytoskeleton to a transmembrane protein(s) at the synapse although we have not yet identified its interacting transmembrane protein(s).
Along this line, we further attempted to isolate other novel F-actin–binding proteins from rat brain. Here, we isolated a novel F-actin–binding protein with a molecular mass of ~205 kD (p205). p205 also had one F-actin–binding domain and one PDZ domain, and was localized at cadherin-based cell-to-cell AJ. During the purification of p205, it was copurified with another protein with a molecular mass of 190 kD (p190) that was a splicing variant of p205 having one PDZ domain but lacking the F-actin– binding domain. Homology search analysis revealed that the amino acid (aa) sequence of p190 showed 90% identity over the entire sequence with the product of the AF-6 gene, which has been found to be fused to the ALL-1 gene (Prasad et al., 1993), known to be involved in acute leukemia (Cimino et al., 1991). We named p205 l-afadin (a large splicing variant of AF-6 protein localized at adherens junction) and p190 s-afadin (a small splicing variant of l-afadin). We describe here isolation and characterization of l-afadin.
Materials and Methods
125I-labeled F-Actin Blot Overlay
125I-labeled F-actin blot overlay was done as described (Chia et al., 1991; Pestonjamasp et al., 1995). Briefly, purified actin monomer (G-actin) was labeled with 125I-Bolton Hunter reagent. 125I-labeled G-actin (1 mg/ml, average specific activity = 63.3 μCi/mg) was polymerized with 18 μg/ml of gelsolin (molar ratio = 100:1; Sigma Chemical Co., St. Louis, MO) by incubation for 10 min at 4°C in a solution containing 20 mM Pipes, pH 7.0, 50 mM KCl, and 2 mM MgCl2. Phalloidin was then added to give a final concentration of 40 μM. The mixture was then incubated for another 15 min at room temperature and stored at 4°C as 125I-labeled F-actin. The sample to be tested was subjected to SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked in TBS containing 5% defatted powder milk. The membrane was then incubated for 1 h at room temperature with 10 μg/ml of 125I-labeled F-actin in TBS containing 5% defatted powder milk and 4 μM phalloidin. After the incubation, the membrane was washed with TBS containing 0.5% Tween 20, followed by autoradiography using an image analyzer (Fujix BAS-2000II; Fuji Photo Film Co., Tokyo, Japan).
For competition experiments, 125I-labeled F-actin was prepared as described above, except that the concentration of gelsolin was reduced to 7.2 μg/ml (molar ratio = 250:1). 20 μg/ml of 125I-labeled F-actin was incubated for 30 min at room temperature with 0.42 mg/ml of myosin subfragment 1 (S1; Sigma Chemical Co.) in a solution containing 20 mM Pipes, pH 7.0, 58 mM KCl, 2 mM MgCl2, 130 μM CaCl2, and 0.8 μM phalloidin. Where indicated, 4 mM MgATP was added to the mixture. After the incubation, the mixture was diluted with an equal volume of TBS containing 10% defatted powder milk, and it was added to the blot membrane, followed by incubation for 1 h at room temperature.
Purification of l-Afadin
All the purification procedures were carried out at 0–4°C. Fetal brains from 60 mother rats (17-d gestation) were homogenized with a solution containing 10 mM Tris/Cl, pH 8.0, 2 mM EDTA, 5 mM EGTA, and a protease inhibitor cocktail (1 mM PMSF, 20 μg/ml of leupeptin, and 1 μg/ml of pepstatin A). The homogenate was mildly stirred for 1 h and centrifuged at 200,000 g for 1 h. The supernatant (600 ml, 2,070 mg of protein) was stored at −80°C until use. The supernatant was applied to a Q-Sepharose FF column (2.6 × 34 cm; Pharmacia Biotech, Inc., Piscataway, NJ) equilibrated with buffer A (20 mM Tris/Cl, pH 8.0, 0.5 mM EDTA, and 1 mM DTT). After the column was washed with 700 ml of buffer A, elution was performed with an 800-ml linear gradient of NaCl (0–0.5 M) in buffer A. 10-ml fractions were collected. l-Afadin appeared in fractions 56–77. These fractions (220 ml, 500 mg of protein) were collected, and 1.2 M (NH4)2SO4 was added to give a final concentration of 0.6 M. The sample was centrifuged at 200,000 g for 20 min, and the supernatant was applied to a phenyl-5PW column (2.15 × 15 cm; Tosoh, Tokyo, Japan) equilibrated with buffer A containing 0.6 M (NH4)2SO4. After the column was washed with 250 ml of the same buffer, elution was performed with a 240-ml linear gradient of (NH4)2SO4 (0.6–0 M) in buffer A. 6-ml fractions were collected. l-Afadin appeared in fractions 28–34. The active fractions (42 ml, 24 mg of protein) were collected and applied to a hydroxyapatite column (0.78 × 10 cm; Koken, Tokyo, Japan) equilibrated with buffer B (10 mM potassium phosphate, pH 7.8, and 1 mM DTT). After the column was washed with 30 ml of buffer B, elution was performed with a 150-ml linear gradient of potassium phosphate (10–500 mM), pH 7.8, in buffer B. 2.0-ml fractions were collected. l-Afadin appeared in fractions 39–41. The active fractions (6 ml, 0.86 mg of protein) were collected and diluted with an equal volume of buffer A, and half of the sample was applied to a Mono Q HR 5/5 column (Pharmacia Biotech, Inc.) equilibrated with buffer A. After the column was washed with 6 ml of buffer A, elution was performed with a 30-ml linear gradient of NaCl (0.15–0.4 M) in buffer A. 0.5-ml fractions were collected. The other half of the sample was subjected to Mono Q column chromatography in a similar manner. l-Afadin appeared in fractions 34–36 (see Fig. Fig.1).1). The active fractions of the two Mono Q column chromatographies (3.0 ml, 0.2 mg of protein) were combined and stored at −80°C until use.
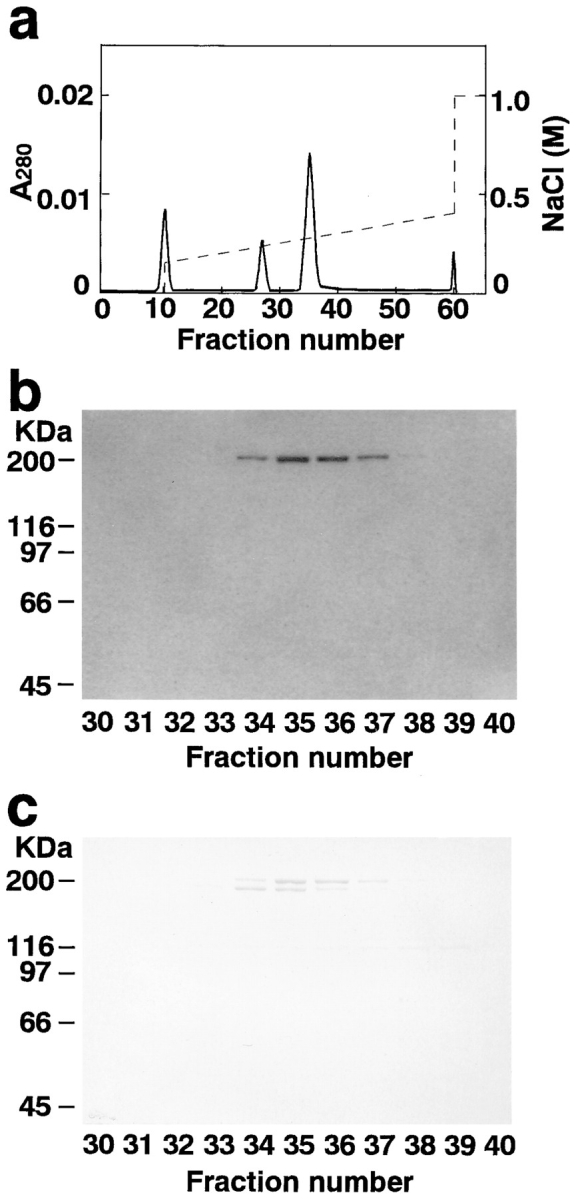
Mono Q column chromatography. (a) Absorbance at 280 nm (A280). (b) 125I-labeled F-actin blot overlay. An aliquot (3 μl) of each fraction was subjected to 125I-labeled F-actin blot overlay. (c) Protein staining with Coomassie brilliant blue. An aliquot (10 μl) of each fraction was subjected to SDS-PAGE (8% polyacrylamide gel), followed by protein staining with Coomassie brilliant blue.
Peptide Mapping of l- and s-Afadins and Molecular Cloning of Their cDNAs
The purified Mono Q sample (~40 μg of protein) was subjected to SDS-PAGE (8% polyacrylamide gel). The protein bands corresponding to p205 (l-afadin) and p190 (s-afadin) were separately cut out from the gel, digested with a lysyl endopeptidase, and the digested peptides were separated by TSKgel ODS-80Ts (4.6 × 150 mm; Tosoh) reverse-phase high pressure liquid column chromatography as described (Imazumi et al., 1994). The aa sequences of the peptides were determined with a peptide sequencer. A rat brain cDNA library in λZAPII (Stratagene, La Jolla, CA) was screened using the oligonucleotide probes designed from the partial aa sequences. DNA sequencing was performed by the dideoxynucleotide termination method using a DNA sequencer (model 373; Applied Biosystems, Inc., Foster City, CA).
