Abstract
Free full text

The Interleukin 7 Receptor Is Required for T Cell Receptor γ Locus Accessibility to the V(D)j Recombinase
Abstract
Defects in the interleukin (IL)-7 signal transduction pathway lead to severe immunodeficiency in humans and in mice. In IL-7 receptor–deficient (IL-7R−/−) mice, lymphoid precursors show a reduced survival rate and variable/diversity/joining region V(D)J recombination is variously affected in different loci, being arrested in the T cell receptor (TCR)-γ locus, aberrant in the immunoglobulin heavy chain (IgH) locus, and delayed in the TCR-β locus. Here, we analyze the recombination defect of the TCR-γ locus. Using ligation-mediated polymerase chain reaction, we sought intermediates of the recombination process. In the absence of the IL-7 signal, no initiation of recombination of the TCR-γ locus was observed, whereas recombination intermediates at the TCR-β locus could be detected. Thus, the failure to rearrange the TCR-γ locus is due to a failure to initiate cleavage rather than a failure to religate broken DNA ends. V(D)J recombination was previously thought to begin at the pro-T2 stage of T cell development after the arrest of IL-7R−/− thymocytes at the pro-T1 stage. However, here we show that both TCR-γ and -β recombination intermediates are readily detectable in normal T1 cells, but only TCR-β intermediates were detected in IL-7R−/− T1 cells, supporting a mechanistic role for IL-7 in TCR-γ locus rearrangement. Since reduced recombination activating gene (rag) expression has been reported in the absence of the IL-7 signal, we directly tested whether the TCR-γ locus is accessible to cleavage by recombinant Rag proteins in vitro. We found a reduction in chromatin accessibility for Rag-mediated cleavage in IL-7R−/− thymocytes compared with wild-type. Thus, IL-7 controls recombination at the TCR-γ locus by regulating locus accessibility.
Introduction
IL-7, a cytokine produced by stromal cells from the thymus or bone marrow, is essential for development of lymphoid cells (for reviews, see references 1, 2). IL-7 binds to the IL-7Rα chain, inducing association with the γc chain, followed by activation of the Janus kinases Jak1 and Jak3. Deletion of any of these components, IL-7, IL-7Rα, γc, Jak1, or Jak3, results in severe inhibition of T and B cell development in mice (for a review, see reference 3). In humans, severe T cell immunodeficiency results from reduced levels of the IL-7Rα chain 4 or from mutations in the genes for γc or Jak3 5 6. The requirement for IL-7 in vivo is partly attributable to trophic effects (IL-7 contributes to the survival of lymphoid precursor cells 7 8 9), and partly due to effects on V(D)J recombination of immune receptor genes. Mice with a deletion of the IL-7Rα chain show a severe reduction of recombination at the TCR-γ locus 10 11 12 or the distal elements of the V IgH locus 13. Mice with a deletion of IL-7 show a delay in rearrangement of the TCR-β, -γ, and -δ loci during fetal development 2.
In this report, we study in detail the recombination defects at the TCR-γ and -β loci and analyze at which stage during recombination the IL-7R−/− thymocytes are arrested by searching for V(D)J recombination reaction intermediates. To test whether the absence of IL-7 leads to a change in chromatin structure, we analyzed directly the accessibility of the TCR-γ locus to recombination-activating gene (Rag)-mediated cleavage in vitro.
Materials and Methods
PCR Analysis.
