Abstract
Free full text

The IKKβ Subunit of IκB Kinase (IKK) is Essential for Nuclear Factor κB Activation and Prevention of Apoptosis
Abstract
The IκB kinase (IKK) complex is composed of three subunits, IKKα, IKKβ, and IKKγ (NEMO). While IKKα and IKKβ are highly similar catalytic subunits, both capable of IκB phosphorylation in vitro, IKKγ is a regulatory subunit. Previous biochemical and genetic analyses have indicated that despite their similar structures and in vitro kinase activities, IKKα and IKKβ have distinct functions. Surprisingly, disruption of the Ikkα locus did not abolish activation of IKK by proinflammatory stimuli and resulted in only a small decrease in nuclear factor (NF)-κB activation. Now we describe the pathophysiological consequence of disruption of the Ikkβ locus. IKKβ-deficient mice die at mid-gestation from uncontrolled liver apoptosis, a phenotype that is remarkably similar to that of mice deficient in both the RelA (p65) and NF-κB1 (p50/p105) subunits of NF-κB. Accordingly, IKKβ-deficient cells are defective in activation of IKK and NF-κB in response to either tumor necrosis factor α or interleukin 1. Thus IKKβ, but not IKKα, plays the major role in IKK activation and induction of NF-κB activity. In the absence of IKKβ, IKKα is unresponsive to IKK activators.
The nuclear factor (NF)-κB1 transcription factor plays a key role in activation of inflammatory and innate immune responses (1, 2). In nonstimulated cells, NF-κB dimers are kept as cytoplasmic latent complexes through binding of specific inhibitors, the IκBs, which mask their nuclear localization signal (NLS). Upon exposure to proinflammatory stimuli, such as bacterial LPS, TNF-α, or IL-1, the IκBs are rapidly phosphorylated at two conserved NH2-terminal serines, a posttranslational modification that is rapidly followed by their polyubiquitination and proteasomal degradation (3–6). This results in unmasking of the NLS of NF-κB dimers followed by their translocation to the nucleus, binding to specific DNA sites (κB sites), and target gene activation. NF-κB target genes include many of the cytokine and chemokine genes, as well as genes coding for adhesion molecules, cell surface receptors, and enzymes that produce secondary inflammatory mediators (7, 8).
The protein kinase that phosphorylates IκBs in response to proinflammatory stimuli has been identified biochemically and molecularly (9–11). Named IKK, this protein kinase is a complex composed of at least three subunits: IKKα, IKKβ and IKKγ (for a review, see reference 12). IKKα and IKKβ are highly similar protein kinases that act as the catalytic subunits of the complex (9, 11, 13, 14). In vitro, both IKKα and IKKβ form homo- and heterodimers that can phosphorylate IκB proteins at their NH2-terminal regulatory serines (15). In mammalian cells, IKKα and IKKβ form a stable heterodimer that is tightly associated with the IKKγ (NEMO) subunit (16, 17). As cell lines that fail to express IKKγ (NEMO) exhibit a major defect in IκB degradation and NF-κB activation in response to proinflammatory stimuli and double-stranded RNA, this regulatory subunit plays an essential function (at least in the examined cell lines) in IKK and NF-κB activation (17). The physiological function of the two catalytic subunits has been less clear. Initially, overexpression of catalytically inactive forms of IKKα and IKKβ that blocked IKK and NF-κB activation suggested that both subunits play similar and possibly redundant roles in IκB phosphorylation and NF-κB activation (13, 14). This hypothesis was fostered by finding that in vitro IKKα and IKKβ can directly phosphorylate IκBα and IκBβ at the serines that trigger their degradation in vivo (15). However, it was also suggested that IKKα rather than IKKβ is responsible for activation of the entire complex in response to certain stimuli, such as the NF-κB inducing kinase, NIK (18). Recently, we found that in addition to an IKKγ subunit with an intact COOH terminus (16), IKK activation requires the phosphorylation of IKKβ at two serines within its activation loop (19). Replacement of these serines, whose phosphorylation is stimulated by proinflammatory stimuli or NIK, with alanines abolishes IKK activation. Interestingly, although the entire activation loop is identical in sequence between IKKα and IKKβ, replacement of the same two serines in IKKα with alanines has no effect on IKK activation (19). These results were further substantiated by gene targeting (knockout) experiments. Cells and tissues from mice that no longer express IKKα (Ikkα−/− mice) exhibit normal IKK activation in response to TNF, IL-1, or LPS (20). Although NF-κB is fully inducible, for an unknown reason, IKKα-deficient fibroblasts exhibit approximately twofold reduction in both basal and induced NF-κB binding activity (20). Thus, IKKα may somehow stimulate NF-κB DNA binding despite not being required for IκB phosphorylation and degradation in most cell types. The gene targeting experiments reveal that, although not involved in activation of IKK by proinflammatory stimuli, IKKα plays an instrumental role in morphogenesis (20). The most important function of IKKα appears to be in the control of keratinocyte differentiation and formation of the epidermis (20). It is not yet clear whether these morphogenetic functions of IKKα are exerted through localized NF-κB activation in response to developmental cues.
