Abstract
Free full text

The Centrosomal Protein C-Nap1 Is Required for Cell Cycle–Regulated Centrosome Cohesion
Abstract
Duplicating centrosomes are paired during interphase, but are separated at the onset of mitosis. Although the mechanisms controlling centrosome cohesion and separation are important for centrosome function throughout the cell cycle, they remain poorly understood. Recently, we have proposed that C-Nap1, a novel centrosomal protein, is part of a structure linking parental centrioles in a cell cycle–regulated manner. To test this model, we have performed a detailed structure–function analysis on C-Nap1. We demonstrate that antibody-mediated interference with C-Nap1 function causes centrosome splitting, regardless of the cell cycle phase. Splitting occurs between parental centrioles and is not dependent on the presence of an intact microtubule or microfilament network. Centrosome splitting can also be induced by overexpression of truncated C-Nap1 mutants, but not full-length protein. Antibodies raised against different domains of C-Nap1 prove that this protein dissociates from spindle poles during mitosis, but reaccumulates at centrosomes at the end of cell division. Use of the same antibodies in immunoelectron microscopy shows that C-Nap1 is confined to the proximal end domains of centrioles, indicating that a putative linker structure must contain additional proteins. We conclude that C-Nap1 is a key component of a dynamic, cell cycle–regulated structure that mediates centriole–centriole cohesion.
Introduction
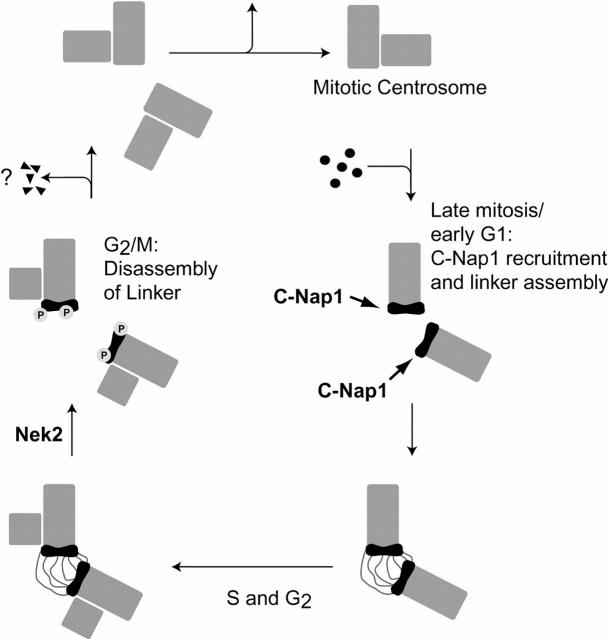
The centrosome, the major microtubule (MT)1-organizing center (MTOC) of animal cells, is composed of two centrioles (hereafter termed parental centrioles) and the surrounding pericentriolar material (PCM) (Kellogg et al. 1994; Andersen 1999). During cell cycle progression, the centrosome undergoes a series of profound structural changes, including duplication, maturation, and separation (Bornens and Moudjou 1999; Mayor et al. 1999; Urbani and Stearns 1999; Zimmerman et al. 1999). The correct execution of this centrosome duplication cycle is important for the formation of a bipolar mitotic spindle and the proper segregation of replicated chromosomes. It has long been recognized that centrosome malfunction may contribute to the development of tumors (Brinkley and Goepfert 1998; Doxsey 1998; Salisbury et al. 1999). Bipolar spindle formation requires that the centrosome duplicates only once per cell cycle. This duplication begins in a semiconservative manner during S phase, when a procentriole buds from the proximal extremity of each parental centriole (Kochanski and Borisy 1990). During G2 phase, procentrioles continue to elongate until the centrosome is composed of two pairs of centrioles. In late G2, the centrosome grows in size, due to the recruitment of several proteins, including γ-tubulin. This maturation step ultimately leads to an increase of MT nucleation activity at the onset of mitosis (Verde et al. 1990; Lane and Nigg 1996). Concomitantly, the two centrosomes separate and migrate apart to form the poles of the mitotic spindle. This separation requires the activity of MT-dependent motor proteins (Vaisberg et al. 1993; Blangy et al. 1995; Walczak et al. 1998). Upon cell division, each daughter cell inherits one centrosome comprising two centrioles. During late mitosis/early G1 phase, the two centrioles move apart and loose their typical orthogonal orientation. The physiological significance of this centriole disorientation and separation has not been definitively established, though it has been considered a prerequisite for centriole duplication (Lacey et al. 1999). Most recently, it has also been proposed to be required for the completion of cell division (Piel et al. 2000).
Recent live-imaging studies have shown that both centrioles are able to nucleate MTs, but that only the mature centriole (the one characterized by appendages) is able to anchor them; conversely, the less mature centriole displays a striking mobility (Piel et al. 2000). These elegant studies highlight the complexity and dynamics of the centrosome structure. Detailed examination of centrosomes in fixed cell preparations confirms that the distances between parental centrioles can vary considerably, not only between different cell types, but also within a given cell population. This notwithstanding, the two parental centrioles generally display a striking pairing, at least for the most part of the cell cycle. Moreover, biochemical isolation of centrosomes readily permits the recovery of paired centrioles. These observations suggest that centrioles may be linked by a cell cycle–regulated cohesive structure. In support of this view, electron microscopic examination of isolated centrosomes has revealed electron-dense material connecting the two parental centrioles (Paintrand et al. 1992). However, in other studies, centrosome cohesion has been attributed primarily to the dynamic properties of the MT cytoskeleton (Jean et al. 1999). Thus, the existence of a structure linking parental centrioles remains controversial. If parental centrioles are indeed connected by a proteinaceous linker in vivo, one would predict that this structure should be highly dynamic, and that mechanisms must exist to regulate its assembly and disassembly. Most importantly, the severing or removal of such a cohesive structure would be expected to represent a prerequisite for centrosome separation at the onset of mitosis.
We have recently characterized a centrosome-associated protein kinase, Nek2, whose overexpression in cultured cells causes the premature splitting of centrosomes (Fry et al. 1998a). Accordingly, we have proposed that centriole–centriole cohesion may be subject to regulation by phosphorylation (Fry et al. 1998a; Mayor et al. 1999). Furthermore, we have identified a 281-kD coiled coil–centrosomal protein, termed C-Nap1, that most likely constitutes a physiological substrate of Nek2 (Fry et al. 1998b). The same protein was independently identified and named Cep250 (Mack et al. 1998). Immunofluorescent staining suggested that C-Nap1 localizes to the interphase centrosome, but not to mitotic spindle poles, and immunoelectron microscopy revealed that the COOH-terminal domain of C-Nap1 colocalizes with Nek2 to the proximal ends of both parental centrioles (Fry et al. 1998b; Mayor et al. 1999). On the basis of these observations, we have put forward the hypothesis that C-Nap1 may be part of a structure connecting the two centrioles.
To test the above model, we have carried out a detailed molecular characterization of the C-Nap1 protein. Specifically, we have performed antibody microinjection experiments to directly interfere with C-Nap1 function. Moreover, we have generated a series of C-Nap1 deletion mutants and examined the consequences of overexpressing these fragments in cultured cells. Finally, we have raised antibodies against different parts of this large protein and used them to study the centrosome association of C-Nap1 by both immunofluorescence and immunoelectron microscopy. Collectively, our results support an important role for C-Nap1 in the cell cycle–dependent regulation of centrosome structure.
Materials and Methods
Microinjection
Microinjection experiments were performed, as described previously (Lane and Nigg 1996), using R63 (C-Ab), an anti–C-Nap1 antibody raised against the COOH-terminal domain (Fry et al. 1998b). This antibody was affinity purified on a histidine tagged–recombinant C-Nap1 protein and isolated using protein A–Sepharose (Amersham Pharmacia Biotech). The affinity-purified C-Ab, as well as nonspecific rabbit IgG (Sigma-Aldrich) used for control, were extensively washed with PBS and concentrated with Ultrafree-0.5 centrifugal filter (Amicon bioseparation; Millipore), to yield a concentration of 2 mg/ml for injections.
Antibody Production
To generate antibodies against two additional domains of C-Nap1, (His)6-tagged fusion proteins were constructed with the NH2 terminus (N-term, amino acid [aa] 67–345) as well as the middle region (Middle, aa 1,098–1,248), using the QIAexpress bacterial expression system (QIAGEN). The plasmid pQE10:N-term was constructed by subcloning an RsaI (bp 867)–BclI (bp 1,702) (blunted with Klenow) fragment from C-Nap1 into the pQE31 vector (QIAGEN) digested with SmaI. The plasmid pQE30:Middle was constructed by subcloning the SacI (bp 3,967)–HindIII (bp 4,410) fragment of C-Nap1 into pQE30 digested with SacI and HindIII. Protein expression was performed in the Escherichia coli strain M15(pREP4) using Luria broth (10 mM Tris-HCl, pH 7.5, 10 g Bacto tryptone, 5 g Bacto yeast extract, 5 g NaCl per liter). Recombinant proteins were expressed for 4–6 h with 1 mM isopropyl-β-d-thiogalactoside (IPTG) and purified on nickel columns under denaturing conditions, as described by the manufacturer (QIAGEN). They were then further purified by preparative gel electrophoresis on a 12% SDS-polyacrylamide gel, as described previously (Fry et al. 1998b). For immunizations, purified proteins (350 μg of the [His])6:N-term and 450 μg of the [His])6:Middle protein) were injected subcutaneously into New Zealand white rabbits (Elevage Scientifique des Dombes) on days 0, 27, 56, 84, 112, 140, and 160 (N-term) and 0, 28, 56, 84, and 112 (Middle). Immune sera were obtained on days 140, 178, and 182 (N-term) and 97, 125, and 132 (Middle). These antibodies are referred to as N-Ab and M-Ab, respectively. For affinity purification of antibodies, 1 mg of each purified (His)6:recombinant protein was coupled covalently to 400 mg cyanogen bromide–activated Sepharose 4B (Amersham Pharmacia Biotech), as recommended by the manufacturer, and antibodies were purified on this affinity matrix, as described previously (Harlow and Lane 1998). Purified antibodies were dialyzed extensively against PBS before use.