Chromosome Preparation and In Situ Hybridization
The fluorescence in situ hybridization (FISH) analysis was performed as described (Matsuda et al., 1992; Matsuda and Chapman, 1995). Briefly, the chromosomes were prepared from rat splenocytes, the R-band was stained with Hoechst 33258, and the chromosomes were exposed to UV light. After the cDNAs of clones 20, 84, and 94 were labeled with biotin 16-dUTP, the chromosomes were hybridized with these labeled cDNAs, and they were incubated with an antibiotin goat antibody and then with an FITC-labeled anti–goat IgG antibody. The chromosomes were then stained with propidium iodide and were viewed using Nikon filter B-2A (450–490 nm; Nikon Inc., Melville, NY).
Expression and Purification of Recombinant l-Afadin
The full-length l-afadin cDNA corresponding to bp 326–5812 (aa 1–1829) and its fragment corresponding to bp 326–5344 (aa 1–1673) were inserted to the pCMV-myc vector using standard molecular biology methods (Sambrook et al., 1989). The constructs were transfected to COS7 cells with the DEAE-dextran method (Hata and Südhof, 1995). The whole-cell extracts were subjected to 125I-labeled F-actin blot overlay and Western blot analysis using an antibody against the myc-epitope as described below. The 597-bp fragment of the l-afadin cDNA corresponding to bp 5216–5812 (aa 1631–1829) was inserted to the pGEX and pQE vectors. These constructs were transformed into Escherichia coli, and the glutathione S-transferase (GST) fusion protein (GST–l-afadin-C) and the six histidine residues (His6)–fusion protein (His6-l-afadin-C) were purified by use of glutathione-Sepharose and Ni-agarose, respectively.
F-Actin–binding and –cross-linking Activities
Low shear viscometry was performed as described (Pollard and Cooper, 1982; Kato et al., 1996). Briefly, α-actinin (recombinant chick lung type) or the Mono Q sample of l-afadin, including s-afadin, in an indicated amount was mixed with 0.28 mg/ml of G-actin in a solution containing 20 mM Tris/Cl, pH 8.0, 0.25 mM EDTA, 2 mM MgCl2, 140 mM NaCl, 0.1 mM ATP, and 1 mM EGTA, and the solution was sucked into a 0.1-ml micropipette. After the incubation for 1 h at 25°C, the time for a stainless steel ball to fall a fixed distance in the pipette was measured.
Cosedimentation of l-afadin with F-actin was performed as follows: G-actin was polymerized by incubation for 30 min at room temperature in a polymerization buffer (20 mM imidazol/Cl, pH 7.0, 2 mM MgCl2, 1 mM ATP, and 90 mM KCl). An indicated amount of GST–l-afadin-C or His6– l-afadin-C (each aa 1631–1829) was incubated for 30 min at room temperature with 0.3 mg/ml of F-actin, and the mixture (100 μl) was placed over a 50-μl cushion of 30% sucrose in the polymerization buffer. After the sample was centrifuged at 130,000 g for 20 min, the supernatant was removed from the cushion, and the pellet was brought to the original volume in an SDS sample buffer. To quantitate cosedimentation of GST–l-afadin-C or His6–l-afadin-C, the comparable amounts of the supernatant and pellet fractions were subjected to SDS-PAGE (15% polyacrylamide gel), transferred to a nitrocellulose membrane, incubated with an antibody against l-afadin as described below, and then incubated with 125I-labeled protein A. Amounts of the free and bound fusion proteins were calculated by determining the radioactivity from the supernatant and pellet fractions with an image analyzer (Fujix BAS-2000II).
Antibodies and Immunofluorescence Microscopy
A rabbit polyclonal antibody against l-afadin, which specifically recognized l-afadin, was directed against a 16-mer synthetic peptide corresponding to aa 1814–1829 (KASRKLTELENELNTK). Another antibody against l-afadin, which recognized both l- and s-afadins, was directed against a 16-mer synthetic peptide corresponding to aa 577–592 (RLDQEQDYRRRESRTQ). Each peptide was coupled via cysteine at the NH2-terminal residue to keyhole limpet hemocyanin and was used to raise the antiserum. The antiserum specific to l-afadin was affinity-purified with GST–l-afadin-C covalently coupled to NHS-activated Sepharose (Pharmacia Biotech, Inc.) and was used as the anti–l-afadin antibody. The antiserum recognizing both l- and s-afadins was purified with the synthetic peptide covalently coupled to EAH-Sepharose (Pharmacia Biotech, Inc.) and was used as the anti–l- and s-afadin antibody. Monoclonal anti–ZO-1 (Itoh et al., 1991) and anti–myc epitope (Takaishi et al., 1995) antibodies were prepared as described. Monoclonal anti–E-cadherin (Takara, Tokyo, Japan), antivinculin (Sigma Chemical Co.), and antidesmoplakin (Progen Biotechnik, Germany) antibodies were purchased from commercial sources.
Immunofluorescence microscopy of frozen sections of various rat or mouse tissues was done as described (Itoh et al., 1991). Briefly, samples of various tissues were frozen using liquid nitrogen, and the frozen sections (~10 μm) were cut in a cryostat. The samples were mounted on glass slides, air dried, and fixed with 95% ethanol for 30 min at 4°C and with 100% acetone for 1 min at room temperature. After being washed with PBS containing 1% BSA, the samples were incubated with the polyclonal anti–l-afadin antibody alone, or with a mixture of the anti–l-afadin antibody and the monoclonal anti–E-cadherin, anti–ZO-1, or antivinculin antibody. The samples were then washed with PBS containing 1% BSA and 0.1% Triton X-100, followed by incubation with rhodamine-conjugated donkey anti–rabbit IgG and FITC-conjugated sheep anti–mouse IgG antibodies. After being washed with PBS, they were embedded and viewed with a confocal imaging system (MRC-1024; Bio-Rad Laboratories, Hercules, CA).
Immunofluorescence microscopy of EL cells was done as described (Itoh et al., 1991). Briefly, cells were cultured on a coverglass and fixed with 1% formaldehyde in PBS. The fixed sample was treated with 0.2% Triton X-100 in PBS and washed three times with PBS. After the sample was soaked with PBS containing 1% BSA, the sample was treated with the rabbit anti–l-afadin antibody and the mouse anti–ZO-1 antibody, and washed with PBS containing 1% BSA, followed by incubation with the rhodamine-conjugated anti–rabbit IgG and FITC-conjugated anti–mouse IgG antibodies. After the incubation, the sample was washed with PBS, embedded in PBS containing 95% glycerol, and viewed with the confocal imaging system (MRC-1024; Bio-Rad Laboratories).
Immunofluorescence microscopy of the bile canaliculi isolated from mouse liver was done as described (Tsukita et al., 1989). Briefly, the fraction rich in the bile canaliculi (Tsukita and Tsukita, 1989) was put on a coverglass, air dried, and fixed with 1% formaldehyde in PBS. The sample was soaked with PBS containing 1% BSA and then processed as described above.
Immunoelectron Microscopy
Immunoelectron microscopy using the silver enhancement technique was done as described (Mizoguchi et al., 1994). Briefly, adult rat small intestine was perfused with 4% paraformaldehyde in PBS. The sample was incubated with the anti–l-afadin antibody, followed by incubation with an anti–rabbit IgG antibody coupled with 1.4-nm gold particles (Nanoprobes Inc.). After the sample was washed, it was fixed with 2% glutaraldehyde in PBS for 15 min, and the sample-bound gold particles were silver enhanced by the HQ-silver kit (Nanoprobes Inc., Stony Brook, NY) for 8 min at 18°C. The sample was again washed and postfixed with 0.5% osmium oxide in a buffer containing 100 mM cacodylate buffer, pH 7.3. They were dehydrated by passage through a graded series of ethanol (50, 70, 90, and 100%) and propylene oxide, and were embedded in epoxy resin. From this sample, ultrathin sections were cut, stained with uranyl acetate and lead citrate, and then observed with an electron microscope (JEM-1200EX; Jeol Ltd., Tokyo, Japan).
Immunoelectron microscopy using the ultrathin cryosection technique was done as described (Tokuyasu, 1980). Briefly, adult rat small intestine was fixed with 4% formaldehyde in 0.1 M phosphate buffer, pH 7.4, for 6 h at room temperature. The fixed sample was infused with 1.8 M sucrose containing 20% polyvinylpyrrolidone at 4°C overnight (Tokuyasu, 1989), rapidly frozen using liquid nitrogen, and then cryosectioned at −110°C using a diamond knife. The ultrathin sections were collected on Formvar-filmed grids and rinsed with PBS containing 10 mM glycine. The samples were treated with a mixture of 3% BSA and 0.5% gelatin for 30 min, and then incubated sequentially with the anti–l-afadin antibody and with a 5-nm colloidal gold-conjugated goat anti–rabbit IgG antibody (Amersham Corp., Arlington Heights, IL). The samples were fixed with 2% glutaraldehyde in PBS, rinsed with distilled water, incubated with 2% methylcellulose and 0.5% uranyl acetate (Griffiths et al., 1984), and air dried. The samples were observed under an electron microscope (JEM-100CX; Jeol Ltd.).
Other Procedures
G-Actin was purified from rabbit skeletal muscle as described (Pardee and Spudich, 1982). Protein concentrations were determined with BSA as a reference protein (Bradford, 1976).