IL-7R−/− mice (C57BL/6 [B6], 129; reference 14) and IL-7R−/− B6 mice (bred onto C57BL/6J from The Jackson Laboratory) were bred as homozygotes in microisolators. Animal care was provided in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication no. 86-23, 1985). For each experiment, 6–10 mice were killed at the age of 4–8 wk and their thymi were pooled for DNA preparation. Each IL-7R−/− thymus 14 yielded ~2 × 105 thymocytes, whereas each control C57BL/6J thymus yielded >108 thymocytes. The IL-7R−/− thymocytes 14 used in this study are 99% negative for the cell surface markers CD4, CD8, or CD25. Fetal thymocytes (C57BL/6J day 14 of gestation) were >98% positive for CD44 and sorted into pro-T1 (95% negative for CD25) and pro-T2 populations (91% positive for CD25). To purify pro-T1 populations from adult animals, C57BL/6J thymocytes were partially predepleted of CD4+ and CD8+ cells using biotinylated CD4 and CD8 antibodies and streptavidin-coated magnetic beads (Dynabeads; Dynal). The predepleted cells were stained with the appropriate antibodies in the presence of the Fc receptor blocking antibody 2.4G2 and sorted into a population termed T1* that is negative for CD4, CD8, CD25, CD3, B220, and Mac-1 and >95% c-kit+ (CD117+) using a FACStarPLUS™ (Becton Dickinson). Genomic DNA (200 ng) was amplified in 50 mM KCl, 10 mM Tris-HCl, pH 8.3, 1.5 mM MgCl2, 0.001% gelatin, 0.5 U Taq polymerase (Perkin-Elmer Corp.), 0.2 pmol of each primer, 0.1 mM dNTP, for 30 cycles. Each cycle consisted of 1 min denaturation at 94°C, 1 min annealing at 60°C, and 1 min elongation at 72°C. The last cycle was followed by a 10-min elongation at 72°C. The DNA fragments were separated by agarose gel electrophoresis, blotted, and hybridized using a specific radiolabeled oligonucleotide. The primers used for detection of Vγ2-Jγ1 and Vγ3-Jγ1 are as in reference 12. The following primers were used: Vβ2 sense, 5′-GCTGGAGCAAAACCCAAGGTGG-3′; Vβ4 sense, 5′-CTGATATGCGAACAGTATCTAG-3′; Vβ8 sense, 5′-GATGACATCATCAGGTTTTGTC-3′; Vβ14 sense, 5′-GCCCTAACCTCTACTGGTAC-3′; Jβ2.1 antisense, 5′-GGACGGTGAGTCGTGTCCCTG-3′; Jβ2.1 probe, 5′-CTATGCTGAGCAGTTCTTCGG-3′; Vγ5 sense, 5′-CAACTTGGAAGAAAGAATAATG-3′; Vγ4 sense, 5′-GGAACGAGTCTCACGTCACCTCTG-3′; Vγ1.1 sense, 5′-GAAGTATCCGTCACCAGAGAG-3′; Jγ4 antisense, 5′-ACTACGAGCTTTGTCCCTTTG-3′; Jγ2 sense, 5′-GGAATTACTATGA-GCTTTG-3′; Vγ1.2 probe, 5′-GCCGGTACCAATGTATAGCTG-3′; and Vγ1.2 antisense, 5′-GTCACCAGAGAGACAGATGAG-3′.
Ligation-mediated PCR Analysis of TCR-β and TCR-γ Recombination Signal Sequence Breaks.
Genomic DNA (1 μg) was subjected to linker ligation as described 15. Individual aliquots of the linker-ligated DNA were analyzed for V(D)J recombinase–generated double strand DNA breaks at recombination signal sequences (RSSs) in the TCR-β and TCR-γ loci in PCR assays using two nested locus-specific primers (Jγ-C and Jγ-D, or Dβ2R2 and Dβ2R1) and the linker primer BW-H 15 16. PCR products were analyzed by gel electrophoresis and Southern blot hybridization using locus-specific oligonucleotide probes (Jγ-E and Dβ2R). Control reactions amplify the nonrearranging CD14 locus as described 17. The following primers and oligonucleotides were used: Dβ2R2, 5′-TGGCGTAATTTCCCATGCATGTACG-3′; Dβ2R1, 5′-GCTGATGGTTAACATGATAAGGGC-3′; Dβ2R, 5′-GAGTTGGTGTTTTTTTGGGTAG-3′; Jγ-C, 5′-TCAATTATCACTAACTCCAGGGAGAACA-3′; Jγ-D, 5′-GAAGATTGCAAGATATCAGATACCACA-3′; and Jγ-E, 5′-GACTTTCTAAGGAACCCAGAAGATCGG-3′.
In Vitro RSS Cleavage Assay.
Nuclear substrates were purified, and in vitro cleavage reactions were performed as described 17. In brief, cleavage reactions contained 100,000 nuclei and were incubated at 30°C for 1 h in the presence of 40 mM Hepes (pH 7.6), 2.5 mM Tris-HCl (pH 8.0), 110 mM KCl, 1 mM MnCl2, 2.5 mM dithiothreitol, 0.2 mM MgCl2, 0.05 mM PMSF, 0.025 mM EDTA, 7% glycerol (vol/vol), 0.6 μg rRAG-1, and 2 μg bovine thymus nuclear extract. Nucleic acid recovered from these reactions was subjected to ligation-mediated (LM)-PCR analysis as described above.