To determine the physiological function(s) of IKKβ, we have used gene targeting to create Ikkβ knockout mice. We now show that the loss of IKKβ results in embryonic lethality at mid-gestation due to extensive apoptosis of the developing liver. This phenotype is similar to that of mice deficient in the RelA (p65) subunit of NF-κB (21). It was recently shown that the lethality of RelA −/− mice is completely suppressed by the loss of TNF-α (22). As NF-κB is required for protection of cells from TNF-α–induced apoptosis (23–25), the apoptotic phenotype of Ikkβ−/− mice strongly suggests that the absence of IKKβ results in a severe defect in NF-κB activation. Indeed, neither IKK nor NF-κB can be activated by TNF-α or IL-1 in IKKβ-deficient cells. Furthermore, we show that in the absence of IKKβ, the IKKα subunit is not responsive to NIK even though it can still associate with the IKKγ subunit.
Materials and Methods
Generation of IKKβ-deficient Mice.
Using a 0.2-kb BstEII-Bsu36I restriction fragment from the 5′ end of human IKKβ cDNA as a probe, three murine IKKβ genomic fragments were isolated from a 129/SvJ mouse genomic library (Stratagene, Inc.). One of the clones contained at least the first three coding exons and was used to construct the targeting vector IKKβKO. A 1.4-kb SacI restriction fragment harboring part of the second exon was used as the short homology arm, and the long arm was a 5.5-kb EcoRV-XhoI restriction fragment containing part of the third intron. The two arms were inserted into the XmnI and SmaI sites, respectively, of pGNA, which contains the G418 resistance gene (Neor) and LacZ (26). As a negative selection marker, a diphtheria toxin gene cassette (DT) was inserted into the KpnI site of pGNA. After cutting with PmeI, 20 μg of the linearized targeting vector was electroporated into 107 mouse embryonic stem (ES) cells (line GS from Genome Systems). After selection with G418 at 0.4 mg/ml, G418-resistant colonies were picked and screened by PCR. The genotype of the PCR-positive clones was confirmed by Southern blotting analysis. Homologous recombinants were karyotyped and analyzed for mycoplasma. Two homologous recombinant ES clones were injected into C57BL/6 blastocysts. Resulting male chimeras were crossed with C57BL/6 females, and germline transmission was scored by coat color. Heterozygous mice were identified by PCR and Southern analysis of mouse tail DNA. Embryos from intercrosses of heterozygous (Ikkβ+/−) mice, as well as mouse embryonic fibroblasts (EFs), were genotyped by PCR and Southern analysis using DNA isolated from a piece of each embryo or a cell pellet, respectively.
PCR and Southern Blotting Analysis.
PCR was performed in the presence of 10% DMSO with Taq DNA polymerase using a Perkin-Elmer 9600 thermocycler programmed for denaturation at 95°C for 5 min, amplification for 35 cycles (94°C for 30 s, 55°C for 30 s, 65°C for 2 min), and elongation at 72°C for 10 min. Primers used were: P1 (5′-AGTCCAACTGGCAGCGAATA-3′) located outside of the homology arm and P2 (5′-CAACATTAAATGTGAGCGAG-3′) located within the LacZ gene. Southern blotting analysis was performed according to a standard protocol (27) except that hybridization was performed in phosphate-SDS buffer (28).
Kinase Assay, Immunoprecipitation, Immunoblotting, and Electrophoretic Mobility Shift Assays.
Ikkβ−/−, Ikkβ+/−, and Ikkβ+/+ ES and EF cells were treated with TNF-α or IL-1 at 20 ng/ml. Kinase assays and immunoprecipitations were performed as described (9). Immunoblotting was performed as described (14, 16). Electrophoretic mobility shift assays (EMSAs) using the consensus κB and NF-1 sequences were performed as described (16, 29).
Histology, In Situ TUNEL Assay, and Transmission Electron Microscopy.
Mouse embryos or embryo livers were fixed in 10% buffered formalin and embedded in paraffin. After routine processing, the sections (5-μm thick) were stained with hematoxylin and eosin (H&E) for histological analysis. In situ TUNEL (terminal deoxynucleotidyl transferase–mediated dUTP nick-end labeling) assay was done using the in situ cell death detection kit according to the manufacturer's instructions (Boehringer Mannheim). For electron microscopy, embryonic day 13 (E13) embryos were removed and the livers were dissected out and fixed for 1 h in 2% formaldehyde and 2% glutaraldehyde in 0.15 M sodium cacodylate buffer (pH 7.4) at 4°C. The remainder of the embryos were placed in PBS for subsequent PCR and Southern analysis. After washing in cacodylate buffer, the livers were postfixed in 1% osmium tetroxide in cacodylate buffer for an additional 1 h. After postfixation, the samples were rinsed in double distilled water, dehydrated in a graded ethanol series, and infiltrated and polymerized in Durcupan ACM resin (Electron Microscopy Sciences). Sections 80-nm thick were stained with Sato lead and examined at 80 keV with either a JEOL 100CX or 2000EX transmission electron microscope.
Results
Generation of Ikkβ Knockout Mice.