Results
Purification of l-Afadin and Molecular Cloning of Its cDNA
To identify novel actin-binding proteins, we attempted to detect actin-binding proteins using a blot overlay method with 125I-labeled F-actin. The soluble fraction of rat fetal brain was subjected to SDS-PAGE followed by the blot overlay. Several radioactive protein bands with various molecular masses were detected. Of these F-actin–binding proteins, the protein band with a molecular mass of ~205 kD (p205) was highly purified by column chromatographies, including Q-Sepharose, phenyl-5PW, hydroxyapatite, and Mono Q column chromatographies. On the final Mono Q column chromatography, the F-actin–binding protein band well coincided with a protein with a molecular mass of ~205 kD that was identified by protein staining with Coomassie brilliant blue (Fig. (Fig.1).1). p205 was copurified with another protein with a molecular mass of ~190 kD (p190), and they could not be separated from each other by further column chromatographies, including Mono S and Superdex 200 column chromatographies (data not shown). These proteins appeared at a position corresponding to a molecular mass of >600 kD on the gel filtration. The Mono Q sample including p205 and p190 was used to study the biochemical properties of p205 as described below.
When the peptide mapping analyses of the Mono Q sample of these two proteins, p205 and p190, were performed, their maps were almost identical, whereas p205 showed the additional peptide peaks. When the aa sequences of the five peptide peaks common between the two proteins were separately determined, all the aa sequences of the p205 and p190 peptides were identical. A computer homology search indicated that the aa sequences of the five peptide peaks were identical with or significantly homologous to those of human AF-6 protein. When the aa sequences of the two peptide peaks specific to p205 were determined, they were not found in the current protein database. These results suggest that p205 and p190 are related to human AF-6 protein, and that p190 is a splicing variant, a homologue, or a degradative product of p205.
A rat brain cDNA library was screened using the degenerated oligonucleotide probes designed from the partial aa sequences of the seven peptide peaks described above. We obtained several overlapping clones (Fig. (Fig.2).2). Among these clones, clone 20 contained a coding region with ~4.9 kb, but it lacked a predicted initiation codon. The deduced aa sequence included all the aa sequences of the peptides of p205. This aa sequence analysis indicated that the aa sequences of the two peptides specific to p205 were both located in the COOH-terminal region. Clone 94 contained a coding region with ~4.5 kb and an in-frame stop codon upstream of the stop codon of clone 20. This clone also lacked a predicted initiation codon. The deduced aa sequence of the coding region included all the aa sequences of the peptides of p190, but not those of the peptides specific to p205. Clone 84 contained a coding region with ~4.2 kb and a predicted initiation codon. To confirm whether these cDNAs were derived from the same locus, we made the chromosomal assignment by FISH analysis using the cDNAs as probes. All the cDNAs were localized on rat chromosome 1q12.2 (data not shown).
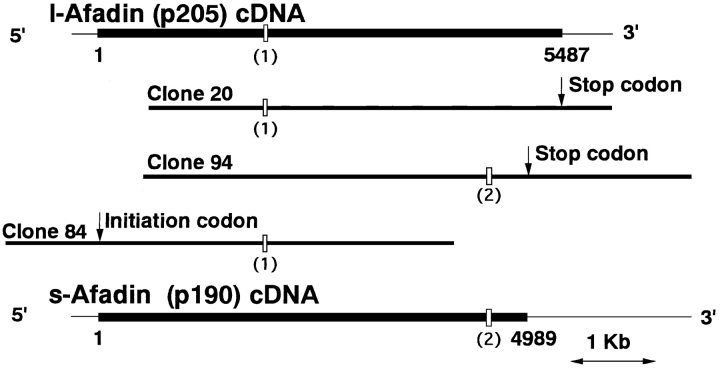
Schematic drawings of l-afadin (p205) and s-afadin (p190) cDNAs. (1) alternative insertion of 21 bp; (2) alternative insertion of 36 bp. The sequence data of the l- and s-afadin genes are available from GenBank/EMBL/DDBJ under accession numbers U83230 and U83231, respectively.
The full-length p205 and p190 cDNAs were constructed from clones 84 and 20, and clones 84 and 94, respectively. When the p205 cDNA was transfected into COS7 cells and the cell extract was subjected to 125I-labeled F-actin blot overlay, the recombinant protein (myc–l-afadin) showed mobility similar to that of native p205 on SDS-PAGE, as well as 125I-labeled F-actin–binding activity (Fig. (Fig.33 a). The deletion mutant of p205 lacking the COOH-terminal 156 aa (myc–l-afadinΔC) did not show the 125I-labeled F-actin– binding activity. A fusion protein of the COOH-terminal region of p205 (199 aa) with GST (GST–l-afadin-C) showed the 125I-labeled F-actin–binding activity.
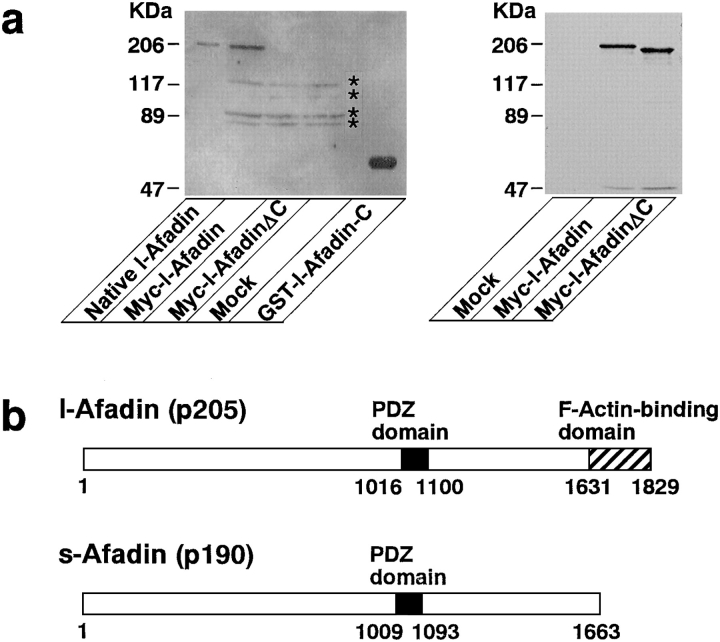
Domain organization of l-afadin (p205) and s-afadin (p190). (a) F-actin–binding activity of various fragments of recombinant l-afadin. Full-length l-afadin (aa 1–1829) (myc–l-afadin) and its deletion mutant (aa 1–1673) (myc–l-afadinΔC) were expressed as myc-tagged proteins in COS7 cells. The COOH-terminal fragment (aa 1631–1829) (GST–l-afadin-C) was expressed as a GST fusion protein in E. coli and was purified with glutathione-Sepharose. The extracts from the COS7 cells and the protein purified from the bacteria were subjected to a 125I-labeled F-actin blot overlay. The extracts from the COS7 cells were subjected to Western blot analysis using the myc epitope antibody to confirm the expression amounts. (Left) 125I-labeled F-actin blot overlay; (right) Western blot analysis using the anti-myc antibody. (Asterisks) endogenous proteins of COS7 cells with 125I-labeled F-actin–binding activity. (b) Schematic drawings of l- and s-afadin structures.
Based on these observations, together with the fact that the Mono Q sample of p205 indeed showed the F-actin– binding activity as described below, we have concluded that the p205 gene encodes a protein of 1,829 aa with a calculated molecular mass of 207,667 kD (Gen Bank/EMBL/ DDBJ accession number U83230), of which the COOH-terminal 199 aa contributes to its F-actin–binding activity, and that the p190 gene encodes a protein lacking ~160 COOH-terminal aa (GenBank/EMBL/DDBJ accession number U83231) and is a splicing variant of the p205 gene. A computer homology search revealed that the aa sequence of p190 showed 90% identity over the entire sequence with that of human AF-6 protein (Prasad et al., 1993), while human AF-6 protein and p190 lacked the COOH-terminal region of p205. p190 is likely to be a rat counterpart of human AF-6 protein. The COOH-terminal, F-actin–binding domain showed no significant homology to any known F-actin–binding protein. Both p205 and p190 had one PDZ domain (Fig. (Fig.33 b). We named p205 and p190 l- and s-afadins, respectively.
Northern blot analysis using a sequence specific to the l-afadin cDNA as a probe detected ~7.5-kb mRNA in all the rat tissues examined, including the heart, brain, spleen, lung, liver, skeletal muscle, kidney, and testis (Fig. (Fig.44 a). A similar result was obtained when a sequence common between the cDNAs of l- and s-afadins was used as a probe (data not shown). Western blot analysis using an antibody specific to l-afadin detected an ~205-kD protein in all the rat tissues examined (Fig. (Fig.44 b1). When an antibody recognizing both l- and s-afadins was used, however, an ~190-kD protein was detected only in the brain (Fig. (Fig.44 b2). These results indicate that l-afadin is expressed ubiquitously while s-afadin is specifically expressed in brain. The reason why the Northern blot analysis did not detect the two bands in the brain may simply be because of the similar length of the mRNAs of l- and s-afadins.