Results and Discussion
Suppressed Recombination at All TCR-γ Clusters in IL-7Rα−/− Thymocytes.
Targeting of the VDJ recombinase to the immune receptor genes is under cell type–specific, stage-specific, and locus-specific control (for reviews, see references 18 19 20). Thus, TCR genes only fully rearrange in T cells (Ig in B cells), and TCR-β locus rearrangement precedes that of the α locus. Furthermore, rearrangement of the TCR-γ gene is strictly regulated during ontogeny such that cluster 1 V regions 3 and 4 recombine first during fetal development followed by recombination of clusters 2 and 4 and the remaining elements in cluster 1 (Fig. 1 a). IL-7 is an extracellular signal that appears to induce certain loci to become accessible for recombination, as IL-7Rα−/− B cells fail to rearrange some IgV genes but succeed with others 13, and IL-7Rα−/− T cells fail to rearrange the TCR-γ locus 10 11 12 but recombine the TCR-β locus.
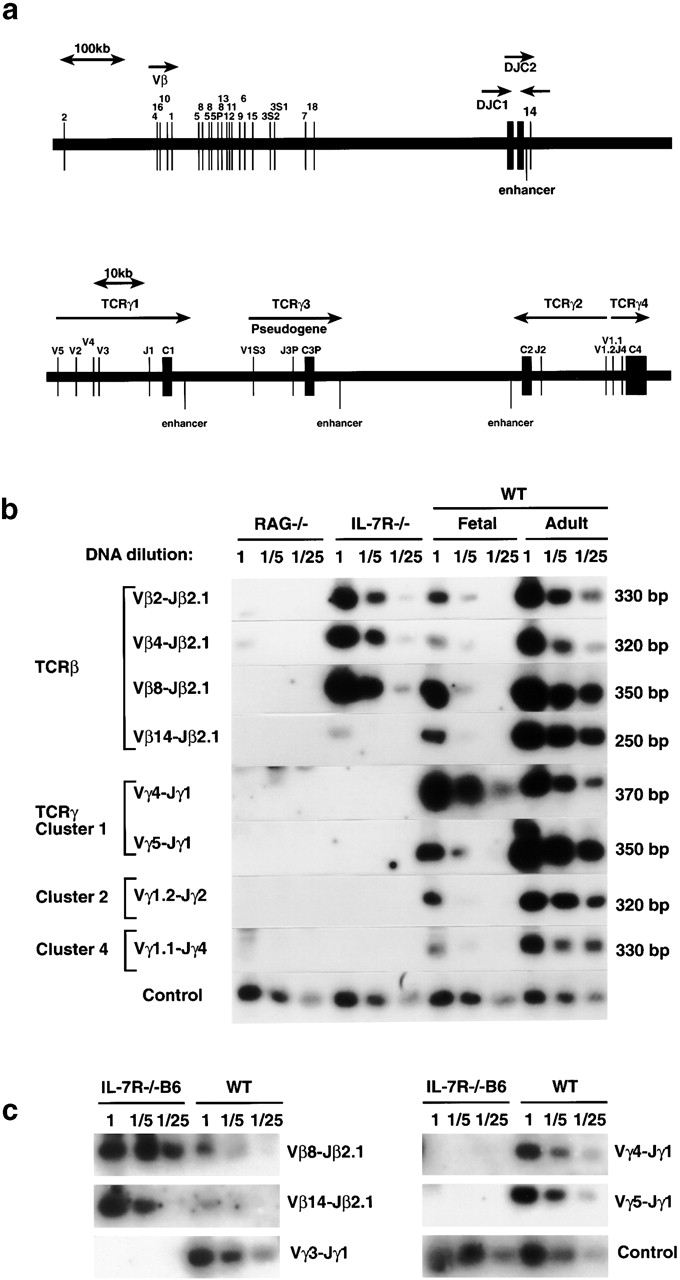
Suppression of recombination at all clusters of the TCR-γ locus. (a) Modified map of the murine TCR-β (top) and TCR-γ (bottom) loci (reference 32). (b) PCR analysis of genomic DNA from thymus of IL-7Rα−/− (reference 14), Rag-2−/−, or wild-type (WT) thymus from C57BL/6J mice derived at day 15 of gestation (Fetal) or at 4–8 wk of age (Adult). Control reaction amplifies V region of the TCR-γ locus (V2) independent of the recombination event. (c) PCR analysis of genomic DNA from thymus of IL-7Rα−/− B6 (IL-7Rα−/− strain cross-bred onto C57BL/6J for more than five generations) compared with wild-type (WT) thymus derived from C57BL/6J mice at day 15 of gestation. Control reaction amplifies V region of the TCR-γ locus (V2) independent of the recombination event.