Cell Culture, Transfections, and Immunochemical Techniques
Human Hs68 fibroblasts, HeLa, U2OS osteosarcoma, and KE37 T-lymphoblastoid cells were grown at 37°C in a 7% CO2 atmosphere in DME supplemented with 10% heat-inactivated FCS and penicillin-streptomycin (100 IU/ml and 100 μg/ml, respectively). For transient transfection studies, U2OS cells were seeded onto hydrochloric acid–treated glass coverslips at a density of 105 cells per 35-mm dish. They were then transfected with 5 μg of plasmid DNA, using calcium phosphate precipitates, as previously described (Seelos 1997), and fixed 24 h later in cold methanol (−20°C) for 6 min. For microinjection, human diploid Hs68 fibroblasts were grown on gelatin- or polylysine-coated coverslips. Where indicated, Hs68 cells were treated with 1.6 μg/ml aphidicolin, 5 μg/ml nocodazole, 5 μM taxol, 3 μg/ml cytochalasin D, or 10 mM 2,3-butanedione monoxime (BDM) (all drugs were from Sigma-Aldrich).
To prepare cell extracts for immunoblotting, cells were harvested, washed once in PBS/1 mM PMSF, and resuspended in RIPA extraction buffer (50 mM Tris-HCl, pH 8, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 150 mM NaCl, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 20 mM β-glycerophosphate) to yield 2 × 104 cells/μl. Samples were left for 30 min on ice, passed 10 times through a 27G needle, and centrifuged at 14,000 rpm for 10 min at 4°C. One volume of protein sample buffer was added to the supernatant and the sample was heated to 95°C for 3 min before analysis by SDS-PAGE on a 7.5% gel. Proteins were electrophoretically transferred to nitrocellulose membranes using a Bio-Rad apparatus (Bio-Rad Laboratories), and membranes were incubated for 30 min in blocking buffer (5% low-fat dried milk in PBS/0.1% Tween-20). All antibody incubations were carried out in blocking buffer for one hour at room temperature (N-Ab and C-Ab, 0.5 μg/ml; M-Ab, 1 μg/ml), and bound IgGs were visualized using alkaline phosphatase–conjugated anti–rabbit IgG secondary antibodies (Promega).
Immunofluorescence and Immunoelectron Microscopy
Immunofluorescence microscopy was performed using a ZEISS Axioplan II microscope and 40× or 63× oil immersion objectives. Photographs were taken using a Quantix 1400 (Photometrics, Inc.) or Micromax (Princeton Instruments) CCD cameras and IP-Lab or Metaview (Universal Imaging Corp.) softwares. Primary antibodies used for immunofluorescence were affinity-purified N-Ab (0.5 μg/ml) and M-Ab (1 μg/ml), anti–γ-tubulin IgG (2 μg/ml), GTU-88 anti–γ-tubulin mAb (1:1,000; Sigma-Aldrich), 9E10 anti-myc mAb (undiluted tissue culture supernatant), and GT335 anti-polyglutamylated tubulin mAb (1:2,000). Secondary antibodies were biotinylated donkey anti–rabbit or goat anti–mouse IgG (1:200; Amersham Pharmacia Biotech), followed by Texas red–conjugated streptavidin (1:100; Amersham Pharmacia Biotech), FITC–conjugated goat anti–mouse Fab fragment (1:100; Sigma-Aldrich), FITC-conjugated goat anti–rabbit (1:100; Sigma-Aldrich), and AlexaFluor488– conjugated goat anti–mouse IgG (1:1,000; Molecular Probes).
Preembedding immunoelectron microscopy of whole U2OS cells was performed after fixation with 3% paraformaldehyde/2% sucrose and permeabilized with 0.5% Triton X-100, as previously described (Fry et al. 1998b).
Plasmid Construction
A full-length C-Nap1 construct was generated by subcloning the fragments P5.71, P4.20, and P2.5 (Fry et al. 1998b) into pBlueScript-SK and fusing them at the restriction sites SalI (bp 2,604) and NheI (bp 3,491). For fusion to the myc tag, EcoRV and PmlI restriction sites were introduced upstream of the first methionine ATG codon (bp 672) to create kC-Nap1. The NH2-terminal fragment of C-Nap1 was amplified by PCR with the high fidelity Taq polymerase (Roche) using two oligonucleotides (5′-TTGATATCACGTGCCGCCATGGAGACAAGAAGC-3′ and 5′-ATGGGCTTCCTTCTCCAGCTCC-3′). The amplified fragment was digested with EcoRV–SmaI (bp 1,116) and fused to the SmaI (bp 1,116)–NotI C-Nap1 fragment. The modified full-length kC-Nap1 was then digested by EcoRV–NotI and ligated to pBS KS+:myc, which was digested with SmaI and NotI. During this procedure, a 10 aa (MNSCPSPRAA) linker was created between the myc tag and C-Nap1. The myc–C-Nap1 insert was then subcloned in the pBK-CMV vector. T1 (aa 1–362) was obtained by cutting KC-Nap1 with EcoRV and BstXI (bp 1,758), T2 (aa 1,982–2,442) was previously described (Fry et al. 1998b), and T3 (aa 243–1,985) was generated using NaeI (bp 1,399) and XhoI (bp 6,624). All fragment were cloned downstream of a myc tag and inserted into pCMV plasmids.
Miscellaneous Techniques
Cold treatment of asynchronously growing U2OS cells was performed by placing tissue culture dishes for 30 min on ice. To induce MT regrowth, prewarmed medium (37°C) was added and the dishes were placed for 60 s at 37°C. Then, cells were fixed with cold methanol. For FACS® analysis, Hs68 fibroblasts were detached by trypsinization and washed with ice-cold PBS/0.3 mM EDTA. They were fixed in 70% ethanol, and then incubated for 30 min in PBS, 20 μg/ml RNase A, and 5 μg/ml propidium iodide. Centrosome purifications from KE37 cells were performed as described previously (Fry et al. 1998a).
Results
Microinjection of C-Nap1 Antibody Induces Centrosome Splitting
To explore a possible involvement of C-Nap1 in a cohesive structure linking centrioles, a highly specific, affinity-purified anti–C-Nap1 antibody (C-Ab) was microinjected into nonimmortalized human fibroblasts (Hs68 cells). This antibody, hereafter referred to as C-Ab, was raised against the COOH-terminal domain of C-Nap1 (aa 1,986–2,442), and has been described previously (R63; Fry et al. 1998b). As shown by Western blotting, C-Ab recognized a single band of the expected molecular mass in Hs68 whole-cell extracts, while the corresponding preimmune serum showed no reactivity (Fig. 1 D). Asynchronously growing Hs68 cells were injected with either C-Ab or nonspecific rabbit IgG for control, fixed with methanol 16 h later, and stained with a mAb anti–γ-tubulin (Fig. 1 A, a′ and b′). To identify injected cells, cultures were counterstained with anti-IgG antibodies (Fig. 1 A, a and b). Whereas the vast majority of the uninjected or control-injected cells showed the expected staining of centrosomes as closely spaced doublets (Fig. 1 A, a′), most cells injected with C-Ab displayed two widely separated γ-tubulin–positive dots (Fig. 1 A, b′). To confirm that these dots contained centrioles (as opposed to merely constituting fragments of PCM), identical experiments were performed using GT335, a mAb that reacts with polyglutamylated tubulin and constitutes a convenient reagent for staining centrioles (Wolff et al. 1992; Bobinnec et al. 1998). As shown in Fig. 1 C (a′ and b′), the split centrosomes clearly contained GT335-positive centrioles. Quantitative analyses revealed that >70% of the cells injected with anti–C-Nap1 antibodies displayed split centrosomes, whereas only 7% was observed in control-injected cells (Fig. 1 B). These results show that antibody-mediated interference with C-Nap1 function causes a disruption of cohesion between centrioles, supporting the hypothesis that C-Nap1 is part of a structure that links centrioles together.