To create a strain of IKKβ-deficient mice, we used gene targeting technology (30). Mouse genomic Ikkβ DNA was cloned from a 129 strain library and, after mapping and sequencing, was used to construct the targeting vector (Fig. (Fig.11 A). To eliminate IKKβ kinase activity, part of the second and the entire third coding exon that specifies an essential part of the kinase domain were replaced with a DNA fragment encoding β-galactosidase (LacZ) and neomycin resistance (Neor). Because the Neor gene contains transcription termination and polyadenylation signals, the COOH-terminal three quarters of IKKβ including its protein interaction motifs are unlikely to be expressed from the targeted allele.

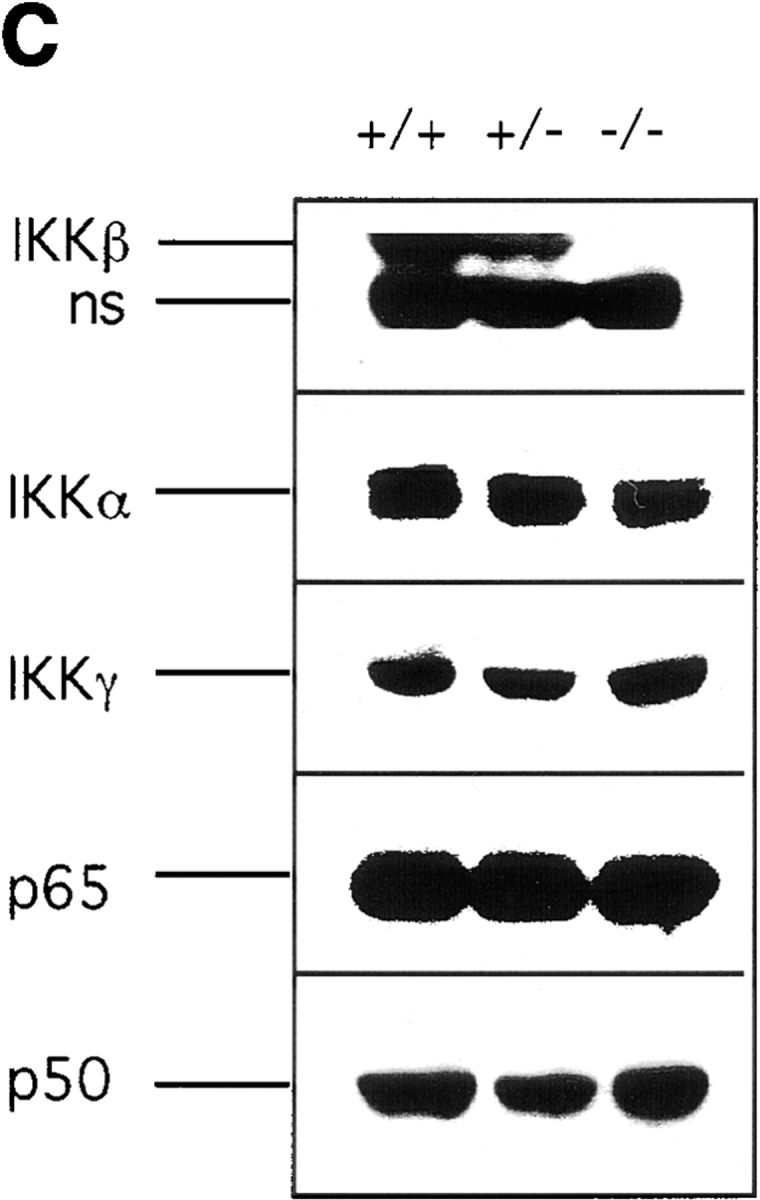
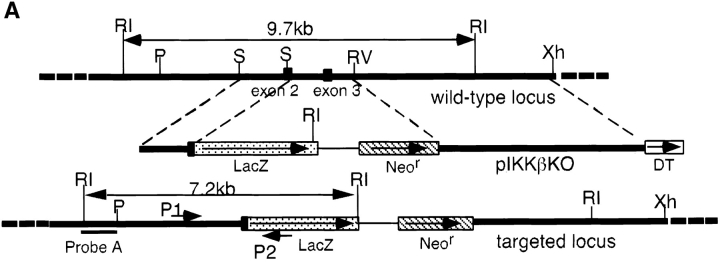
Generation of IKKβ-deficient mice. (A) The mouse Ikkβ locus and the targeting vector. Map of the Ikkβ genomic fragment used for gene targeting is shown. The exons are indicated by solid black boxes, the introns are indicated by bold lines, and the selection markers, lengths of restriction fragments, restriction enzyme sites, the probes used for Southern analysis, and the location of primers used in PCR screening are also shown. RI, EcoRI; P, PstI; S, SacI; RV, EcoRV; Xh, XhoI. (B) Southern blot analysis of mouse genomic DNA. Mouse genomic DNA was digested with EcoRI and probed with probe A (1.2-kb HindIII-PstI fragment of Ikkβ). After homologous recombination, the 9.7-kb EcoRI fragment of wild-type Ikkβ is replaced by a 7.2-kb EcoRI fragment, as indicated in panel A. (C) Western blot analysis of mouse proteins using antibody H470 specific for IKKβ. Location of the IKKβ band is indicated. The lower band is nonspecific (ns). The same blot was also probed with antibodies to IKKα, IKKγ, p65(RelA), and p50(NF-κB1). The genotypes are as indicated.