Recent evidence demonstrates that there is also specific control for recombination within one locus. Mice with a deletion of the T early α (TEA) element in the TCR-α locus are only able to perform recombination for the most proximal Jα elements, but fail to do so at the distal Jα regions 21. IL-7Rα−/− precursor B cells successfully recombine proximal V regions of their IgH locus but not the distal V regions 13, and pax5−/− B cells recombine D to J but not V to DJ regions in the IgH locus 22. Since control of TCR-β rearrangement has many similarities with control of the IgH locus (e.g., both loci undergo D–J joining before V–D and both display allelic exclusion), we analyzed whether IL-7Rα−/− thymocytes might show deficient recombination of the distal V regions of TCR-β using the IL-7Rα–deficient strain originated by Peschon's laboratory 14. As shown in Fig. 1 b, Vβ8 and Vβ4 genes (located ~520 and 620 kb upstream of the D region elements, respectively) were recombined normally in IL-7Rα−/− thymocytes compared with wild-type thymus (fetal or adult). The most distal V region Vβ2 (150 kb upstream of the bulk of V regions) also showed a normal recombination frequency. However, successful recombination involving Vβ14, which is located downstream of the constant region C2 (and is rearranged by an inversion process), was less frequently detected in IL-7Rα−/− thymocytes.
In contrast to the relatively normal TCR-β pattern, the TCR-γ locus showed a severely reduced frequency of recombination at all three clusters (clusters 1, 2, and 4, as shown in Fig. 1 b). Clusters 1, 2, and 3 are 83–92% homologous in the regions consisting of J, C, and enhancer elements, and their enhancers are 98% identical 23 24. However, cluster 1 recombines before cluster 2 during fetal development (cluster 3 is a pseudogene). Thus, it is unlikely that the identified TCR-γ enhancer alone would be responsible for regulation of accessibility at the γ locus. Moreover, cluster 4 does not share the identified enhancer with the other clusters and cannot be recombined in the absence of IL-7 either. This suggests that the identified enhancer may be necessary but not sufficient for regulation of recombination and that as yet unidentified cis-acting control elements of the TCR-γ locus may be responsible for mediating the IL-7 effect for recombination.
The IL-7Rα–deficient mice used in this study show a severe phenotype such that the thymocytes are arrested at the pro-T1 cell stage 14. However, backcrossing these mice onto C57BL/6J strain results in partial restoration of α/β T cell development although at greatly reduced numbers (for a review, see reference 2). Thus, it was important to examine the effect of IL-7R signaling on TCR-γ locus recombination in the backcrossed strain. As shown in Fig. 1 c, severe suppression of TCR-γ gene rearrangements was also found in IL-7Rα−/− B6 mice. In contrast, Vβ14 rearrangement at the TCR-β locus was easily detectable in IL-7Rα−/− B6 thymocytes. Similarly, recombination intermediates at the Vβ14 locus could be detected using thymocytes from the IL-7Rα−/− B6 backcross but were absent in the original IL-7Rα−/− strain (data not shown). Thus, the control of recombination at the Vβ14 locus does not appear to depend solely on IL-7R signaling and can be substituted by as yet unidentified genetic factors. In contrast, control of recombination at the TCR-γ locus requires the IL-7R signal, and thus was chosen for further, more detailed studies.
Absence of γ Locus Recombination Intermediates in IL-7Rα−/− Thymocytes.
Successful V(D)J recombination involves the targeting of the recombinase to the appropriate site, the cleavage of the signal sequence, the generation of hairpin coding ends and blunt signal ends, and the ligation of these ends. To understand which of these recombination processes is controlled by the IL-7R signal, we analyzed at which stage during recombination of the TCR-γ locus the arrest occurred.