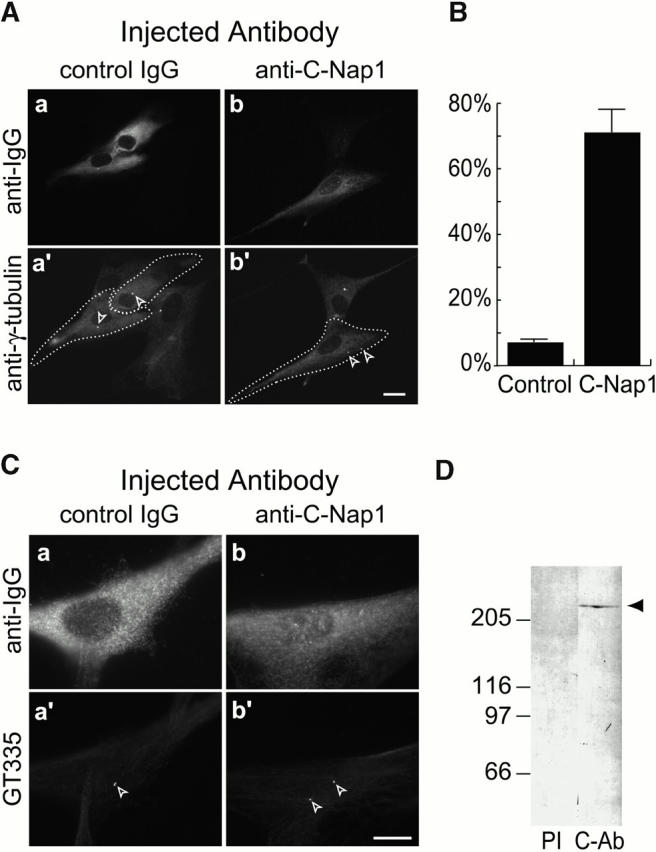
C-Nap1 antibody injection causes centrosome splitting in Hs68 cells. (A) Representative examples of Hs68 cells injected with control IgG (a and a′) or C-Ab (b and b′). Injected cells were visualized using an anti–rabbit IgG secondary antibody (a and b), and the centrosomes were stained for γ-tubulin (a′ and b′). For illustrative purposes, injected cells were surrounded by a dotted line and arrowheads in b′ point to a typical split centrosome. Bar, 10 μm. (B) Asynchronously growing Hs68 cells were injected with control or anti–C-Nap1 antibodies and analyzed 16 h later by immunofluorescence microscopy, as described above. The histogram indicates the percent of cells with split centrosomes. A total of 167 cells were injected with C-Ab and 158 cells with control IgG. Cells were scored as having split centrosomes whenever the distance between the two γ-tubulin dots exceeded 2 microns, i.e., two times the diameter of these dots. Results were averaged from two independent experiments. (C) Representative examples of asynchronously growing Hs68 cells injected with control IgG (a and a′) or C-Ab (b and b′). Injected cells were visualized using an anti–rabbit IgG secondary antibody (a and b), and the centrioles were stained with GT335, a mAb specific for polyglutamylated tubulin (a′ and b′). Bar, 10 μm. (D) Total protein from Hs68 cells was separated by SDS-PAGE and probed by immunoblotting with preimmune serum (PI) or affinity purified antibody against the COOH-terminal domain of C-Nap1 (C-Ab). The positions of molecular weight markers are indicated (in kD); the arrowhead marks C-Nap1.
Cell Cycle Analysis of Centrosome Splitting
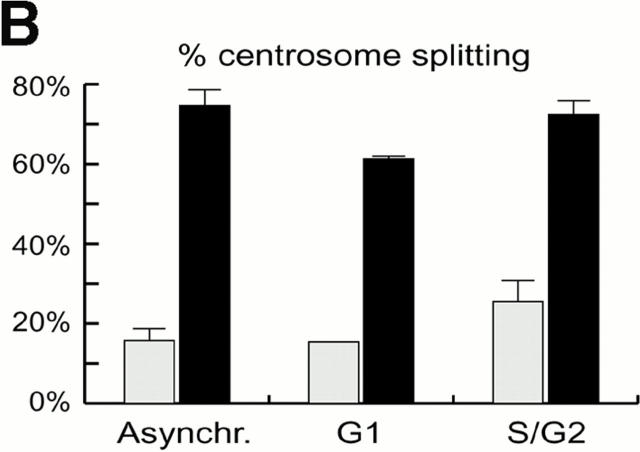
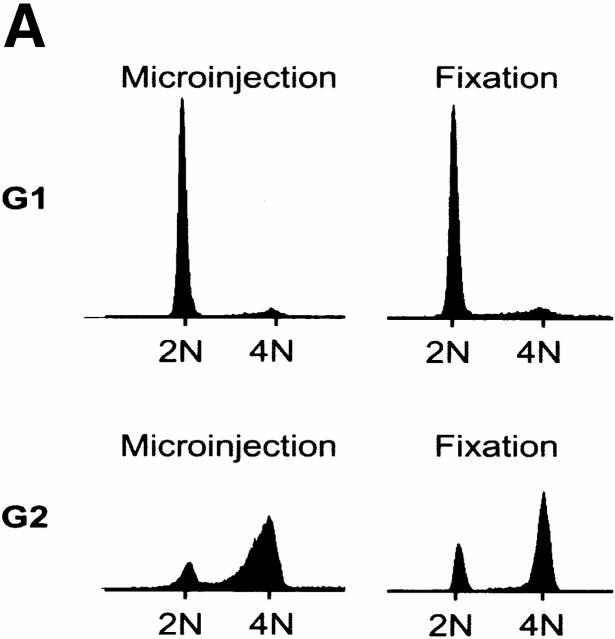
To determine whether centrosomes were susceptible to splitting at different cell cycle stages, we microinjected anti–C-Nap1 antibodies into cells that had been synchronized in either G1 or G2 phase. For injection of G1 phase cells, Hs68 fibroblasts were arrested in G0 by serum deprivation, and then they were released into the cell cycle by the addition of 20% FCS. For synchronization in G2, cells were released from an aphidicolin-induced block at the G1/S boundary. Synchronized cells, as well as asynchronous populations for control, were injected with either control IgG or anti–C-Nap1 antibodies, and the extent of centrosome splitting was determined by immunofluorescent staining with anti–γ-tubulin antibodies or GT335, as described above. Cultures were subjected to flow cytometric analysis of cell cycle profiles, in parallel. As shown in Fig. 2 A, the G1 population of cells showed a 2N DNA content both at the time of injection and at the time of fixation. The G2 population showed predominantly a 4N DNA content at the time of injection, though some cells had progressed through mitosis by the time of fixation 4 h later. Compared with results for asynchronous cell populations, anti–C-Nap1 antibodies caused centrosome splitting to similar extents, regardless of whether cells were in G1 or in G2 (Fig. 2 B). Furthermore, in both cases, the distances between split centrosomes showed a very broad distribution, with an average distance of about 10 μm (with a range of 2 to >20 μm). This indicates that interference with C-Nap1 function induces the separation of parental centrioles, whether or not they are associated with growing procentrioles.
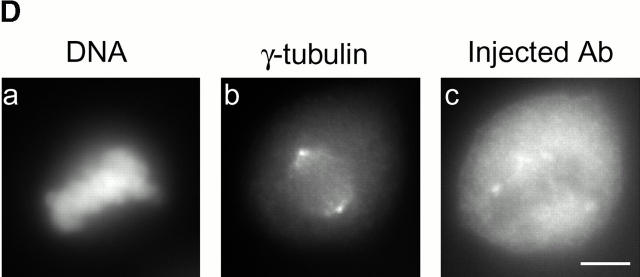
Cell cycle analysis of centrosome splitting. (A) FACS® analysis of synchronized Hs68 cells. For cell cycle synchronization, Hs68 cells were serum starved for 36 h and then released into medium containing 20% FCS and 1.6 μg/ml aphidicolin. G1 cells were microinjected 16 h after serum addition. To microinject a G2 population, cells were released for 5 h from a 36 h block in aphidicolin. Aliquots of cells were analyzed by FACS® before microinjection as well as before fixation 4 h after injection. (B) Centrosome splitting occurs regardless of cell cycle position. Hs68 cells were injected with anti–C-Nap1 antibodies at the indicated cell cycle stages and analyzed 4 h later by immunofluorescence microscopy, as described in the legend to Fig. 1 B. The histogram shows the percent of cells with split centrosomes. For C-Ab, a total of 252, 371, and 309 injected cells were counted for asynchronous, G1, and G2 populations, respectively. Similarly, 150, 318, and 366 cells were counted after injection with control IgG. Rare cells showing obvious chromosome condensation were excluded from the analysis. Results were averaged from two independent experiments. (C) Centrosome splitting does not prevent cell division. Single scattered Hs68 cells were injected with control (n = 50) or anti–C-Nap1 antibodies (n = 58) and analyzed 30 h later by immunofluorescence microscopy. Cell doubling was assessed by counting cell numbers in clusters originating from single injected cells. The histograms indicate the percent of clusters containing 1, 2, 3, or 4 cells. B and C: Black bars refer to cells injected with C-Ab, gray bars to control-injected cells. (D) Double immunofluorescent staining of a representative mitotic cell, injected with anti–C-Nap1 antibody in the preceding interphase. a, DNA; b, γ-tubulin; c, the injected anti–C-Nap1 antibody. Bar, 5 μm.