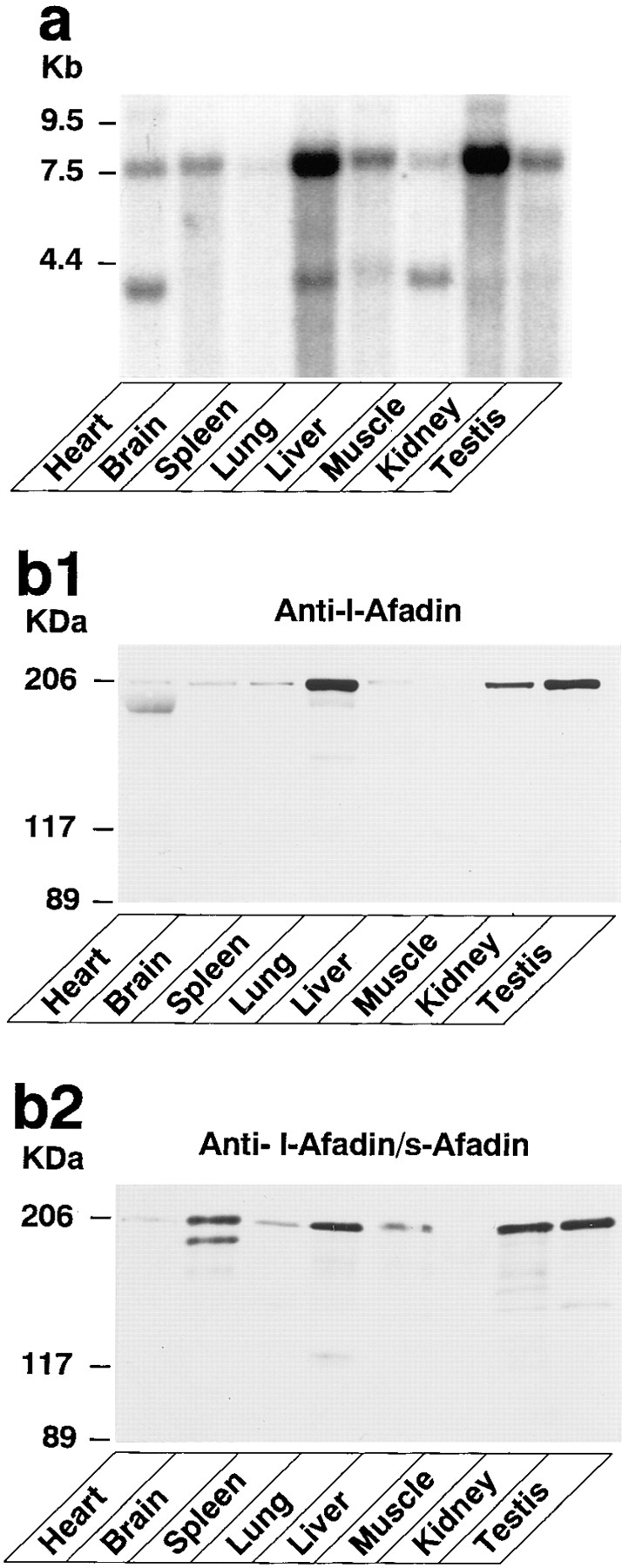
Tissue distribution of l-afadin. (a) Northern blot analysis. A RNA blot membrane (CLONTECH, Palo Alto, CA) was hybridized with the 32P-labeled fragment (bp 5327–5812) of the l-afadin cDNA according to the manufacturer's protocol. In addition to the ~7.5-kb mRNA of l-afadin, a smaller mRNA of ~4.2 kb was detected in several tissues, but significance of this is unknown. (b1 and b2) Western blot analyses. Various rat tissue homogenates (10 μg of protein each) were subjected to SDS-PAGE (8% polyacrylamide gel), followed by immunoblot using the anti–l-afadin (b1) or the anti–l- and –s-afadin antibody (b2). The lower band in the heart lane with the anti–l-afadin antibody was nonspecific, since the treatment with the peptide that was used to raise the antibody did not quench this lower band, while it quenched the band with a molecular mass of ~205 kD (l-afadin).
Biochemical Properties of l-Afadin
It was examined by competition experiments whether the Mono Q sample of l-afadin (including s-afadin) bound along the sides of F-actin or at its ends. The binding of 125I-labeled F-actin to l-afadin was completely inhibited by an excessive amount of myosin S1, a well-characterized protein that binds along the sides of F-actin (Rayment et al., 1993; Schröder et al., 1993) (Fig. (Fig.55 a). This inhibition was reversed by the addition of MgATP because MgATP dissociates the actin–myosin complex (Fraser et al., 1975). These results indicate that l-afadin binds along the sides of F-actin. The effect of the Mono Q sample of l-afadin on F-actin was examined using the falling ball method for low shear viscometry. This effect was compared to that of α-actinin, a well-characterized protein that shows F-actin–cross-linking activity (Burridge and Feramisco, 1981). l-Afadin increased the viscosity of F-actin in a dose-dependent manner, and the viscosity became maximal at approximately threefold (Fig. (Fig.55 b). α-Actinin also increased the viscosity, but the viscosity became unmeasurably high. This effect of the Mono Q sample of l-afadin on F-actin was further assessed by transmission electron microscopy of negatively stained specimens. l-Afadin hardly caused F-actin to associate into bundles and meshworks under conditions where α-actinin showed a marked F-actin–cross-linking activity (data not shown). This result was consistent with that of the low shear viscometry. The stoichiometry of the binding of l-afadin to actin and the affinity constant were examined using His6– l-afadin-C. The stoichiometry was calculated to be 1 His6– l-afadin-C molecule per ~500 actin molecules (Fig. (Fig.55 c). The kilodalton value was calculated to be the order of 10−7. A similar result was obtained with GST–l-afadin-C (data not shown). The effect of the Mono Q sample of l-afadin on actin polymerization was finally examined using pyrene-conjugated actin. Pyrene-conjugated actin is known to increase its fluorescent intensity with the actin polymerization (Cooper and Pollard, 1982). l-Afadin did not affect the actin polymerization under the conditions where gelsolin stimulated it (data not shown).
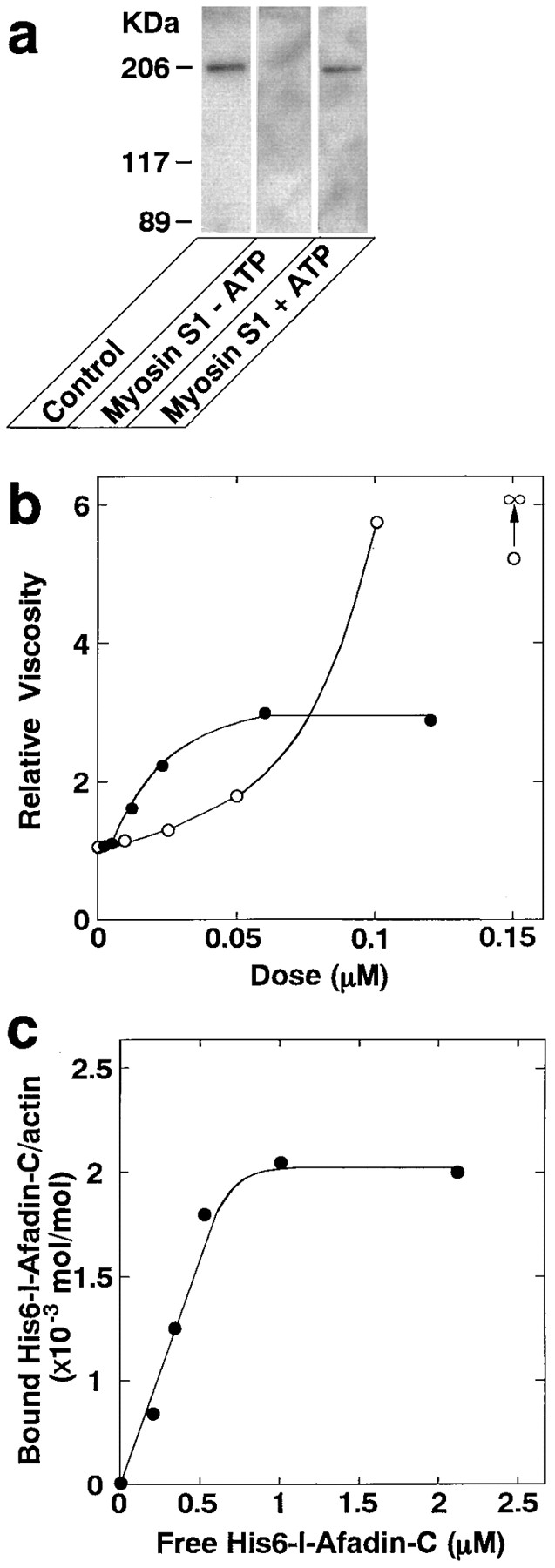
Biochemical properties of l-afadin. (a) Inhibition by myosin S1 of the binding of l-afadin to 125I-labeled F-actin. The Mono Q sample of l-afadin (0.1 μg of protein) was subjected to SDS-PAGE (8% polyacrylamide gel), followed by the blot overlay with 125I-labeled F-actin pretreated with myosin S1 in the presence or absence of ATP. (b) Increase by l-afadin of the viscosity of F-actin. The viscosity of F-actin was measured by low shear viscometry. •, With l-afadin; ○, with α-actinin. (c) Binding of His6–l-afadin-C to F-actin. The binding curve was generated by the cosedimentation of His6–l-afadin-C with F-actin.