After selection and screening by Southern blotting, six ES cell clones with homologous integration of the targeting vector into the Ikkβ locus were isolated, and two of them were used to generate chimeric mice. Chimeric mice derived from these clones transmitted the targeted Ikkβ allele to their progeny (Fig. (Fig.11 B). Although Ikkβ+/− male and female mice appeared normal and were fertile, upon intercrossing they did not give rise to live Ikkβ−/− progeny.
Analysis of protein extracts of Ikkβ+/+, Ikkβ+/−, and Ikkβ−/− cells revealed that, as expected, no IKKβ protein was expressed from the targeted allele (Fig. (Fig.11 C). In addition, Ikkβ+/− cells expressed approximately half the dose of IKKβ present in wild-type cells. No compensatory increases in IKKα, IKKγ, p65(RelA), or p50(NF-κB1) expression were observed.
Phenotype of Ikkβ−/− Mice.
Given the expected importance of IKKβ for NF-κB activation and the embryonic lethality of RelA −/− mice (21), we suspected that the loss of IKKβ would result in a similar phenotype. Therefore, we analyzed embryos from timed pregnancies of Ikkβ+/− intercrosses. Although Ikkβ−/− embryos isolated at E11.5 were alive and had perfectly normal appearance (data not shown), Ikkβ−/− embryos isolated at E13.5 were no longer alive and were rather anemic in appearance (Fig. (Fig.2).2). Even external examination suggested that the liver of E13.5 Ikkβ−/− embryos had degenerated. Notably, however, the limbs and head of Ikkβ−/− embryos were normally developed, unlike those of Ikkα−/− E13.5 embryos (20). Histochemical examination of transverse sections of normal and mutant E13.5 mouse embryos stained with H&E revealed massive cell death in livers of Ikkβ−/− embryos (Fig. (Fig.33 A). Essentially, no viable hepatocytes could be detected, and the numbers of dead cells with highly condensed and fragmented nuclei were markedly increased. However, hematopoietic precursors retained their normal appearance in Ikkβ−/− livers. TUNEL staining revealed that the observed cell death is most likely due to apoptosis, whose rate was increased manyfold (Fig. (Fig.33 B). Examination of E13 Ikkβ−/− embryos revealed close to normal external appearance (data not shown), but electronmicroscopic examination of ultrathin sections from their livers revealed massive numbers of dead hepatocytes with highly condensed nuclei characteristic of apoptotic cell death (Fig. (Fig.4).4). The livers of Ikkβ+/+ or Ikkβ+/− littermates had perfectly normal appearance.
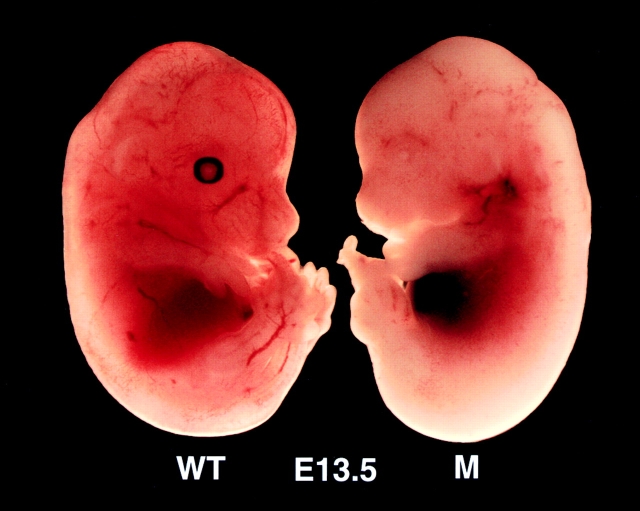
Appearance of an Ikkβ−/− E13.5 embryo and a normal littermate. Wild-type (Ikkβ+/+, WT) and mutant (Ikkβ−/−, M) embryos were isolated at E13.5 and photographed. The genotypes of the embryos were later determined by PCR and Southern blot analysis.
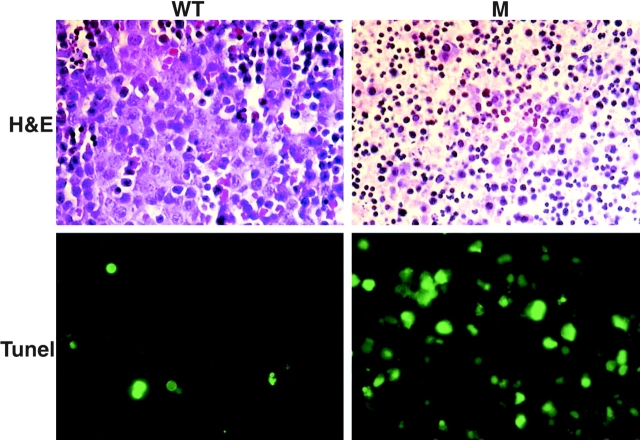
Analysis of wild-type (WT) and mutant (M) livers. E13.5 embryos were fixed and sectioned. Paraffin-embedded transverse sections at the area of the liver were subjected to H&E (top; original magnification: 400×) or TUNEL (bottom; original magnification: 600×) staining. The stained sections were photographed.