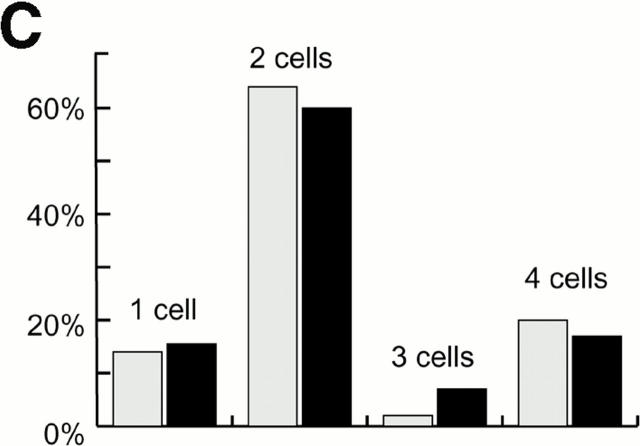
It was of interest to determine whether anti–C-Nap1 antibody–induced centrosome splitting adversely affected cell division. To address this question, single cells were injected with C-Ab or control IgG and their progeny were analyzed 30 h later (Lane and Nigg 1996). This assay revealed no significant difference between C-Ab–injected cells and control-injected cells in their ability to divide: in both cases, ~60% of the cells had gone through one division at the time of analysis, and ~20% had even completed a second division (Fig. 2 C). Furthermore, we carefully examined mitotic cells arising after the injection of C-Nap1 antibodies during the preceding interphase and found that such cells had assembled normal-looking bipolar spindles (Fig. 2 D). We conclude from these results that cells harboring split centrosomes are not prevented from undergoing cell division. Although this conclusion may appear surprising at first, it is in complete agreement with previous findings (Fry et al. 1998a).
Overexpression of Wild-type C-Nap1 and Deletion Mutants
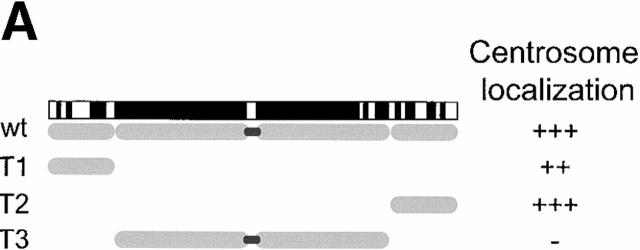
To provide further evidence for a role of C-Nap1 in centrosome cohesion, independent of antibody microinjection, we asked whether particular regions of C-Nap1 might cause centrosome splitting by acting as dominant-negative mutants. To this end, three C-Nap1 deletion mutants were designed on the basis of the C-Nap1 sequence, which predicts the existence of small NH2- and COOH-terminal end domains, separated by a large central coiled-coil structure. The mutants T1 and T2 were thus generated to represent the NH2- and COOH-terminal end domains, respectively, whereas T3 was made to span the central, putative coiled-coil region (Fig. 3 A). Full-length C-Nap1 and the three C-Nap1 deletion mutants were then expressed in U2OS cells, under the control of a cytomegalovirus (CMV) promoter. The localization of the ectopically expressed proteins was determined by immunofluorescence microscopy, taking advantage of an NH2-terminal–myc tag. In parallel, the distribution and morphology of centrosomes was examined by staining with antibodies against γ-tubulin, and the extent of centrosome splitting was scored as described above.
Mutational domain analysis of C-Nap1. (A) Schematic view of C-Nap1 domain structure and different C-Nap1 deletion mutant constructs. The dark gray boxes designate the predicted coiled-coil domains in C-Nap1. A semiquantitative assessment of the ability of C-Nap1 mutants to localize to centrosomes is given as follows: −, no centrosomal signal; ++, the ability to localize to centrosomes in ~2/3 of transfected cells; +++, centrosome association in all transfected cells. (B) Illustration of the phenotypes observed after overexpression of three different C-Nap1 deletion mutants: T1 (a and a′), T2 (b and b′), and T3 (c and c′). U2OS cells were analyzed by double immunofluorescence microscopy, by using anti-myc (9E10) mAb to visualize C-Nap1 deletion mutants (a–c) and anti–γ-tubulin (a′–c′). Arrowheads mark the positions of centrosomes. Bar, 10 μm. (C) Overexpression of C-Nap1 deletion mutants induces centrosome splitting. The histogram indicates the percent of transfected cells with split centrosomes. Cells transfected with the full-length C-Nap1 (wt) as well as nontransfected cells (−) are shown for control, and the extent of splitting observed with the protein kinases Nek2 and Nek3 is shown for comparison. A myc tag was present in all constructs. Errors bars indicate standard deviations for data collected from three independent experiments.
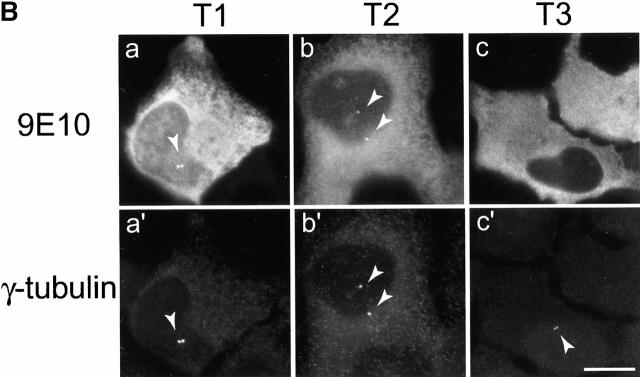
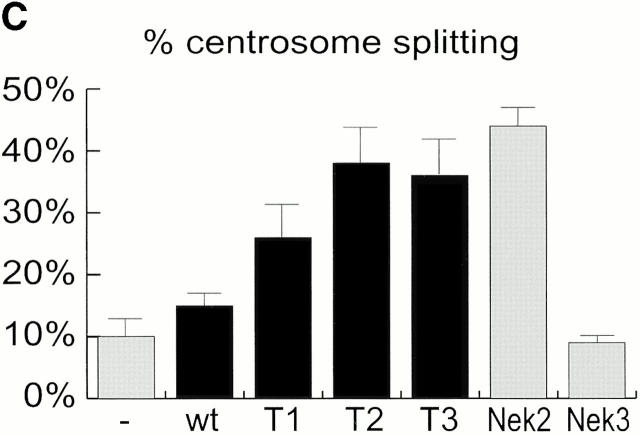
In cells expressing low levels of exogenous full-length C-Nap1, the protein localized correctly, and almost exclusively, to the centrosome, but in cells expressing higher protein levels, one or several compact C-Nap1–positive aggregates became visible (data not shown). This probably reflects a propensity of C-Nap1 to oligomerize. However, under no circumstances did overexpression of full-length C-Nap1 cause any significant centrosome splitting (Fig. 3 C). In contrast, all three C-Nap1 deletion mutants caused substantial centrosome splitting, albeit to a different extent. Although the mutants T2 and T3 were almost as effective in causing centrosome splitting as the active-Nek2 kinase, T1 was only half as effective. Nek3, a Nek2-related kinase analyzed for a control, did not induce any splitting above background. It is noteworthy that the ability of C-Nap1 mutants to cause centrosome splitting was not directly related to their ability to localize to centrosomes (Fig. 3a and Fig. c). Whereas T1 and T2 were efficiently targeted to centrosomes, T3 was almost completely cytoplasmic. Yet, T3 was able to cause a substantial amount of splitting similar to T2. This suggests that C-Nap1 truncation mutants most likely cause centrosome splitting by sequestering one or more centrosomal components in the cytoplasm, thereby preventing them from performing a cohesive function. One protein likely to be titrated by C-Nap1–deletion mutants is C-Nap1 itself. In support of this view, we have observed that endogenous C-Nap1 was displaced from centrosomes when these were split by overexpression of C-Nap1 mutants (data not shown). Experiments aimed at identifying other C-Nap1–interacting proteins are currently in progress.
Centrosome Splitting and Cytoskeletal Dynamics
The mechanisms underlying centrosome cohesion remain largely unknown. On the one hand, it is plausible that centrioles may be held together by a proteinaceous but flexible and dynamic structure. This hypothesis is supported by both electron microscopic evidence (Bornens et al. 1987; Paintrand et al. 1992) and recent live-imaging studies (Piel et al. 2000). On the other hand, it has been argued that centrioles may localize in close proximity primarily as a result of MT dynamics (Jean et al. 1999). To explore a possible critical contribution of the cytoskeleton to the phenomena described here, we asked whether anti–C-Nap1 antibodies were able to induce centrosome splitting in cells in which cytoskeletal dynamics had been disturbed by drugs. MT dynamics were profoundly altered by treatment with either nocodazole or taxol, whereas the functions of the microfilament (MF) system were impaired by treatments with either cytochalasin D or BDM, an inhibitor of myosin ATPase.
We found that depolymerization of MTs by nocodazole roughly doubled the proportion of control-injected cells with split centrosomes, whereas treatment of cells with the MT-stabilizing drug taxol did not significantly alter this proportion (Fig. 4 A, gray bars). These results are in agreement with the observation that MT dynamics can influence the positioning of centrioles within cells (Jean et al. 1999). Most importantly, however, microinjection of anti–C-Nap1 antibodies still induced centrosome splitting even in nocodazole- or taxol-treated cells (Fig. 4 A, black bars). Similarly, although centrosome splitting in control-injected cells was slightly affected by drugs acting on the MF system (Fig. 4 B, gray bars), neither cytochalasin D nor BDM prevented the splitting induced by anti–C-Nap1 antibodies (Fig. 4 B, black bars). As we observed in all our experiments, the distributions of distances between split centrioles were very broad (with a range of 2 to ≥20 μm), with no clear peaks. Quantitative analyses of control cells and drug-treated cells did not allow us to uncover statistically significant differences, indicating that the drug treatments described above exerted at most a minor influence on the average distances between split centrioles (data not shown). Thus, we conclude that anti–C-Nap1 antibody–induced centrosome splitting does not require an intact MT or MF system.