Recombination intermediates (signal sequences that have been cleaved but not yet religated) for the TCR-γ locus were examined using a previously described LM-PCR approach 15. Fig. 2 b shows that IL-7Rα−/− T cells had no detectable broken DNA ends at the RSSs within the TCR-γ locus, whereas the β locus serving as a control did have such broken ends. This result was observed using either the IL-7Rα−/− mixed background strain 14 and the IL-7Rα−/− B6 backcross. This indicates that IL-7Rα−/− thymocytes fail to initiate cleavage at the signal sequences in the TCR-γ locus, rather than failing in subsequent steps of recombination such as end modification or ligation. This is in contrast to the recently reported TCR-β enhancer knockout mouse 25 that failed to rearrange the TCR-β locus, but for different reasons: cleavage was initiated, although at reduced levels, but ligation of coding ends could not be successfully completed 16.
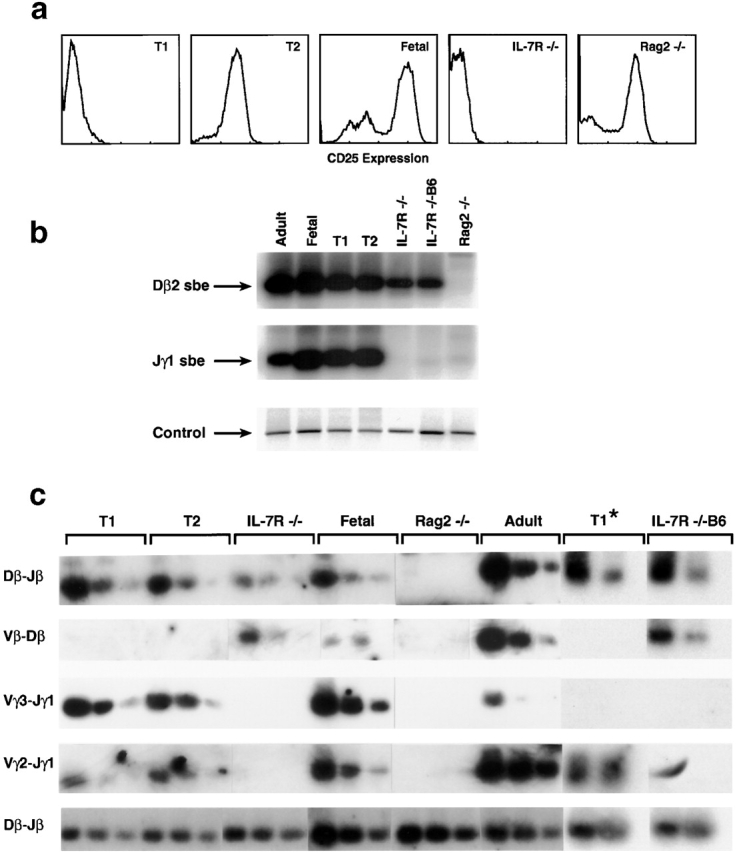
Failure to initiate recombination at the TCR-γ locus. (a) FACS® analysis for CD25 cell surface expression for thymocytes of IL-7Rα−/− mice (reference 14), Rag-2−/− thymocytes, or wild-type thymocytes from C57BL/6 mice at day 15 of gestation (Fetal). Pro-T1 and -T2 thymocytes were sorted by CD44/CD25 cell surface expression from C57BL/6J mice at day 14 of gestation. (b) To detect broken DNA ends in the TCR-γ or -β loci, genomic DNA from IL-7Rα−/− thymocytes 14, thymocytes from IL-7Rα−/− B6 backcrossed mice, Rag-2−/− thymocytes, or wild-type thymocytes from C57BL/6J mice at day 15 of gestation (Fetal) or 4–8 wk old (Adult) or pro-T1 or -T2 cells were linker ligated and analyzed by PCR. Control reactions amplify the CD14 gene that does not undergo recombination. sbe, signal broken end. (c) To detect complete rearrangements for the gene segments indicated to the right, PCR was performed using genomic DNA that was titered for better comparison in 1:5 dilution steps. T1 and T2 populations were purified from day 14 fetal thymocytes. T1* populations were purified as a CD4−CD8−CD25−c-kit+ population from adult thymus and directly compared with IL-7Rα−/− B6 backcrossed thymocytes using consecutive PCR analysis (20, 40, and 60 cycles).