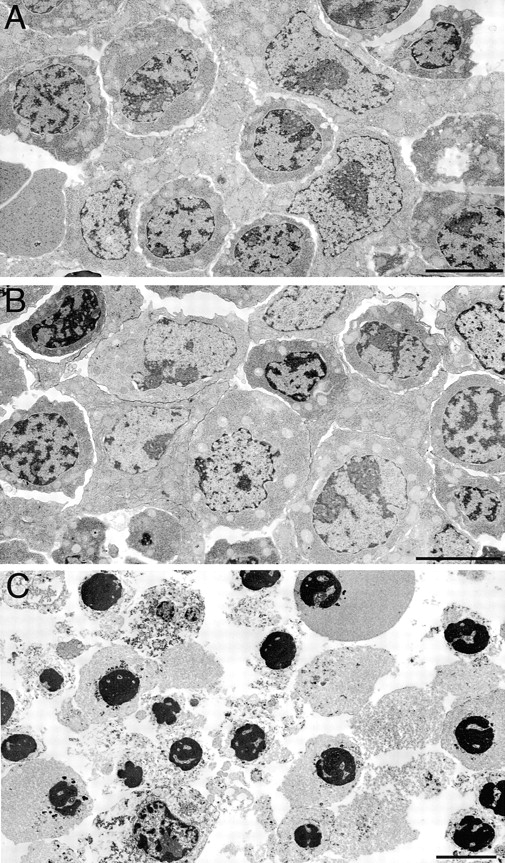
Electron microscopic analysis of livers from E13 Ikkβ+/+, Ikkβ+/−, and Ikkβ−/− embryos. Both E13 Ikkβ+/+ (A) and Ikkβ+/− (B) livers exhibited normal morphology. The Ikkβ−/− liver (C) exhibited varying degrees of apoptosis characterized by collapsed and condensed nuclei and general cellular degeneration. Bars = 5 μm.
Defective NF-κB Activation in Ikkβ−/− Cells.
We used two different approaches to determine the consequences of the loss of IKKβ expression on IKK and NF-κB activation. First, we prepared Ikkβ−/− ES cell lines by subjecting Ikkβ+/− ES cells to selection at higher G418 concentration. One Ikkβ−/− cell line was identified. As shown in Fig. Fig.55 A, stimulation of these cells with either TNF-α or IL-1 did not result in IKK activation, whereas a normal activation response was observed in Ikkβ+/− cells. Note, however, that Ikkβ+/− cells had ~50% of the IKK activity of wild-type (Ikkβ+/+) ES cells, consistent with the reduced amount of IKKβ protein (data not shown). In addition to the defect in IKK activation, hardly any induction of NF-κB DNA binding activity was observed in Ikkβ−/− cells after stimulation with either IL-1 or TNF-α (Fig. (Fig.55 B). Even the basal level of NF-κB DNA binding activity was considerably reduced in Ikkβ−/− cells, despite no detectable changes in p65(RelA) or p50(NF-κB1) abundance (data not shown). The second approach to evaluate the function of IKKβ was to prepare cultures of EFs from E11.5 mouse embryos of all three genotypes. As shown in Fig. Fig.6,6, essentially no induction of IKK or NF-κB activity could be detected in Ikkβ−/− EF cells treated with either IL-1 or TNF-α. Interestingly, Ikkβ+/− EF cells exhibited an ~50% reduction in IKK activity (consistent with the reduction in IKKβ expression) but a much larger decrease in NF-κB DNA binding activity.
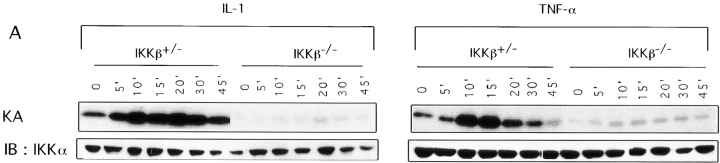
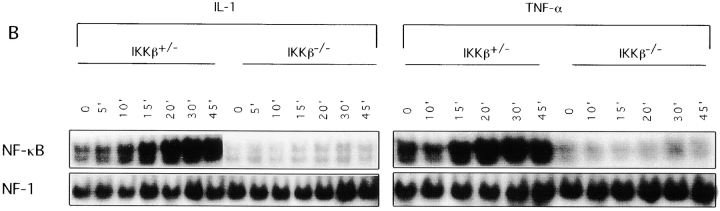
Defective IKK and NF-κB activation in IKKβ- deficient ES cells. (A) IKK activity. Lysates of TNF-α– or IL-1–treated Ikkβ+/− and Ikkβ−/− cells were prepared at the indicated time points (in min) after stimulation and immunoprecipitated with antibody M280 to IKKα. IKK activity (KA) was measured by an immunecomplex kinase assay using GST-IκBα(1-54) as a substrate. The kinase assay products were separated by SDS-PAGE, transferred to nitrocellulose membrane, and autoradiographed. The membrane was reprobed with antibody M280 (IB: IKKα) for loading control. (B) NF-κB binding activity. Nuclear extracts of Ikkβ+/− and Ikkβ−/− cells stimulated with IL-1 or TNF-α for the indicated times (in min) were incubated with 32P-labeled κB oligonucleotide probe and subjected to EMSA. Binding to an NF-1 probe was used to control the quality and amount of nuclear protein extracts.