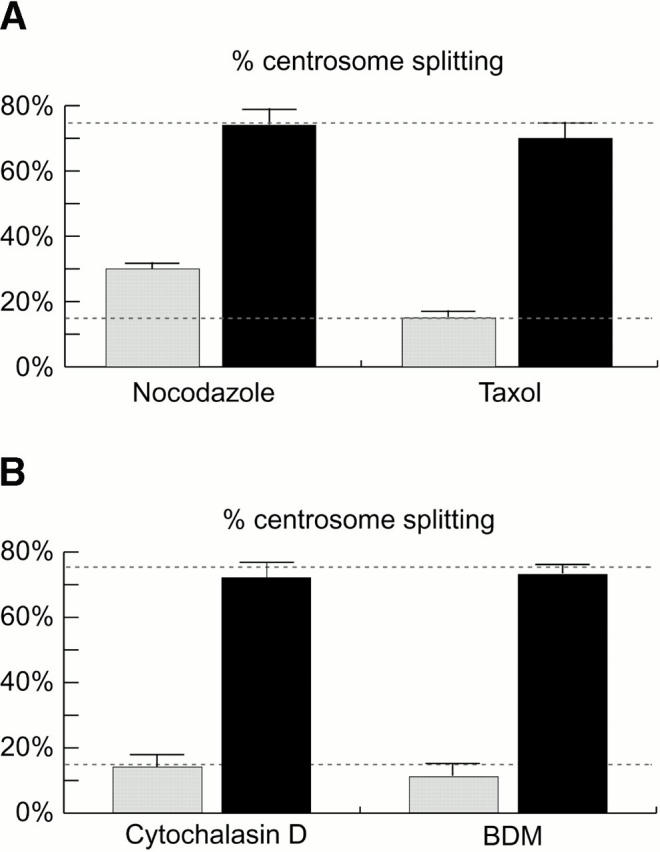
Anti–C-Nap1 antibody-induced centrosome splitting does not require an intact cytoskeleton. (A) Centrosome splitting caused by C-Ab injection is independent of MTs. The histogram indicates the percent of antibody-injected cells with split centrosomes when treated with nocodazole (5 μg/ml) and taxol (5 μM). Drug treatment was initiated 1–1.5 h before antibody injection and the cells were maintained throughout the injection and the following 4 h of incubation before fixation. In two independent experiments, 213 and 259 cells injected with C-Ab were counted (black bars), along with 183 and 217 cells injected with control IgG (gray bars). (B) Centrosome splitting is independent of the MF network. The histogram indicates the percent of injected cells with split centrosomes after treatment with either cytochalasin D (3 μg/ml) or BDM (10 mM). To avoid cell rounding, BDM was applied only 30 min before microinjection and cytochalasin D was applied right after injection. Cells were incubated for 4 h in the presence of drugs before they were fixed and analyzed by immunofluorescence microscopy. In two independent experiments, 150 and 211 cells injected with C-Ab were counted (black bars), along with 196 and 212 cells injected with control IgG (gray bars). (A and B) The percent of centrosome splitting observed in injected cells in the absence of drugs are indicated by dotted lines (lower line for control IgG and upper line for C-Nap1 antibodies).
We also asked whether centrosome splitting would interfere with the ability of separated centrioles to nucleate MTs. To this end, MT depolymerization and regrowth assays were performed on anti–C-Nap1–injected cells. We observed many cells harboring spit centrosomes in which both separated centrioles were at the centers of MT asters (data not shown). This indicates that centrosome splitting does not prevent MT nucleation, in line with the fact that both separated centrioles are readily stained with anti–γ-tubulin antibodies, implying that they are associated with PCM. Also, these results are in agreement with data showing that both parental centrioles are competent to nucleate MTs through their associated PCM, although they differ in their ability to retain them (Jean et al. 1999; Piel et al. 2000).
The data described so far show that C-Nap1 plays a critical role in mediating centrosome cohesion throughout interphase of the cell cycle. This conclusion is based on both antibody-mediated interference with C-Nap1 function and the analysis of dominant-negative–acting C-Nap1 mutants. Furthermore, we have shown that interference with C-Nap1 function causes centrosome splitting by a mechanism that is largely independent of an intact MT or MF network, and that centrosome splitting does not encumber cell division.
Reversible Dissociation of C-Nap1 from Mitotic Spindle Poles
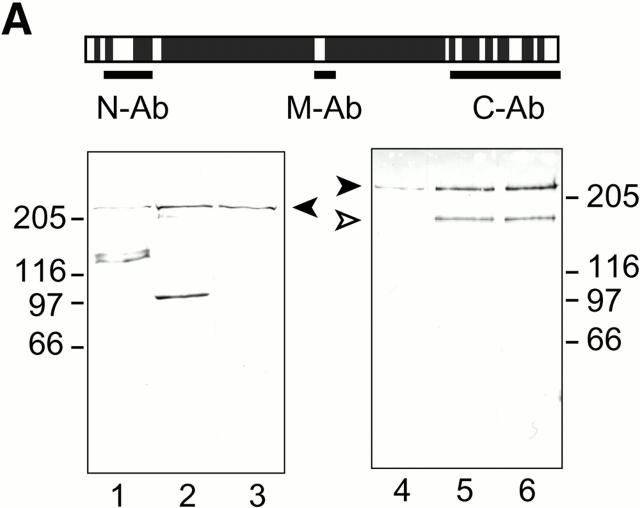
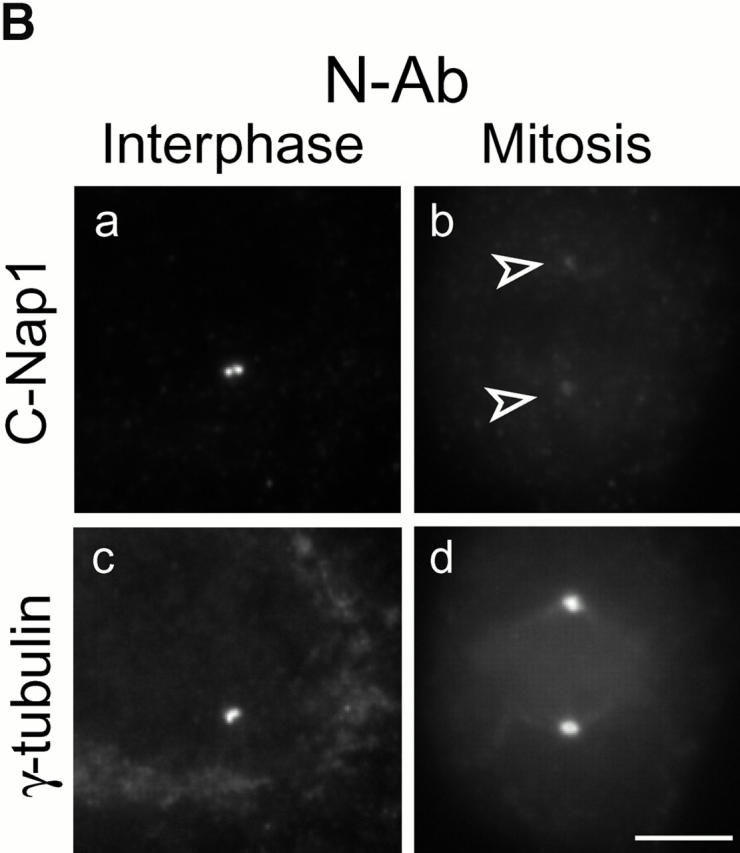
As an important consequence of the proposed role of C-Nap1 in centrosome cohesion, one would predict that a C-Nap1–containing cohesive structure should be transiently dismantled (or functionally inactivated) during mitosis when parental centrioles separate from each other to form the spindle poles. In line with this prediction, we have observed previously that C-Nap1 staining appeared to fade away upon formation of mitotic spindle poles (Fry et al. 1998b). However, since our former data were obtained using a single antibody reagent directed against the COOH terminus of C-Nap1, C-Ab, it was impossible to exclude the caveat of epitope masking. To rigorously test whether C-Nap1 dissociates from centrosomes during mitosis, we raised two additional antibodies, one directed against the NH2-terminal end domain, the other against the middle domain (the putative “hinge” region; Fig. 5 A). These reagents are referred to as N-Ab and M-Ab, respectively. As shown by Western blotting, both N-Ab and M-Ab readily recognized a protein of the expected mobility in U2OS total-cell extracts (Fig. 5 A). M-Ab was about as reactive as C-Ab, whereas N-Ab was slightly less reactive. In addition, both N-Ab and M-Ab recognized a few smaller proteins. The same antibodies were also used for Western blots on centrosomes purified from KE37 lymphoid cells (Fig. 5 A). As reported previously, KE37 cells exhibit two forms of C-Nap1 (Fry et al. 1998b). Of these, the slower migrating form was recognized by all three antibodies, but the faster migrating one was only recognized by M-Ab and C-Ab, indicating that it lacks the NH2 terminus (Fig. 5 A). More importantly, we may also conclude that the additional bands seen in U2OS whole-cell lysates represent noncentrosomal, cross-reactive proteins, as they were not present in purified centrosomes. When tested by immunofluorescence microscopy on methanol-fixed U2OS cells, both new antibodies readily labeled the centrosome, as shown by costaining for γ-tubulin (Fig. 5 B, and data not shown). Furthermore, both N-Ab and M-Ab showed staining of two distinct dots, as reported previously for C-Ab. This suggests that C-Nap1 is unlikely to span the entire distance between the two centrioles, a conclusion strengthened further by immunoelectron microscopy (see below). Most importantly, both N-Ab and M-Ab produced at most very faint staining of mitotic spindle poles (Fig. 5 B, and data not shown), confirming and extending the result obtained previously with C-Ab. These data are important in that they strongly argue against epitope masking, and instead indicate that the bulk of C-Nap1 truly dissociates from centrosomes during mitosis.