IL-7Rα−/− thymocytes appear to be arrested at the pro-T1 stage based on their CD44+CD25− surface phenotype; this is the stage of the earliest thymic immigrants from bone marrow or fetal liver. Early thymic development then proceeds through pro-T2 (CD44+CD25+), pro-T3 (CD44−CD25+), and pro-T4 (CD44−CD25−) stages, followed by further intermediate and late stages. T cells with a deletion of either Rag-1 or Rag-2, which are unable to rearrange TCR genes, are arrested at the pro-T3 stage with high levels of CD25 expression (see Fig. 2 a). Thus, progression beyond pro-T3 requires gene rearrangement, and normal pro-T3 cells indeed show extensive rearrangements. It is not clear, however, at which stage rearrangement begins. As the TCR-β locus is rearranged in IL-7Rα−/−, it implies that this locus can undergo rearrangement at the pro-T1 stage. However, a previous study using Southern blot hybridization failed to detect TCR-β locus rearrangement at this stage 26. Determining the onset of TCR-γ rearrangement has relevance to the IL-7R mechanism, in that if it occurred later than the pro-T1 stage, IL-7Rα−/− thymocytes might fail to rearrange the γ locus because they fail to mature to the stage at which rearrangement normally occurs.
As shown in Fig. 2 b, recombination intermediates for both TCR-γ and -β loci were found in both pro-T1 and -T2 populations sorted from fetal thymocytes, indicating that the onset of rearrangement of both loci clearly begins in pro-T1 cells. The recombination is not only initiated at pro-T1 and -T2 but also successfully completed, since V–J joins can be detected in the γ locus and D–J (but not V–D) joins in the β locus in T1 as well as T2 populations sorted from fetal thymus (as shown in Fig. 2 c). For additional comparison, pro-T1 cells were purified from adult thymocytes as a CD4−CD8−CD25−c-kit+ population. Similarly to the fetal T1 populations, these T1 cells derived from adult mice also showed complete D–J joining (but not V–D joining) at the TCR-β locus as well as completed V–J joining at the TCR-γ locus. Another study failed to demonstrate D–J joining at the β locus in “adult” T1 populations using distinct purification techniques that may have depleted the recombined fraction we were able to detect 27. However, the T1 population we purified from fetal or adult thymocytes does not appear to contain mature T cell contaminants, since only D–J but not V–D joining could be detected at the TCR-β locus, indicating a discrete stage of T cell differentiation.
The finding that at least some rearrangements normally occur at the pro-T1 stage has two implications. First, it explains how IL-7Rα−/− thymocytes, whose surface markers correspond to pro-T1 cells 14, could initiate TCR-β rearrangements (which then become complete V–DJ joinings without expressing the usual T3 surface markers). Second, it shows that the TCR-γ locus can fully rearrange in pro-T1 cells; therefore, the failure of the TCR-γ locus to rearrange in IL-7Rα−/− cells is due to a selective failure to initiate cleavage of the TCR-γ locus, rather than to a failure to progress to later stages of development.
Reduced TCR-γ Locus Accessibility in IL-7Rα−/− Thymocytes.
IL-7Rα−/− thymocytes are unable to initiate cleavage at the TCR-γ locus (this paper), and we previously showed a reduction of sterile transcripts from the Vγ and Cγ regions 12. Recently, it has been demonstrated that the transcription factor signal transducer and activator of transcription 5 (Stat5) can partially replace the IL-7 signal for both germline transcription and rearrangement from the TCR-γ locus 28. Transcription is thought to be an indicator of “open” chromatin, which would also be accessible to the VDJ recombinase 29. However, recent evidence suggests that transcription and recombination may not always strictly correlate. Deletion of the TCR-β enhancer suppresses transcription of the locus but cleavage by the VDJ recombinase can still be performed (albeit at a reduced rate [16, 25]). Thus, we tested directly whether the TCR-γ locus in IL-7Rα−/− thymocytes was accessible to Rag-mediated in vitro cleavage 17. Nuclei purified from IL-7Rα−/− thymocytes were incubated with exogenous Rag protein. Nuclei from Rag-2−/− thymocytes were used as a control because they also fail to initiate cleavage at the γ locus, but have a normal accessibility to Rag cleavage at this site. Fig. 3 a shows that no broken DNA ends can be detected at the TCR-γ locus in the chromatin of IL-7Rα−/−or Rag-2−/− thymocytes before addition of the Rag proteins. However, after incubation with the Rag proteins, only the chromatin from Rag-2−/− thymocytes could be efficiently cleaved in contrast to chromatin from IL-7Rα−/− thymocytes. As shown in Fig. 3 b, the cleavage rate in vitro is greatly reduced for IL-7Rα−/− compared with Rag-2−/− nuclei. A small degree of cleavage that was detectable in IL-7Rα−/− thymocytes may indicate a minor pathway independent of IL-7R signaling that controls chromatin accessibility at the TCR-γ locus. However, this minor pathway does not lead to substantial Rag-mediated cleavage rates in vivo (Fig. 2 b). Thus, IL-7 is the main effector in controlling the accessibility of the chromatin at the TCR-γ locus for the V(D)J recombinase.