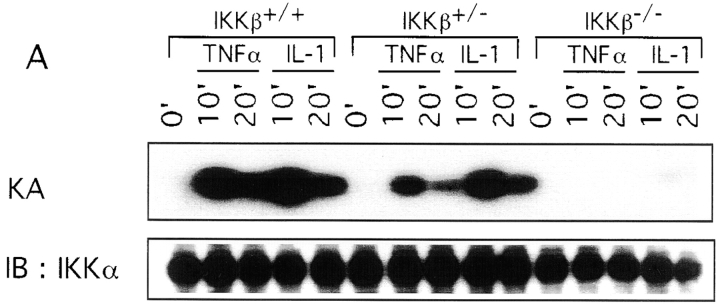
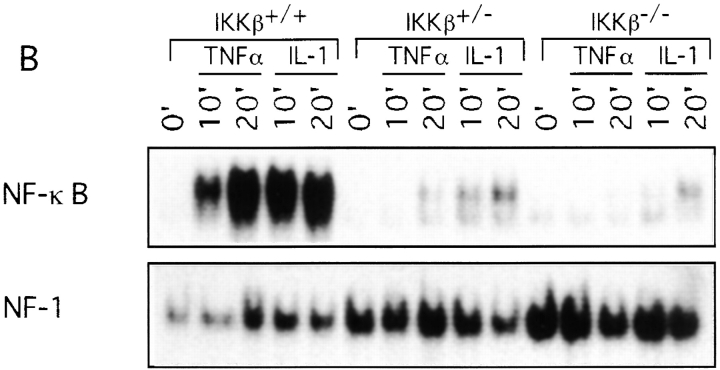
Defective IKK and NF-κB activation in IKKβ-deficient EF cells. Second passage EFs from E11.5 Ikkβ+/+, Ikkβ+/−, and Ikkβ−/− embryos were stimulated with TNF-α or IL-1. At the indicated times, whole cell extracts were prepared and used to measure (A) IKK activity (KA), and (B) NF-κB DNA binding activity. IB, immunoblotting.
IKKα Cannot Be Activated by NIK in the Absence of IKKβ.
The results described above indicate that IKKα, which is expressed in normal levels in Ikkβ−/− cells, cannot be activated by either TNF-α or IL-1. To further examine this point, we cotransfected an HA epitope–tagged IKKα expression vector into Ikkβ−/− ES cells in the absence or presence of an NIK expression vector. NIK is the most potent IKK activator identified to date (31) and was suggested to be a direct IKKα kinase (18). Recently, however, we obtained results that suggested that NIK-induced IKKα phosphorylation is not direct and is likely to be dependent on IKKβ (19). Consistent with this hypothesis, we found no increase in IKK activity towards IκBα(1-54) substrate upon coexpression of HA-IKKα with NIK in Ikkβ−/− cells (Fig. (Fig.77 A). Yet, when an IKKβ expression vector was included in these transfections, NIK elicited a clear increase in IKK activity. As shown previously, NIK coexpression efficiently stimulates IKKα-associated IKK activity in IKKβ-expressing cells (19).
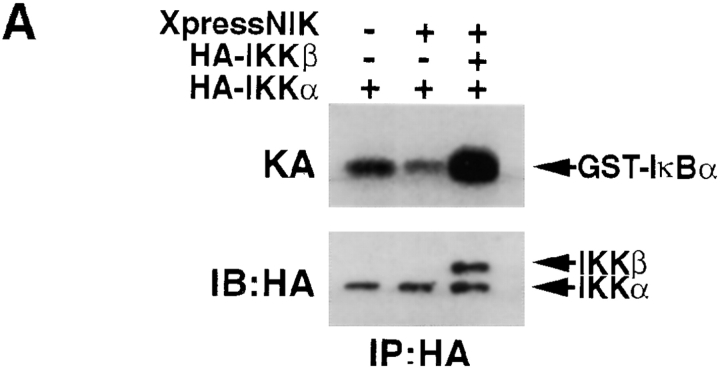
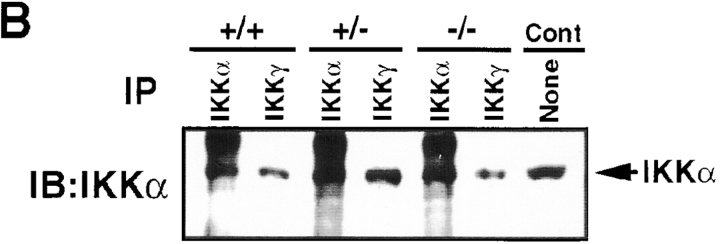
IKKα is refractory to activation in Ikkβ−/− cells despite its association with IKKγ. (A) Ikkβ−/− ES cells were transiently transfected by electroporation with an HA-IKKα expression vector alone or together with XpressNIK or HA-IKKβ and XpressNIK expression vectors. 24 h after transfection, HA-IKK proteins were immunoprecipitated (IP) with anti-HA antibody and their associated IKK activity (KA) was determined using GST-IκBα(1-54) as a substrate. Protein expression levels were determined by immunoblotting (IB) with anti-HA. (B) Lysates of Ikkβ+/+, Ikkβ+/−, and Ikkβ−/− cells were immunoprecipitated (IP) with either anti-IKKα or anti-IKKγ antibodies as indicated. The immunecomplexes were dissolved in SDS loading buffer and separated by SDS-PAGE. After transfer to an Immobilon membrane, the proteins were analyzed by immunoblotting (IB) with anti-IKKα antibody. A lysate of 3T3 cells was used as a control (Cont).