Cell cycle–dependent association of C-Nap1 with centrosomes. (A) Total protein was prepared from exponentially growing U2OS cells (lanes 1–3), and centrosomes purified from KE37 cells (lanes 4–6), separated by SDS-PAGE, and probed by immunoblotting with N-Ab (lanes 1 and 4), M-Ab (lanes 2 and 5), or C-Ab (lanes 3 and 6). The positions of molecular weight markers are indicated (in kD). The C-Nap1 fragments used for immunization to produce N-Ab, M-Ab, and C-Ab are illustrated schematically on top. (B) Double-indirect immunofluorescent staining of U2OS cells. Centrosomes were labeled with antibodies against γ-tubulin (c and d), and C-Nap1 with N-Ab (a and b). (a and c) Interphasic cell. (b and d) Mitotic cell. Bar, 5 μm. (C) C-Nap1 reassociates with the centrosome upon exit from mitosis. U2OS cells were stained as indicated. (a, c, and e) Telophase cell. (b, d, and f) Early G1 cell. DNA was stained with Hoechst dye 33258, showing condensed (a) and decondensed chromatin (b). Midbody (c) and postmitotic bridge (d) were stained with antibodies against α-tubulin; C-Nap1 was stained with N-Ab (e and f). Bar, 10 μm. (D) Double-indirect immunofluorescent staining of a dividing HeLa cell. The shape of the dividing cell is shown by differential interference contrast microscopy (a), and chromatin is visualized by staining with Hoechst dye 33258 (b). Centrioles were labeled with γ-tubulin (c and d) and C-Nap1 with N-Ab (e and f). Note that the pictures shown in c and e and d and f were taken in two different focal plains to allow visualization of both centrioles. In this particular cell, the two parental centrioles are separated over a very large distance. Bar, 10 μm.
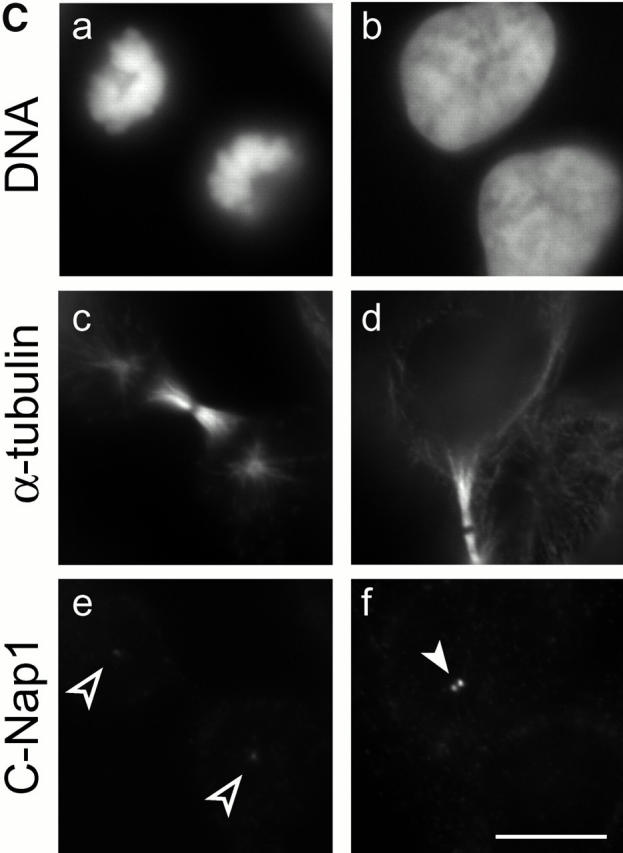
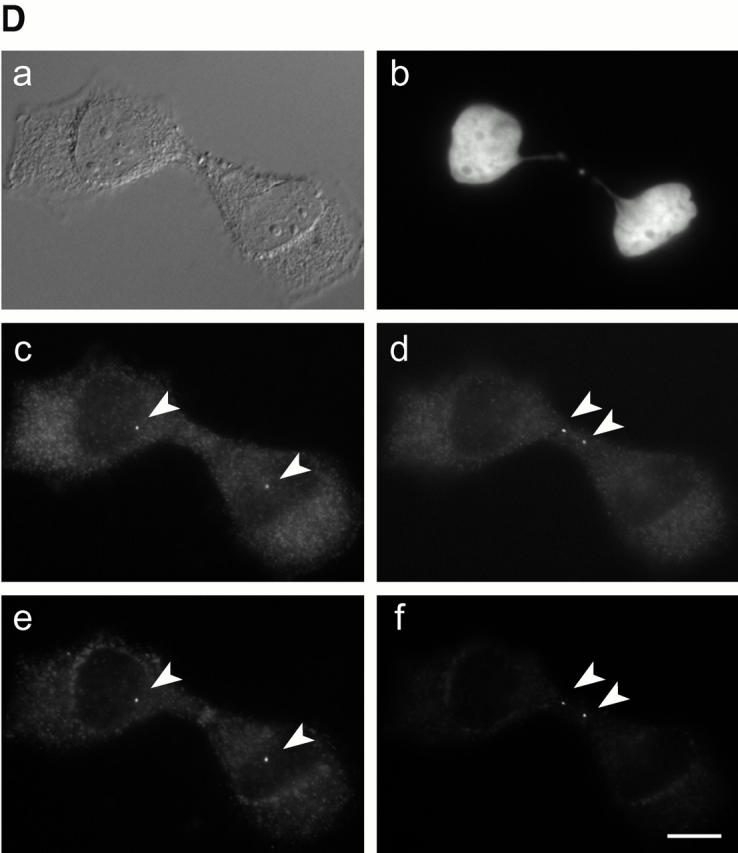
We also asked when exactly C-Nap1 reaccumulates at centrosomes and found that this occurs at the very end of cell division (Fig. 5 C). Up to telophase, when chromatin was still condensed (Fig. 5 C, a), only traces of C-Nap1 could be seen on spindle poles (Fig. 5 C, e). However, C-Nap1 staining then increased progressively and approached interphase levels already in very early G1 cells (Fig. 5 C, f and d; in d, note the postmitotic bridge still connecting these cells). Of particular interest, recent live imaging studies have revealed that parental centrioles frequently separate over very large distances in dividing cells (Piel et al. 2000). Fig. 5 D shows an example in a dividing HeLa cell. Clearly, under these conditions, C-Nap1 is associated with both parental centrioles, that is, with the one located in the cell body (d and f), and the one located close to the future cleavage site (c and e).
Immunoelectron Microscopic Localization of C-Nap1 Domains
Using C-Ab, we have previously localized the COOH-terminal end domain of C-Nap1 to the proximal ends of both parental centrioles (Fry et al. 1998b). However, in the absence of antibodies directed against other parts of the molecule, we had no previous information on how the remainder of the protein was distributed within the centrosome. To further study the exact disposition of C-Nap1, we have therefore used the two new reagents, N-Ab and M-Ab, for immunoelectron microscopy. Specifically, we sought to determine, first, whether C-Nap1 spans the entire distance between parental centrioles, and second, whether C-Nap1 might also be located at the contact site between parental centrioles and nascent procentrioles. With regard to the first question, we found that both N-Ab and M-Ab stained the proximal tips of centrioles (Fig. 6), similar to results obtained with C-Ab (Fry et al. 1998b). However, immunogold particles were slightly more distant from the centriole tips in the case of N-Ab (Fig. 6 a). Preimmune IgG produced no specific labeling and mitotic centrioles showed negligible staining (data not shown), in agreement with the immunofluorescence data shown above. With regard to the second question, we were able to exploit occasional favorable sections to examine the distribution of C-Nap1 with regard to protruding procentrioles (Fig. 6 b, the procentriole is marked by thin double arrows). In no case could any C-Nap1 staining be seen in the contact area between procentrioles and centrioles (arrow). This confirms our postulate that C-Nap1 is part of a cohesive structure associated with parental centrioles, but is unlikely to mediate the interaction between centrioles and procentrioles.