Localization of l-Afadin at Cadherin-based Cell-to-Cell AJ
The localization of l-afadin was first examined by immunofluorescence microscopy of the frozen sections of various rat or mouse tissues using the anti–l-afadin antibody. In the liver, l-afadin was localized at the beltlike junctional complex region along the bile canaliculi (Fig. (Fig.66 a). When the frozen sections of the small intestine were doubly stained with the monoclonal anti–E-cadherin, l-afadin was concentrated with E-cadherin at the junctional complex region of intestine absorptive epithelia, but l-afadin was more highly concentrated at the junctional complex region than E-cadherin (Fig. (Fig.6,6, b1–3 and c1–3). The frozen sections of the heart were doubly stained with the monoclonal antivinculin antibody. Vinculin is known to be a marker, not only for cell-to-cell AJ, but also for cell-to-matrix AJ (Geiger, 1979, 1983). l-Afadin was colocalized with vinculin at the intercalated disc (Fig. (Fig.66 d1–3). Vinculin was also periodically located along the lateral borders of cardiac muscle cells (costamere) while l-afadin was not. When the cultured EL cells expressing E-cadherin (Nagafuchi et al., 1987) were doubly stained with the anti–ZO-1 antibody, l-afadin showed localization similar to that of ZO-1 (Fig. (Fig.7,7, a and b). ZO-1 is known to be concentrated at cadherin-based, spotlike cell-to-cell AJ in fibroblasts (Itoh et al., 1991, 1993). These results suggest that l-afadin is localized at cadherin-based cell-to-cell AJ.
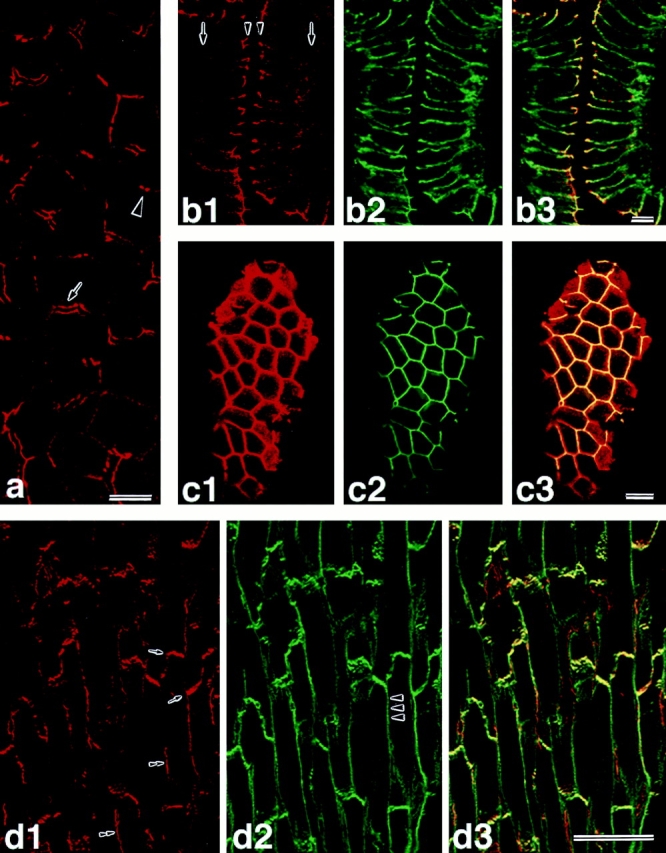
Localization of l-afadin, E-cadherin, and vinculin in various rat or mouse tissues. (a) The staining of rat liver with the anti– l-afadin antibody. Arrow, longitudinal-section view of bile canaliculi; arrowhead, cross-section view of bile canaliculi. (b1–b3) The double staining of mouse small intestinal epithelial cells with the anti–l-afadin and anti–E-cadherin antibodies: (b1) l-afadin; (b2) E-cadherin; (b3) l-afadin and E-cadherin. Arrows, basal level; arrowheads, apical level. (c1–c3) The double staining of the tangential section of mouse small intestinal cells at the level of the apical surface with the anti–l-afadin and anti–E-cadherin antibodies: (c1) l-afadin; (c2) E-cadherin; (c3) l-afadin and E-cadherin. (d1–d3) The double staining of mouse heart with the anti–l-afadin and antivinculin antibodies. It should be noted that the staining of l-afadin is also associated with the blood vessels: (d1) l-afadin; (d2) vinculin; (d3) l-afadin and vinculin. arrows, intercalated disc; arrowhead, costamere; double arrowheads, blood vessel. Bars: (a and b) 10 μm; (c) 5 μm; (d) 50 μm.
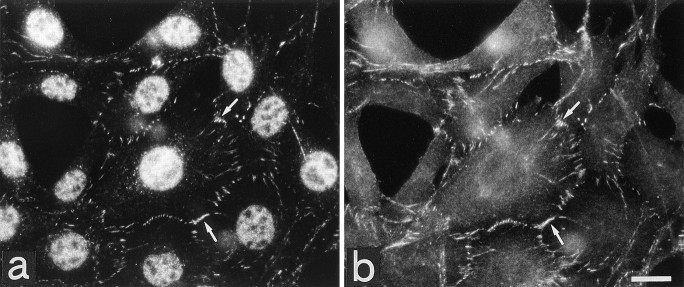
Localization of l-afadin and ZO-1 in the EL cells expressing E-cadherin. The EL cells expressing E-cadherin were doubly stained with the anti–l-afadin and anti–ZO-1 antibodies. l-Afadin was concentrated at cell-to-cell AJ. There were perinuclear and nuclear stainings with this anti–l-afadin antibody, but its significance is not clear. (a) l-afadin; (b) ZO-1. arrows, cell-to-cell AJ. Bar, 25 μm.
To examine the precise localization of l-afadin at the junctional complex region, the frozen sections of small intestine were doubly stained with the monoclonal anti–ZO-1 antibody, and the bile canaliculi isolated from liver were doubly stained with the monoclonal antidesmoplakin antibody. ZO-1 and desmoplakin are known to be markers for tight junction (Stevenson et al., 1986; Itoh et al., 1993) and desmosome (Skerrow and Matoltsy, 1974; Franke et al., 1983; Mueller and Franke, 1983; Miller et al., 1987), respectively. In the absorptive epithelia, l-afadin was localized slightly more at the basal side than ZO-1 (Fig. (Fig.88 a1–a3). In the canaliculi, the localization of l-afadin did not coincide with that of desmoplakin (Fig. (Fig.88 b1–b3). These results indicate that l-afadin is localized at cell-to-cell AJ rather than at tight junction or desmosome. The localization of l-afadin at cell-to-cell AJ was finally confirmed by immunoelectron microscopy of intestine absorptive epithelia using the silver enhancement and ultrathin cryosection techniques. l-Afadin was associated with the undercoat of cell-to-cell AJ (Fig. (Fig.9,9, a and b).
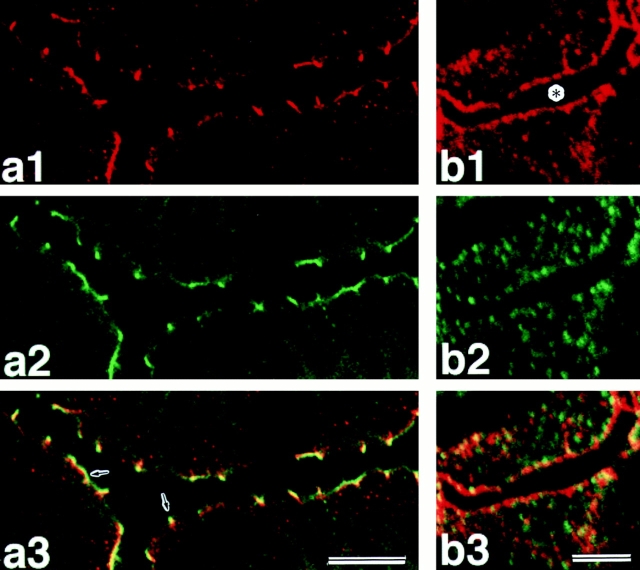
Different localization of l-afadin, ZO-1, and desmoplakin. (a1–a3) Localization of l-afadin and ZO-1 in mouse small intestine: (a1) l-afadin; (a2) ZO-1; (a3) l-afadin and ZO-1. arrows, the sites where green (ZO-1), yellow (the mixture of l-afadin and ZO-1), and red (l-afadin) signals occurred in this order from the apical side to the basal side. (b1–b3) Localization of l-afadin and desmoplakin in the isolated bile canaliculi: (b1) l-afadin; (b2) desmoplakin; (b3) l-afadin and desmoplakin. Asterisk, the inner space of the bile canaliculi. Bars: (a) 15 μm; (b) 5 μm.
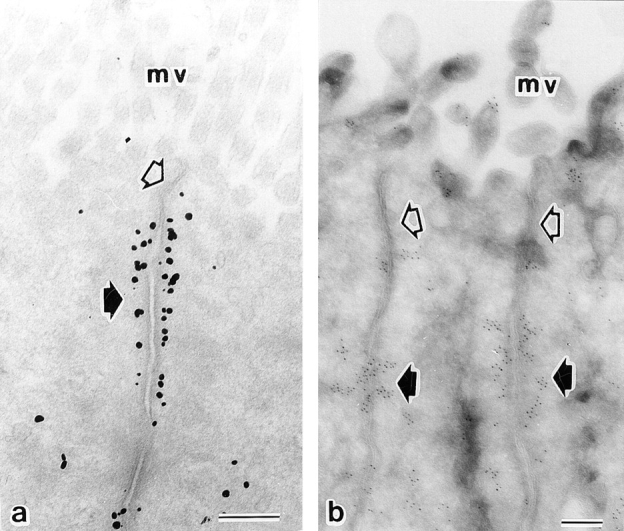
Ultrastructural localization of l-afadin in rat small intestine. Intestinal epithelial cells were labeled singly with the anti–l-afadin antibody. (a) silver enhancement technique; (b) ultrathin cryosection technique. Open arrow, tight junction; closed arrow, cell-to-cell AJ; mv, microvilli. Bars, 0.2 μm.