One reason for the inability of IKKα to respond to proinflammatory stimuli or NIK in the absence of IKKβ could be its inability to directly associate with IKKγ, the regulatory subunit of the IKK complex. Previous experiments indicate that IKKγ is essential for recruitment of upstream activators to IKK (16). In addition, using recombinant proteins, it was found that IKKβ directly interacts with IKKγ much more efficiently than does IKKα (16, 17). Having available IKKβ-deficient cells, we reexamined the ability of IKKα to interact with IKKγ. In contrast to the results obtained with recombinant proteins, very efficient coprecipitation of IKKα by anti-IKKγ antibodies was observed using lysates of Ikkβ−/− cells as a starting material (Fig. (Fig.77 B). Therefore, the refractoriness of IKKα to IKK activators in IKKβ-deficient cells is not due to its inability to associate with IKKγ.
Discussion
The enzymatic activity of the IKK complex, composed of two catalytic subunits, IKKα and IKKβ, and one regulatory subunit, IKKγ, is rapidly stimulated by proinflammatory cytokines and LPS (for a review, see reference 12). Activated IKK phosphorylates the different IκBs at the two NH2-terminal serines that trigger their polyubiquitination and proteasome-mediated degradation. Once the IκBs are degraded, the freed NF-κB dimers migrate to the nucleus and activate target gene transcription. Based on their similar primary structures (11, 13, 14) and substrate specificities (15), IKKα and IKKβ were expected to play redundant and interchangeable roles in proinflammatory signaling to NF-κB. Therefore, it was rather surprising that only IKKβ was found to be involved in IKK activation. Alanine substitutions of the two serines in the activation loop of IKKβ, whose phosphorylation is stimulated by either TNF-α treatment or NIK overexpression, prevented IKK activation. Yet, the same mutations introduced into the activation loop of the IKKα subunit had no effect on the response of IKK to TNF-α or NIK (19). These results were confirmed by the analysis of IKKα-deficient cells and tissues which revealed no defect in IKK activation and IκBα degradation in response to TNF-α, IL-1, or LPS (20). However, it remained possible that the function of IKKα in IκB phosphorylation in response to proinflammatory stimuli can be fully replaced by IKKβ. The results described here indicate that IKKβ and IKKα have different physiological functions and that IKKα cannot substitute for IKKβ.
To determine the physiological function of IKKβ, we generated Ikkβ−/− knockout mice and cell lines. The loss of IKKβ results in embryonic death at mid-gestation due to massive hepatocyte apoptosis. This phenotype is remarkably similar to that of RelA knockout mice (21), with one exception: while Ikkβ−/− embryos die around E13, RelA −/− embryos die around E15. The earlier death of Ikkβ−/− embryos is likely to be due to a more extensive reduction in NF-κB activity, as embryos that are deficient in both the p65 (RelA) and the p50 (NF-κB1) subunits of NF-κB die at E12.5, the same time as IKKβ-deficient embryos, from massive hepatocyte apoptosis (32). Thus, IKKβ and RelA are genetically proven to be components of the same pathway. Accordingly, cells that lack IKKβ are completely defective in IKK and NF-κB activation in response to either TNF-α or IL-1. Therefore, the IKKβ subunit is absolutely essential for mounting a response to proinflammatory stimuli. This function is not replaced by IKKα, whose expression is not diminished in the absence of IKKβ. In addition, as indicated by the normal morphology of the head and limbs of E13.5 Ikkβ−/− embryos, IKKα can carry out its developmental function (20) in the complete absence of IKKβ. Interestingly, a 50% reduction in IKKβ expression, as in Ikkβ+/− cells, results in a similar decrease in IKK activity but a much more severe defect in NF-κB activation. These results underscore the importance of the IKKβ subunit and indicate that the NF-κB activation response does not follow a simple linear relationship to the magnitude of IKK activation. It also appears from these results that a low level of NF-κB activity may be sufficient for protecting the liver from TNF-α–induced apoptosis.