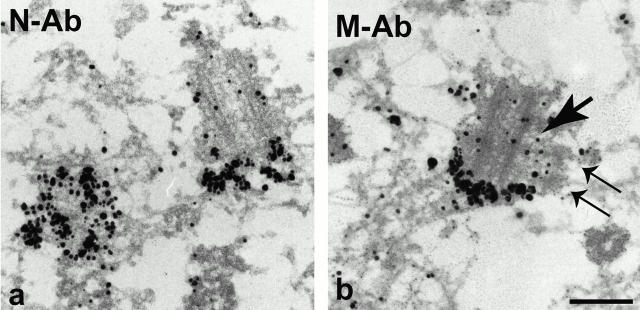
Immunoelectron microscopic localization of C-Nap1 domains. U2OS cells were fixed with 3% paraformaldehyde/2% sucrose and subjected to immunoelectron microscopic analysis with silver-enhanced Nanogold®. Both N-Ab (a) and M-Ab (b) strongly labeled the proximal ends of both centrioles. Note that no C-Nap1 staining could be seen at the interface (b, arrow) between procentriole (b, thin double arrows) and centriole. Bar, 250 nm.
Taken together, our immunocytochemical analyses at both the light- and electron-microscopic levels indicate that the entire C-Nap1 protein is located close to the tips of both parental centrioles. However, although the NH2 terminus could be detected at a greater distance from the tips than the COOH terminus, no C-Nap1 protein could be seen spanning the entire region between the two centrioles. Thus, we conclude that any cohesive structure extending from one centriole to the other is almost certain to contain additional, as yet unidentified proteins (Fig. 7). Furthermore, the association between parental centriole and procentriole is unlikely to be mediated by C-Nap1, and finally, C-Nap1 is almost completely absent from mitotic spindle poles.
Discussion
The centrosomal protein C-Nap1 was originally identified by virtue of its ability to interact with a centrosome-associated protein kinase, Nek2 (Fry et al. 1998b; Mack et al. 1998). We have now performed a detailed analysis of C-Nap1 structure, function, and distribution. Our results support the hypothesis that C-Nap1 functions as part of a dynamic, cell cycle–regulated structure linking parental centrioles to each other. In the first part of this study, we showed that microinjection of highly specific anti–C-Nap1 antibodies caused extensive splitting of centrosomes, and that a similar phenotype can be induced by overexpression of truncated C-Nap1 mutants, but not full-length protein. Centrosome splitting, as induced by interference with C-Nap1 function, was largely independent of the MT and MF networks. In the second part of our study, we used antibodies directed against three distinct domains of C-Nap1 to show that this protein is displaced from mitotic spindle poles, consistent with the prediction that a structure connecting parental centrioles should be dismantled at the onset of mitosis. The same antibodies were also used to show, by immunoelectron microscopy, that C-Nap1 localizes exclusively to the tip of each parental centriole. Importantly, C-Nap1 does not span the entire distance between parental centrioles and cannot be found at the contact site between parental centriole and growing procentriole. This implies that centrosome cohesion during interphase of the cell cycle requires additional, as yet unidentified, proteins.
Split Centrosomes Are Functional
Our data indicate that interference with C-Nap1 function severely affects centrosome structure. Although split centrosomes are commonly seen in a small proportion of untreated cells, the frequency of cells with split centrosomes increases drastically when C-Nap1 function is disrupted. Moreover, this splitting occurs even when the MT or MF networks are disturbed. This argues that, in the absence of a connective structure, split parental centrioles can drift apart passively. Splitting occurs between parental centrioles and each centriole remains associated with the surrounding PCM. Thus, cells harboring split centrosomes are able to nucleate MTs. Furthermore, it is remarkable that centrosome splitting does not interfere with the ability of antibody-injected cells to divide. Spindles formed in the presence of anti–C-Nap1 antibodies appeared normal when stained with anti–γ-tubulin antibodies, suggesting the presence of functional poles. However, live-imaging studies and quantitative analyses of chromosome segregation would be required to determine whether such spindles are fully functional in the error-free segregation of chromosomes (for further discussion see Fry et al. 1998a).
The Centrosome Association of C-Nap1 Is Cell Cycle Regulated
From the proposed function of C-Nap1, we predicted that this protein should be displaced from centrosomes, or functionally inactivated, when parental centrioles are separated at the onset of mitosis. Our earlier studies had indeed suggested that C-Nap1 levels may be diminished at mitotic spindle poles, but it was impossible to exclude epitope masking as an alternative explanation (Fry et al. 1998b). Furthermore, though C-Nap1 protein levels in total cell lysates are detectably diminished during mitosis (Mayor, T., and E.A. Nigg, unpublished results), these results are difficult to interpret in view of a large cytoplasmic C-Nap1 pool. Thus, the use of antibodies directed against distinct parts of C-Nap1 proved critical for examining the fate of C-Nap1 during the cell cycle. We found that antibodies directed against widely separated C-Nap1 domains all produced negligible staining of mitotic spindle poles, strongly arguing against the caveat of epitope masking, and confirming that C-Nap1 truly dissociates from centrosomes during mitosis.
C-Nap1 Shows a Restricted Localization within the Centrosome
Using antibodies raised against the NH2 terminus, COOH terminus, and middle domain of C-Nap1 for immunoelectron microscopy, we found that all three domains are located close to the proximal tips of centrioles. Although the NH2-terminal end domain was slightly more distant from the centriole than either the middle or the COOH-terminal domain, we obtained no evidence to indicate that C-Nap1 oligomers could span the entire distance between the two parental centrioles. We conclude, therefore, that additional proteins must participate in a centriole–centriole linkage. The identity of these proteins is presently unknown, although δ-tubulin (Chang and Stearns 2000), as well as a putative calcium-binding protein (Moudjou et al. 1991), represent candidates. Of equal importance, careful examination of centrioles in the process of duplication revealed a conspicuous absence of C-Nap1 from the contact area between parental centrioles and procentrioles. This strongly suggests that the structure connecting a procentriole to its parental centriole is of a different nature than the one proposed to exist between the two parental centrioles.
Models for Centrosome Cohesion
Transient splitting of centrosomes is a physiological event that has been observed under a variety of conditions, including treatment of cells with mitogenic growth factors (Sherline and Mascardo 1982) and chemotactic stimulation (Schliwa et al. 1982). However, what mechanisms control centrosome cohesion remain largely unknown. Two distinct models have been put forward to explain the close proximity of centrioles observed under most circumstances. On the one hand, it has been proposed that parental centrioles are connected by a proteinaceous structure (Bornens et al. 1987; Paintrand et al. 1992). If sufficiently slender and flexible, such a structure would appear compatible with the strikingly dynamic behavior described recently for centrioles in living cells (Piel et al. 2000). On the other hand, the close proximity between parental centrioles has been proposed to result predominantly from the intrinsic dynamics of the MT network (Jean et al. 1999). Clearly, our present data are relevant to these models. The centrosome splitting observed in response to both C-Nap1 antibody injection and the overexpression of dominant-negative–C-Nap1 mutants strongly argues that centrosome cohesion depends on a proteinaceous structure that is part of the centrosome proper. Furthermore, centrosome splitting can be induced also in cells in which MTs have been either depolymerized by nocodazole or stabilized by taxol, and similar results were obtained after disturbing the MF network. At first glance, these findings may appear difficult to reconcile with proposals that attribute a major role to the cytoskeleton in determining the close proximity of parental centrioles (Jean et al. 1999). However, this conflict is more apparent than real. In fact, MT dynamics (particularly minus-end–directed motility) is likely to be important for determining the local concentrations of both structural and regulatory proteins at the centrosome. Considering that a proteinaceous linker between parental centrioles is predicted to be a dynamic structure, any change in the local concentration of either linker subunits or regulators of linker subunit assembly (such as kinases or phosphases) would be expected to profoundly affect centrosome integrity. Thus, there is no reason to doubt that centrosome structure is indeed intimately linked to cytoskeletal dynamics. Most importantly, though we have shown here that anti–C-Nap1 antibody-induced centrosome splitting does not require an intact cytoskeleton, this should not distract from the fact that the physiological event of centrosome separation, as it occurs at the onset of mitosis, clearly requires the cytoskeleton and MT-dependent motor proteins.
In summary, we propose that C-Nap1 is part of a dynamic structure that links parental centrioles in a cell cycle–dependent manner (Fig. 7). The existence of such a structure has previously been inferred from careful electron microscopic studies on isolated centrosomes (Paintrand et al. 1992). Our model predicts that C-Nap1 provides the docking sites at the proximal tips of parental centrioles, with which as yet unidentified proteins can interact during interphase of the cell cycle. This interaction appears to be important for centriole–centriole cohesion, as interference with C-Nap1 causes centrosome splitting. In contrast, C-Nap1 is not detectable at the contact sites between parental centrioles and procentrioles, and it is unlikely to play a role in the association between these latter organelles. Most importantly, the association of C-Nap1 with centrioles is cell cycle–regulated. C-Nap1 is conspicuously reduced at mitotic spindle poles, and, as proposed previously, this may be a consequence of phosphorylation by Nek2 (Fry et al. 1998b). We favor the view that phosphorylation may regulate centriole–centriole cohesion, but emphasize that the detailed mechanism regulating this process remains to be determined. Finally, it is interesting to consider that the orthogonal link between the two mitotic centrioles is broken at the end of mitosis. At about the same time, C-Nap1 reaccumulates at the proximal ends of the two centrioles. Although this temporal correlation does not necessarily imply a cause and effect relationship, it is tempting to speculate that the separation between the two mitotic centrioles may be necessary for the recruitment of C-Nap1 to what will become the two parental centrioles in the subsequent cell cycle. Alternatively, it is equally possible that C-Nap1 recruitment may actually promote the disorientation between the orthogonally oriented mitotic centrioles. Whatever the precise mechanism, one would predict that the orthogonal linkage between mitotic centrioles needs to be broken in order to establish a more flexible connection between parental centrioles in the subsequent cell cycle.