Discussion
Here, we isolated a novel F-actin–binding protein with a molecular mass of ~205 kD (p205). This protein was copurified with another protein with a molecular mass of 190 kD (p190) that lacked the F-actin–binding activity on various column chromatographies. The molecular cloning of the cDNAs of these two proteins revealed that the nucleotide sequence of the p190 cDNA was identical to that of the p205 cDNA, except for the two splicing regions. FISH analysis revealed that the genes of these two proteins were localized at the same locus. These results suggest that p205 and p190 are splicing variants derived from the same gene. Because a computer homology search revealed that the aa sequence of p190 was almost identical to that of human AF-6 protein, we theorize that p190 may be a rat counterpart of human AF-6 protein. We named p205 and p190 l- and s-afadins, respectively. Further purification steps of l-afadin, including Mono S column and Superdex 200 column chromatographies, did not separate l- and s-afadins from each other, and their elution profiles on these column chromatographies were similar but not identical. These proteins appeared at a position corresponding to a molecular mass of >600 kD on the gel filtration. These results suggest that 1- and s-afadins form a complex, but we cannot exclude the possibility that these two proteins do not form a complex, but instead are incidentally eluted at similar fractions on these column chromatographies. It is, therefore, possible that the complex is composed of a heterooligomer alone, a homooligomer alone, or the mixture. Western blot analysis indicated that the tissues other than brain expressed l-afadin alone. It is likely that l-afadin forms a homooligomer in these tissues.
Recently, Luna's group (Pestonjamasp et al., 1995) has shown that a protein with a molecular mass of ~205 kD in bovine neutrophils has the 125I-labeled F-actin–binding activity. When human neutrophils were subjected to SDS-PAGE, followed either by Western blot analysis using the antibody specific to l-afadin or by 125I-labeled F-actin blot overlay, an immunoreactive band with a molecular mass of ~200 kD was detected, and this band showed the 125I-labeled F-actin–binding activity (data not shown). This protein band is likely to be l-afadin, although its molecular mass was slightly smaller than that of rat l-afadin. The different molecular mass values may result from the species' differences. The exact relationship between the 205-kD protein described by Luna's group and l-afadin is not known, since the primary structure of the 205-kD protein has not been determined, but the 205-kD protein may be l-afadin.
l-Afadin showed 125I-labeled F-actin–binding activity. Neither s-afadin nor the deletion mutant of l-afadin lacking the COOH-terminal 156 aa (myc–l-afadinΔC) showed this activity, whereas GST–l-afadin-C corresponding to the COOH-terminal 199 aa showed the 125I-labeled F-actin– binding activity. The Mono Q sample of l-afadin increased the viscosity of F-actin on the low shear viscometry. Furthermore, His6–l-afadin-C corresponding to the COOH-terminal 199 aa was cosedimented with F-actin. These results indicate that l-afadin shows F-actin–binding activity and that the domain responsible for this activity is located at the COOH-terminal 199 aa. Many actin-binding protein families have been isolated and characterized. Of these families, for instance, the α-actinin/spectrin family members have been shown to usually form oligomers, such as a homodimer or a heterotetramer, and thereby show the F-actin–cross-linking activity. α-Actinin made the viscosity of F-actin unmeasurably high on the low shear viscometry, and it caused F-actin to associate into bundles and meshworks, as estimated by transmission electron microscopy. The stoichiometry is one α-actinin molecule per about seven actin molecules, and the kilodalton value is the order of 10 −7 (Meyer and Aebi, 1990). In contrast, the effect of the Mono Q sample of l-afadin on F-actin was small on the low shear viscometry, and the sample hardly showed F-actin–cross-linking activity, as estimated by transmission electron microscopy. The stoichiometry was calculated to be 1 His6–l-afadin-C molecule per ~500 actin molecules. The kilodalton value was calculated to be the order of 10−7. These results indicate that l-afadin belongs to a family different from the α-actinin/spectrin family. The ERM family, including ezrin, radixin, and moesin, belongs to the band 4.1 superfamily, an F-actin–binding protein superfamily (Bretscher, 1993; Tsukita et al., 1997), and there have been several reports that purified ezrin and moesin are hardly cosedimented with F-actin (Bretscher, 1983; Pestonjamasp et al., 1995; Shuster and Herman, 1995). Luna's group (Pestonjamasp et al., 1995) has recently shown by the 125I-labeled F-actin blot overlay that ezrin and moesin exhibit the F-actin–binding activity. They have also shown by competition experiments using the 125I-labeled F-actin blot overlay that ezrin and moesin bind along the sides of F-actin. The biochemical properties of l-afadin are apparently similar to those of ezrin and moesin. l-Afadin may bind F-actin in a manner similar to those of ezrin and moesin.
An immunofluorescence microscopic study by use of the antibody specific to l-afadin revealed that l-afadin was localized at the junctional complex region. Further microscopic and electron microscopic studies revealed that l-afadin was concentrated at cadherin-based cell-to-cell AJ. Taken together with the biochemical properties of l-afadin, l-afadin is likely to be involved in the linkage between the actin cytoskeleton and cell-to-cell AJ. Both l- and s-afadins have one PDZ domain. The PDZ domain has been found in a variety of proteins that are typically localized at cell-to-cell junctions (Saras and Heldin, 1996) and shown to bind to the unique COOH-terminal motifs of target proteins, which are found in a large number of transmembrane proteins (Kim et al., 1995; Kornau et al., 1995; Niethammer et al., 1996). l-Afadin may be recruited to cell-to-cell AJ through its PDZ domain. To understand the mechanism by which l-afadin is localized at cell-to-cell AJ, it is of crucial importance to identify its interacting molecule(s).
Many F-actin–binding proteins, including α-actinin, talin, and vinculin, have been shown to serve as linkers between the actin cytoskeleton and integrin at cell-to-matrix AJ (Jockusch et al., 1995). The cytoplasmic domain of the β-subunit of integrin interacts directly with α-actinin and talin and indirectly with vinculin through α-actinin and talin (Jockusch et al., 1995). At cell-to-cell AJ, the cytoplasmic domain of cadherin interacts with α-catenin through β-catenin (Ozawa et al., 1989; Nagafuchi et al., 1991; Takeichi, 1991; Tsukita et al., 1992). α-Catenin interacts directly with F-actin (Rimm et al., 1995), α-actinin (Knudsen et al., 1995), and ZO-1 (Itoh et al., 1997). Vinculin is highly concentrated at cell-to-cell AJ, but its interacting molecule has not yet been identified (Geiger and Ginsberg, 1991; Tsukita et al., 1992). We showed here that l-afadin was localized at cell-to-cell AJ. Therefore, the molecular linkage mechanism between the actin cytoskeleton and cadherin is apparently similar to that between the actin cytoskeleton and integrin in the sense that F-actin is linked to each transmembrane protein through multiple F-actin–binding proteins.
The AF-6 gene has originally been identified as a fusion partner of the ALL-1 gene (Prasad et al., 1993), which is known to be involved in acute leukemia (Cimino et al., 1991). It has recently been shown that the AF-6 gene maps outside the minimal region of deletion in ovarian cancers with similar chromosomal aberrations (Saha et al., 1995; Saito et al., 1996). However, neither the significance of the fusion of these two genes nor the role of s-afadin in carcinogenesis has been clarified. s-Afadin has recently been shown to have significant homology to the product of the Drosophila canoe gene (Miyamato et al., 1995). The canoe gene interacts genetically with the Notch signal cascade in the eye, bristle, and wing development (Miyamato et al., 1995). Moreover, the canoe gene interacts genetically with the armadillo (β-catenin) gene (Miyamato et al., 1995). The products of the Notch and armadillo genes have been implicated in adhesive cell-to-cell communications (Peifer et al., 1993; Artavanis-Tsakonas et al., 1995). l- and s-Afadins may also play a role in the development of various tissues by mediating interactions between the adhesive molecule–initiated signal transduction pathways.
The AF-6 protein (s-afadin) has recently been shown to interact specifically with the GTP-bound active form of Ras small G protein (Kuriyama et al., 1996); however, we have not been able to reproduce this earlier finding. We found that both the GDP- and GTP-bound forms of Ki-Ras interacted with l- and s-afadins to similar extents and that the stoichiometry of these interactions was negligible (data not shown). Moreover, an immunofluorescence microscopic study of Madin-Darby canine kidney cells stably expressing an myc-tagged dominant active mutant of Ki-Ras (N12V) or an myc-tagged wild type of Ki-Ras showed that these two types of Ras are not concentrated at cell-to-cell AJ (Takaishi, K., and Y. Takai, unpublished observations). All of these results do not support the specific interaction of Ras with s-afadin, and the relationship between Ras and s-afadin is currently unknown.
Acknowledgments
We thank Dr. M. Takeichi (Kyoto University, Kyoto, Japan) for providing the EL cells expressing E-cadherin, and Dr. M. Imamura (National Institute of Neuroscience, Kodaira, Japan) for providing us the recombinant chick lung type α-actinin.