One possible cause for the inability of IKKα to substitute for IKKβ was its relatively lower affinity to IKKγ, the regulatory subunit that is absolutely required for IKK activation (17). Using recombinant proteins, it was observed that IKKα does not form a stable complex with IKKγ in vitro, whereas IKKβ readily associates with IKKγ (16, 17). However, immunoprecipitation experiments indicate that a similar amount of IKKα is precipitated by IKKγ antibodies from Ikkβ−/− cells as from Ikkβ+/+ cells. Despite its ability to associate with IKKγ in the absence of IKKβ, IKKα is refractory to upstream activators involved in proinflammatory signaling, including the most potent IKK activator identified so far, NIK, in IKKβ-deficient cells. These results underscore the differences in regulation of IKKα and IKKβ activities.
In summary, together with the previous analysis of IKKα-deficient mice, the analysis of IKKβ-deficient mice, described here, indicates that the two catalytic subunits of the IKK complex, although similar in structure, have very different functions. Although IKKβ is responsible both for activation of the entire complex in response to proinflammatory stimuli, through phosphorylation at its activation loop, and for activation of NF-κB, through IκB phosphorylation, IKKα is assigned the control of epidermal and skeletal morphogenesis. Although the stimuli that activate IKKβ and the substrates that mediate its biological activity are known, the stimuli and the relevant substrates for IKKα remain to be identified.
Acknowledgments
We thank G. Hageman for the gift of mouse TNF-α, D.M. Rothwarf for advice and technical assistance, and B. Thompson for assistance with manuscript preparation.
Abbreviations used in this paper
| EF | embryonic fibroblast |
| EMSA | electrophoretic mobility shift assay |
| ES | embryonic stem |
| H&E | hematoxylin and eosin |
| IKK | IκB kinase |
| NF | nuclear factor |
| NIK | NF-κB inducing kinase |
| TUNEL | terminal deoxynucleotidyl transferase–mediated dUTP nick-end labeling |
Footnotes
Y. Hu was supported by a postdoctoral fellowship from the Arthritis Foundation. This work was supported by grants from the National Institutes of Health (AI43477, ES04151, AG05131, and RR04050) and the Department of Energy (DE-FG03-86ER60429). M. Karin is the Frank and Else Schilling-American Cancer Society Research Professor.
References
Articles from The Journal of Experimental Medicine are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1084/jem.189.11.1839
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jem/article-pdf/189/11/1839/1120114/99-0594.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1084/jem.189.11.1839
Article citations
NFκB signalling in colorectal cancer: Examining the central dogma of IKKα and IKKβ signalling.
Heliyon, 10(12):e32904, 12 Jun 2024
Cited by: 0 articles | PMID: 38975078
Review
Amicis Omnia Sunt Communia: NF-κB Inhibition as an Alternative to Overcome Osteosarcoma Heterogeneity.
Pharmaceuticals (Basel), 17(6):734, 05 Jun 2024
Cited by: 1 article | PMID: 38931401
Review
Mechanisms and Cardiorenal Complications of Chronic Anemia in People with HIV.
Viruses, 16(4):542, 30 Mar 2024
Cited by: 1 article | PMID: 38675885 | PMCID: PMC11053456
Review Free full text in Europe PMC
Implications of neonatal absence of innate immune mediated NFκB/AP1 signaling in the murine liver.
Pediatr Res, 95(7):1791-1802, 23 Feb 2024
Cited by: 1 article | PMID: 38396130
Exploring the association between rosacea and acne by integrated bioinformatics analysis.
Sci Rep, 14(1):3065, 06 Feb 2024
Cited by: 0 articles | PMID: 38321132 | PMCID: PMC10847114
Go to all (591) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Regulation and function of IKK and IKK-related kinases.
Sci STKE, 2006(357):re13, 17 Oct 2006
Cited by: 758 articles | PMID: 17047224
Review
NEMO-binding domains of both IKKalpha and IKKbeta regulate IkappaB kinase complex assembly and classical NF-kappaB activation.
J Biol Chem, 284(40):27596-27608, 07 Aug 2009
Cited by: 22 articles | PMID: 19666475 | PMCID: PMC2785688
Distinct roles of the Ikappa B kinase alpha and beta subunits in liberating nuclear factor kappa B (NF-kappa B) from Ikappa B and in phosphorylating the p65 subunit of NF-kappa B.
J Biol Chem, 277(6):3863-3869, 03 Dec 2001
Cited by: 256 articles | PMID: 11733537
Funding
Funders who supported this work.
NCRR NIH HHS (1)
Grant ID: P41 RR004050
NIA NIH HHS (2)
Grant ID: AG05131
Grant ID: P50 AG005131
NIAID NIH HHS (3)
Grant ID: AI43477
Grant ID: R37 AI043477
Grant ID: R01 AI043477
NIEHS NIH HHS (2)
Grant ID: R37 ES004151
Grant ID: ES04151

 Laboratory of Gene Regulation and Signal Transduction, ‡Department of Biology, and §Department of Neuroscience, University of California, San Diego, La Jolla, California 92093-0636
Laboratory of Gene Regulation and Signal Transduction, ‡Department of Biology, and §Department of Neuroscience, University of California, San Diego, La Jolla, California 92093-0636