Acknowledgments
We thank Bernard Eddé (Collège de France, Paris, France) for a kind gift of the GT335 antibody, Jorgos Pyrowolakis (Max-Planck-Institute for Biochemistry [MPIB], Martinsried, Germany) for assistance with FACS® analysis, and the Ullrich laboratory (MPIB) for the use of their microinjection setup. We also thank Monika Matzner for technical assistance, and many colleagues in our laboratory and beyond for stimulating discussions.
In its initial stage, this work was supported by the Swiss National Science Foundation (31-50576.97) and the Canton of Geneva.
Footnotes
T. Mayor's present address is Department of Cell Biology, Max-Planck-Institute for Biochemistry, D-82152 Martinsried, Germany.
K. Tanaka's present address is School of Biological Science, University of Manchester, Manchester M13 9PT, United Kingdom.
A.M. Fry's present address is Department of Biochemistry, University of Leicester, Leicester LE1 7 RH, United Kingdom.
Abbreviations used in this paper: aa, amino acid; BDM, 2,3-butanedione monoxime; MF, microfilament; MT, microtubule; PCM, pericentriolar material.
References
- Andersen S.S. Molecular characteristics of the centrosome. Int. Rev. Cytol. 1999;187:51–109. [Abstract] [Google Scholar]
- Blangy A., Lane H.A., d'Herin P., Harper M., Kress M., Nigg E.A. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. [Abstract] [Google Scholar]
- Bobinnec Y., Moudjou M., Fouquet J.P., Desbruyeres E., Edde B., Bornens M. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil. Cytoskeleton. 1998;39:223–232. [Abstract] [Google Scholar]
- Bornens M., Paintrand M., Berges J., Marty M.C., Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskeleton. 1987;8:238–249. [Abstract] [Google Scholar]
- Bornens M., Moudjou M. Studying the composition and function of centrosomes in vertebrates. Methods Cell Biol. 1999;61:13–34. [Abstract] [Google Scholar]
- Brinkley B.R., Goepfert T.M. Supernumerary centrosomes and cancerBoveri's hypothesis resurrected. Cell Motil. Cytoskeleton. 1998;41:281–288. [Abstract] [Google Scholar]
- Chang P., Stearns T. delta-Tubulin and varepsilon-tubulintwo new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat. Cell Biol. 2000;2:30–35. [Abstract] [Google Scholar]
- Doxsey S. The centrosome—a tiny organelle with big potential. Nat. Genet. 1998;20:104–106. [Abstract] [Google Scholar]
- Fry A.M., Meraldi P., Nigg E.A. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators EMBO (Eur. Mol. Biol. Organ.) J. 17 1998. 470 481a [Europe PMC free article] [Abstract] [Google Scholar]
- Fry A.M., Mayor T., Meraldi P., Stierhof Y.D., Tanaka K., Nigg E.A. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle–regulated protein kinase Nek2 J. Cell Biol. 141 1998. 1563 1574b [Europe PMC free article] [Abstract] [Google Scholar]
- Harlow E., Lane D. Antibodies 1998. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: pp. 283–318 [Google Scholar]
- Jean C., Tollon Y., Raynaud-Messina B., Wright M. The mammalian interphase centrosometwo independent units maintained together by the dynamics of the microtubule cytoskeleton. Eur. J. Cell Biol. 1999;78:549–560. [Abstract] [Google Scholar]
- Kellogg D.R., Moritz M., Alberts B.M. The centrosome and cellular organization. Annu. Rev. Biochem. 1994;63:639–674. [Abstract] [Google Scholar]
- Kochanski R.S., Borisy G.G. Mode of centriole duplication and distribution. J. Cell Biol. 1990;110:1599–1605. [Europe PMC free article] [Abstract] [Google Scholar]
- Lacey K.R., Jackson P.K., Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA. 1999;96:2817–2822. [Europe PMC free article] [Abstract] [Google Scholar]
- Lane H.A., Nigg E.A. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 1996;135:1701–1713. [Europe PMC free article] [Abstract] [Google Scholar]
- Mack G.J., Rees J., Sandblom O., Balczon R., Fritzler M.J., Rattner J.B. Autoantibodies to a group of centrosomal proteins in human autoimmune sera reactive with the centrosome. Arthritis Rheum. 1998;41:551–558. [Abstract] [Google Scholar]
- Mayor T., Meraldi P., Stierhof Y.D., Nigg E.A., Fry A.M. Protein kinases in control of the centrosome cycle. FEBS Lett. 1999;452:92–95. [Abstract] [Google Scholar]
- Moudjou M., Paintrand M., Vigues B., Bornens M. A human centrosomal protein is immunologically related to basal body- associated proteins from lower eucaryotes and is involved in the nucleation of microtubules. J. Cell Biol. 1991;115:129–140. [Europe PMC free article] [Abstract] [Google Scholar]
- Paintrand M., Moudjou M., Delacroix H., Bornens M. Centrosome organization and centriole architecturetheir sensitivity to divalent cations. J. Struct. Biol. 1992;108:107–128. [Abstract] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C.L., Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 2000;149:317–330. [Europe PMC free article] [Abstract] [Google Scholar]
- Salisbury J.L., Whitehead C.M., Lingle W.L., Barrett S.L. Centrosomes and cancer. Biol. Cell. 1999;91:451–460. [Abstract] [Google Scholar]
- Schliwa M., Pryzwansky K.B., Euteneuer U. Centrosome splitting in neutrophilsan unusual phenomenon related to cell activation and motility. Cell. 1982;31:705–717. [Abstract] [Google Scholar]
- Seelos C. A critical parameter determining the aging of DNA-calcium-phosphate precipitates. Anal. Biochem. 1997;245:109–111. [Abstract] [Google Scholar]
- Sherline P., Mascardo R.N. Epidermal growth factor induces rapid centrosomal separation in HeLa and 3T3 cells. J. Cell Biol. 1982;93:507–512. [Europe PMC free article] [Abstract] [Google Scholar]
- Urbani L., Stearns T. The centrosome. Curr. Biol. 1999;9:R315–R317. [Abstract] [Google Scholar]
- Vaisberg E.A., Koonce M.P., McIntosh J.R. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J. Cell Biol. 1993;123:849–858. [Europe PMC free article] [Abstract] [Google Scholar]
- Verde F., Labbe J.C., Doree M., Karsenti E. Regulation of microtubule dynamics by cdc2 protein kinase in cell-free extracts of Xenopus eggs. Nature. 1990;343:233–238. [Abstract] [Google Scholar]
- Walczak C.E., Vernos I., Mitchison T.J., Karsenti E., Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 1998;8:903–913. [Abstract] [Google Scholar]
- Wolff A., de Nechaud B., Chillet D., Mazarguil H., Desbruyeres E., Audebert S., Edde B., Gros F., Denoulet P. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 1992;59:425–432. [Abstract] [Google Scholar]
- Zimmerman W., Sparks C.A., Doxsey S.J. Amorphous no longerthe centrosome comes into focus. Curr. Opin. Cell Biol. 1999;11:122–128. [Abstract] [Google Scholar]
Articles from The Journal of Cell Biology are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1083/jcb.151.4.837
Read article for free, from open access legal sources, via Unpaywall:
http://jcb.rupress.org/content/151/4/837.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Loss of ninein interferes with osteoclast formation and causes premature ossification.
Elife, 13:e93457, 05 Jun 2024
Cited by: 3 articles | PMID: 38836552
Time-series reconstruction of the molecular architecture of human centriole assembly.
Cell, 187(9):2158-2174.e19, 10 Apr 2024
Cited by: 5 articles | PMID: 38604175 | PMCID: PMC11060037
Prolonged overexpression of PLK4 leads to formation of centriole rosette clusters that are connected via canonical centrosome linker proteins.
Sci Rep, 14(1):4370, 22 Feb 2024
Cited by: 1 article | PMID: 38388511 | PMCID: PMC10883960
Insulin signaling and pharmacology in humans and in corals.
PeerJ, 12:e16804, 31 Jan 2024
Cited by: 1 article | PMID: 38313028 | PMCID: PMC10838073
A potential patient stratification biomarker for Parkinson´s disease based on LRRK2 kinase-mediated centrosomal alterations in peripheral blood-derived cells.
NPJ Parkinsons Dis, 10(1):12, 08 Jan 2024
Cited by: 2 articles | PMID: 38191886 | PMCID: PMC10774440
Go to all (136) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2.
J Cell Biol, 141(7):1563-1574, 01 Jun 1998
Cited by: 268 articles | PMID: 9647649 | PMCID: PMC2133000
Rootletin forms centriole-associated filaments and functions in centrosome cohesion.
J Cell Biol, 171(1):27-33, 03 Oct 2005
Cited by: 205 articles | PMID: 16203858 | PMCID: PMC2171225
Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion.
J Cell Sci, 120(pt 24):4321-4331, 27 Nov 2007
Cited by: 131 articles | PMID: 18042621
Organisation and functional regulation of the centrosome in animal cells.
Prog Cell Cycle Res, 3:285-299, 01 Jan 1997
Cited by: 12 articles | PMID: 9552423
Review