Abbreviations used in this paper
| aa | amino acid |
| AJ | adherens junction |
| F-actin | actin filament |
| FISH | fluorescence in situ hybridization |
| G-actin | actin monomer |
| GST | glutathione S-transferase |
| His6 | six histidine residues |
| l-afadin | a large splicing variant of AF-6 protein localized at adherens junction |
| S1 | subfragment 1 |
| s-afadin | a small splicing variant of l-afadin |
Footnotes
Address all correspondence to Y. Takai, Department of Molecular Biology and Biochemistry, Osaka University Medical School, Suita 565, Osaka, Japan. Tel.: 81-6-879-3410. Fax: 81-6-879-3419. E-mail: pj.ca.u-akaso.dem.oiblom@iakaty
References
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science (Wash DC) 1995;268:225–232. [Abstract] [Google Scholar]
- Bentley D, O'Connor TP. Cytoskeletal events in growth cone steering. Curr Opin Neurobiol. 1994;4:43–48. [Abstract] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. [Abstract] [Google Scholar]
- Bretscher A. Purification of an 80,000-dalton protein that is a component of the isolated microvillus cytoskeleton, and its localization in nonmuscle cells. J Cell Biol. 1983;97:425–432. [Europe PMC free article] [Abstract] [Google Scholar]
- Bretscher A. Microfilaments and membranes. Curr Opin Cell Biol. 1993;5:653–660. [Abstract] [Google Scholar]
- Burridge K, Feramisco JR. Non-muscle α-actinins are calcium-sensitive actin-binding proteins. Nature (Lond) 1981;294:565–567. [Abstract] [Google Scholar]
- Chia CP, Hitt AL, Luna EJ. Direct binding of F-actin to ponticulin, an integral plasma membrane glycoprotein. Cell Motil Cytoskel. 1991;18:164–179. [Abstract] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophiladisc large tumor suppressor protein. Neuron. 1992;9:929–942. [Abstract] [Google Scholar]
- Cimino G, Moir DT, Canaani O, Williams K, Crist WM, Katzav S, Cannizzaro L, Lange B, Nowell PC, Croce CM, Canaani E. Cloning of ALL-1, the locus involved in leukemias with the t(4;11)(q21;q23), t(9;11) (p22;q23), and t(11;19)(q23;p13) chromosome translocations. Cancer Res. 1991;51:6712–6714. [Abstract] [Google Scholar]
- Cooper JA, Pollard TD. Methods to measure actin polymerization. Methods Enzymol. 1982;85:182–210. [Abstract] [Google Scholar]
- Franke WW, Kapprell HP, Mueller H. Isolation and symmetrical splitting of desmosomal structures in 9 M urea. Eur J Cell Biol. 1983;32:117–130. [Abstract] [Google Scholar]
- Fraser AB, Eisenberg E, Kielley WW, Carlson FD. The interaction of heavy meromyosin and subfragment 1 with actin. Physical measurements in the presence and absence of adenosine triphosphate. Biochemistry. 1975;14:2207–2214. [Abstract] [Google Scholar]
- Geiger B. A 130K protein from chicken gizzard: its localization at the termini of microfilament bundles in cultured chicken cells. Cell. 1979;18:193–205. [Abstract] [Google Scholar]
- Geiger B. Membrane-cytoskeleton interaction. Biochim Biophys Acta. 1983;737:305–341. [Abstract] [Google Scholar]
- Geiger B. Cytoskeleton-associated cell contacts. Curr Opin Cell Biol. 1989;1:103–109. [Abstract] [Google Scholar]
- Geiger B, Ginsberg D. The cytoplasmic domain of adherens-type junctions. Cell Motil Cytoskel. 1991;20:1–6. [Abstract] [Google Scholar]
- Griffiths G, McDowall A, Back R, Dubochet J. On the preparation of cryosections for immunocytochemistry. J Ultrastruct Res. 1984;89:65–78. [Abstract] [Google Scholar]
- Gumbiner B, Lowenkopf T, Apatira D. Identification of a 160-kDa polypeptide that binds to the tight junction protein ZO-1. Proc Natl Acad Sci USA. 1991;88:3460–3464. [Europe PMC free article] [Abstract] [Google Scholar]
- Hata Y, Südhof TC. A novel ubiquitous form of Munc-18 interacts with multiple syntaxins. Use of the yeast two-hybrid system to study interactions between proteins involved in membrane traffic. J Biol Chem. 1995;270:13022–13028. [Abstract] [Google Scholar]
- Imazumi K, Sasaki T, Takahashi K, Takai Y. Identification of a rabphilin-3A-interacting protein as GTP cyclohydrolase I in PC12 cells. Biochem Biophys Res Commun. 1994;205:1409–1416. [Abstract] [Google Scholar]
- Itoh M, Yonemura S, Nagafuchi A, Tsukita Sa, Tsukita Sh. A 220-kD undercoat-constitutive protein: its specific localization at cadherin-based cell-cell adhesion sites. J Cell Biol. 1991;115:1449–1462. [Europe PMC free article] [Abstract] [Google Scholar]
- Itoh M, Nagafuchi A, Yonemura S, Kitani-Yasuda T, Tsukita Sa, Tsukita Sh. The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J Cell Biol. 1993;121:491–502. [Europe PMC free article] [Abstract] [Google Scholar]
- Itoh M, Nagafuchi A, Moroi S, Tsukita Sh. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α-catenin and actin filaments. J Cell Biol. 1997;138:181–192. [Europe PMC free article] [Abstract] [Google Scholar]
- Jesaitis LA, Goodenough DA. Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and the Drosophiladisc-large tumor suppressor protein. J Cell Biol. 1994;124:949–961. [Europe PMC free article] [Abstract] [Google Scholar]
- Jockusch BM, Bubeck P, Giehl K, Kroemker M, Moschner J, Rothkegel M, Rudiger M, Schluter K, Stanke G, Winkler J. The molecular architecture of focal adhesions. Annu Rev Cell Dev Biol. 1995;11:379–416. [Abstract] [Google Scholar]
- Kato M, Sasaki T, Ohya T, Nakanishi H, Nishioka H, Imamura M, Takai Y. Physical and functional interaction of rabphilin-3A with α-actinin. J Biol Chem. 1996;271:31775–31778. [Abstract] [Google Scholar]
- Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+channels by interaction with a family of membrane-associated guanylate kinases. Nature (Lond) 1995;378:85–88. [Abstract] [Google Scholar]
- Kistner U, Wenzel BM, Veh RW, Cases-Langhoff C, Garner AM, Appeltauer U, Voss B, Gundelfinger ED, Garner CC. SAP90, a rat presynaptic protein related to the product of the Drosophila tumor suppressor gene dlg-A. . J Biol Chem. 1993;268:4580–4583. [Abstract] [Google Scholar]
- Knudsen KA, Soler AP, Johnson KR, Wheelock MJ. Interaction of α-actinin with the cadherin/catenin cell-cell adhesion complex via alpha-catenin. J Cell Biol. 1995;130:67–77. [Europe PMC free article] [Abstract] [Google Scholar]
- Kornau HC, Schenker LT, Kennedy MB, Sheeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science (Wash DC) 1995;269:1737–1740. [Abstract] [Google Scholar]
- Kuriyama M, Harada N, Kuroda S, Yamamoto T, Nakafuku M, Iwamatsu A, Yamamoto D, Prasad R, Croce C, Canaani E, Kaibuchi K. Identification of AF-6 and canoe as putative targets for Ras. J Biol Chem. 1996;271:607–610. [Abstract] [Google Scholar]
- Lin CH, Thompson CA, Forscher P. Cytoskeletal reorganization underlying growth cone motility. Curr Opin Neurobiol. 1994;4:640–647. [Abstract] [Google Scholar]
- Luna EJ, Hitt AL. Cytoskeleton-plasma membrane interactions. Science (Wash DC) 1992;258:955–964. [Abstract] [Google Scholar]
- Matsuda Y, Harada YN, Natsuume-Sakai S, Lee K, Shiomi T, Chapman VM. Location of the mouse complement factor H gene(cfh) by FISH analysis and replication R-banding. Cytogenet Cell Genet. 1992;61:282–285. [Abstract] [Google Scholar]
- Matsuda Y, Chapman VM. Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis. 1995;16:261–272. [Abstract] [Google Scholar]
- Meyer RK, Aebi U. Bundling of actin filaments by α-actinin depends on its molecular length. J Cell Biol. 1990;110:2013–2024. [Europe PMC free article] [Abstract] [Google Scholar]
- Miller K, Mattey D, Measures H, Hopkins C, Garrod D. Localization of the protein and glycoprotein components of bovine nasal epithelial desmosomes by immunoelectron microscopy. EMBO (Eur Mol Biol Organ) J. 1987;6:885–889. [Europe PMC free article] [Abstract] [Google Scholar]
- Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1:761–772. [Abstract] [Google Scholar]
- Miyamoto H, Nihonmatsu I, Kondo S, Ueda R, Togashi S, Hirata K, Ikegami Y, Yamamoto D. canoe encodes a novel protein containing a GLGF/DHR motif and functions with Notch and scabrous in common developmental pathways in Drosophila. . Genes Dev. 1995;9:612–625. [Abstract] [Google Scholar]
- Mizoguchi A, Yano Y, Hamaguchi H, Yanagida H, Ide C, Zahraoui A, Shirataki H, Sasaki T, Takai Y. Localization of Rabphilin-3A on the synaptic vesicle. Biochem Biophys Res Commun. 1994;202:1235–1243. [Abstract] [Google Scholar]
- Mueller H, Franke WW. Biochemical and immunological characterization of desmoplakins I and II, the major polypeptides of the desmosomal plaque. J Mol Biol. 1983;163:647–671. [Abstract] [Google Scholar]
- Nagafuchi A, Shirayoshi Y, Okazaki K, Yasuda K, Takeichi M. Transformation of cell adhesion properties by exogenously introduced E-cadherin cDNA. Nature (Lond) 1987;329:341–343. [Abstract] [Google Scholar]
- Nagafuchi A, Takeichi M, Tsukita Sh. The 102 kd cadherin-associated protein: similarity to vinculin and posttranscriptional regulation of expression. Cell. 1991;65:849–857. [Abstract] [Google Scholar]
- Nakanishi, H., H. Obaishi, A. Satoh, M. Wada, K. Mandai, K. Satoh, H. Nishioka, Y. Matsuura, A. Mizoguchi, and Y. Takai. 1997. Neurabin: a novel neural tissue-specific actin filament–binging protein involved in neurite formation. J. Cell Biol. In press. [Europe PMC free article] [Abstract]
- Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16:2157–2163. [Europe PMC free article] [Abstract] [Google Scholar]
- Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO (Eur Mol Biol Organ) J. 1989;8:1711–1717. [Europe PMC free article] [Abstract] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. [Abstract] [Google Scholar]
- Peifer, M., S. Orsulic, L.M. Pai, and J. Loureiro. 1993. A model system for cell adhesion and signal transduction in Drosophila. Development (Camb.). (Suppl.)163–176. [Abstract]
- Pestonjamasp K, Amieva MR, Strassel CP, Nauseef WM, Furthmayr H, Luna EJ. Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes. Mol Biol Cell. 1995;6:247–259. [Europe PMC free article] [Abstract] [Google Scholar]
- Pollard TD, Cooper JA. Methods to characterize actin filament networks. Methods Enzymol. 1982;85:211–233. [Abstract] [Google Scholar]
- Prasad R, Gu Y, Alder H, Nakamura T, Canaani O, Saito H, Huebner K, Gale RP, Nowell PC, Kuriyama K, et al. Cloning of the ALL-1 fusion partner, the AF-6gene, involved in acute myeloid leukemias with the t(6;11) chromosome translocation. Cancer Res. 1993;53:5624–5628. [Abstract] [Google Scholar]
- Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA. Structure of the actin-myosin complex and its implications for muscle contraction. Science (Wash DC) 1993;261:58–65. [Abstract] [Google Scholar]
- Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci USA. 1995;92:8813–8817. [Europe PMC free article] [Abstract] [Google Scholar]
- Saha V, Lillington DM, Shelling AN, Chaplin T, Yaspo ML, Ganesan TS, Young BD. Genes Chromosomes Cancer. 1995;14:220–222. [Abstract] [Google Scholar]
- Saito S, Sirahama S, Matsushima M, Suzuki M, Sagae S, Kudo R, Saito J, Noda K, Nakamura Y. Definition of a commonly deleted region in ovarian cancers to a 300-kb segment of chromosome 6q27. Cancer Res. 1996;56:5586–5589. [Abstract] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Saras J, Heldin CH. PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem Sci. 1996;21:455–458. [Abstract] [Google Scholar]
- Schröder RR, Manstein DJ, Jahn W, Holden H, Rayment I, Holmes KC, Spudich JA. Three-dimensional atomic model of F-actin decorated with Dictyosteliummyosin S1. Nature (Lond) 1993;364:171–174. [Abstract] [Google Scholar]
- Shuster CB, Herman IM. Indirect association of ezrin with F-actin: isoform specificity and calcium sensitivity. J Cell Biol. 1995;128:837–848. [Europe PMC free article] [Abstract] [Google Scholar]
- Skerrow CJ, Matoltsy AG. Isolation of epidermal desmosomes. J Cell Biol. 1974;63:515–523. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith SJ. Neuronal cytomechanics: the actin-based motility of growth cones. Science (Wash DC) 1988;242:708–715. [Abstract] [Google Scholar]
- Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;103:755–766. [Europe PMC free article] [Abstract] [Google Scholar]
- Takaishi K, Sasaki T, Kameyama T, Tsukita Sa, Tsukita Sh, Takai Y. Translocation of activated Rho from the cytoplasm to membrane ruffling area, cell-cell adhesion sites and cleavage furrows. Oncogene. 1995;11:39–48. [Abstract] [Google Scholar]
- Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development (Camb) 1988;102:639–655. [Abstract] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science (Wash DC) 1991;251:1451–1455. [Abstract] [Google Scholar]
- Tanaka E, Sabry J. Making the connection: cytoskeletal rearrangements during growth cone guidance. Cell. 1995;83:171–176. [Abstract] [Google Scholar]
- Tokuyasu KT. Immunochemistry on ultrathin frozen sections. Histochem J. 1980;12:381–403. [Abstract] [Google Scholar]
- Tokuyasu KT. Use of poly(vinylpyrrolidone) and poly(vinyl alcohol) for cryoultramicrotomy. Histochem J. 1989;21:163–171. [Abstract] [Google Scholar]
- Tsukita Sa, Yonemura S, Tsukita Sh. ERM proteins: head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem Sci. 1997;22:53–58. [Abstract] [Google Scholar]
- Tsukita Sh, Tsukita Sa. Isolation of cell-cell adherens junctions from rat liver. J Cell Biol. 1989;108:31–41. [Europe PMC free article] [Abstract] [Google Scholar]
- Tsukita Sh, Itoh M, Tsukita Sa. A new 400-kD protein from isolated adherens junctions: its localization at the undercoat of adherens junctions and at microfilament bundles such as stress fibers and circumferential bundles. J Cell Biol. 1989;109:2905–2915. [Europe PMC free article] [Abstract] [Google Scholar]
- Tsukita Sh, Tsukita Sa, Nagafuchi A, Yonemura S. Molecular linkage between cadherins and actin filaments in cell-cell adherens junctions. Curr Opin Cell Biol. 1992;4:834–839. [Abstract] [Google Scholar]
- Tsukita Sh, Itoh M, Nagafuchi A, Yonemura S, Tsukita Sa. Submembranous junctional plaque proteins include potential tumor suppressor molecules. J Cell Biol. 1993;123:1049–1053. [Europe PMC free article] [Abstract] [Google Scholar]
- Turner CE, Burridge K. Transmembrane molecular assemblies in cell-extracellular matrix interactions. Curr Opin Cell Biol. 1991;3:849–853. [Abstract] [Google Scholar]
- Willott E, Balda MS, Fanning AS, Jameson B, Van Itallie C, Anderson JM. The tight junction protein ZO-1 is homologous to the Drosophiladisc-large tumor suppressor protein of septate junctions. Proc Natl Acad Sci USA. 1993;90:7834–7838. [Europe PMC free article] [Abstract] [Google Scholar]
- Woods DF, Bryant PJ. The discs-large tumor suppressor gene of Drosophilaencodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. [Abstract] [Google Scholar]
Articles from The Journal of Cell Biology are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1083/jcb.139.2.517
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jcb/article-pdf/139/2/517/1272790/15006.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The Nectin family ligands, PVRL2 and PVR, in cancer immunology and immunotherapy.
Front Immunol, 15:1441730, 02 Aug 2024
Cited by: 0 articles | PMID: 39156900 | PMCID: PMC11327090
Review Free full text in Europe PMC
Association of connexin36 with adherens junctions at mixed synapses and distinguishing electrophysiological features of those at mossy fiber terminals in rat ventral hippocampus.
Int J Physiol Pathophysiol Pharmacol, 16(3):28-54, 15 Jun 2024
Cited by: 0 articles | PMID: 39021415 | PMCID: PMC11249852
The actin binding sites of talin have both distinct and complementary roles in cell-ECM adhesion.
PLoS Genet, 20(4):e1011224, 25 Apr 2024
Cited by: 1 article | PMID: 38662776 | PMCID: PMC11075885
C. elegans Afadin is required for epidermal morphogenesis and functionally interfaces with the cadherin-catenin complex and RhoGAP PAC-1/ARHGAP21.
Dev Biol, 511:12-25, 29 Mar 2024
Cited by: 1 article | PMID: 38556137
The Dilute domain in Canoe is not essential for linking cell junctions to the cytoskeleton but supports morphogenesis robustness.
J Cell Sci, 137(6):jcs261734, 21 Mar 2024
Cited by: 0 articles | PMID: 38323935 | PMCID: PMC11006394
Go to all (299) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (2 citations) ENA - U83230
- (2 citations) ENA - U83231
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Ponsin/SH3P12: an l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions.
J Cell Biol, 144(5):1001-1017, 01 Mar 1999
Cited by: 153 articles | PMID: 10085297 | PMCID: PMC2148189
Nexilin: a novel actin filament-binding protein localized at cell-matrix adherens junction.
J Cell Biol, 143(5):1227-1238, 01 Nov 1998
Cited by: 47 articles | PMID: 9832551 | PMCID: PMC2133087
Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein.
J Cell Biol, 145(3):539-549, 01 May 1999
Cited by: 327 articles | PMID: 10225955 | PMCID: PMC2185068
Nectin and afadin: novel organizers of intercellular junctions.
J Cell Sci, 116(pt 1):17-27, 01 Jan 2003
Cited by: 346 articles | PMID: 12456712
Review