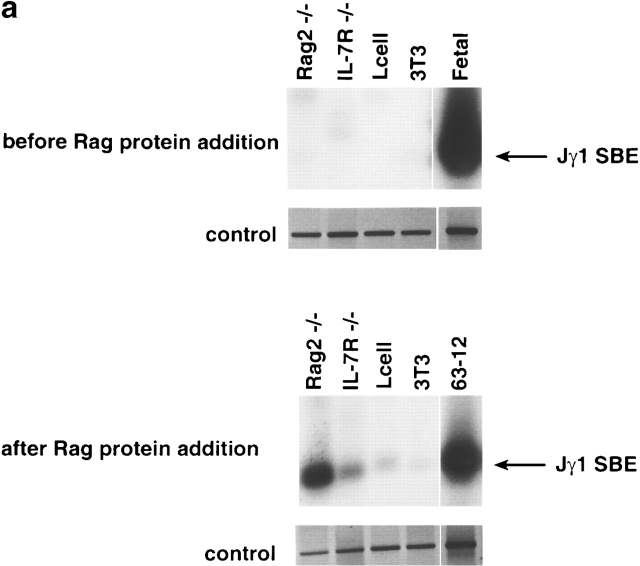
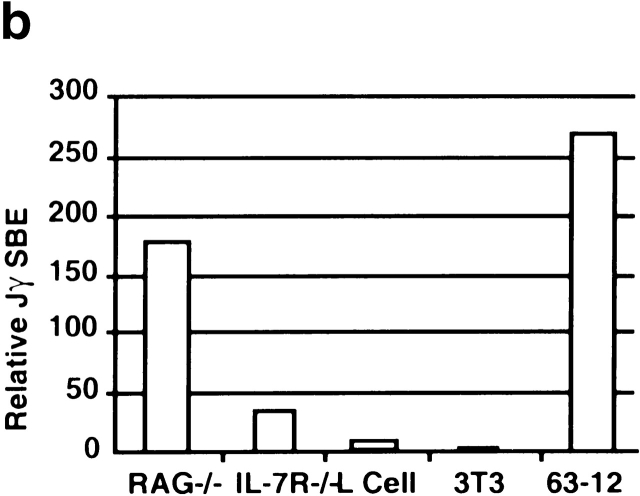
Reduced accessibility of the TCR-γ locus in IL-7Rα−/− thymocytes to Rag-mediated cleavage. (a) Nuclei were prepared from IL-7Rα−/− 14 or Rag-2−/− thymus, from fibroblast cell lines (3T3, L cells), or from the pro-B cell line 63-12 and incubated with recombinant Rag-1 and fetal cow thymus nuclear extract for 60 min. Genomic DNA was extracted before or after the addition of Rag proteins, linker ligated, and the PCR reaction was performed for detection of broken DNA ends. Control reactions amplify the CD14 gene that does not undergo recombination. SBE, signal broken end. (b) The results of the Rag-mediated cleavage experiment were analyzed by PhosphorImager® (Molecular Dynamics) and expressed as relative breakage at the TCR-γ locus.
Mononucleosomes, the smallest units of chromatin structure, have been shown in vitro to inhibit cleavage by the Rag proteins 30 31 and can be modulated by high mobility group 1 (HMG1) proteins 31. Several histone modifications have been suggested to relieve the inhibitory effects of mononucleosomes on transcription, among them histone acetylation. Trichostatin A, a specific inhibitor of histone deacetylase, can overcome the block caused by the absence of the IL-7 signal, suggesting that histone acetylation can relieve the block caused by mononucleosomes 12. However, the acetylation of histone tails only minimally influences the mononucleosomal structure directly. Thus, it is thought that histone acetylation may rather attract other factors that can interrupt mononucleosomal arrays. The sucrose nonfermenting (SNF) or nucleosome-remodeling factor (NURF) complexes, which contain SNF2 factors thought to be the ATP-driven motors of chromatin remodeling, are candidates. However, no recombination-specific mononucleosomal remodeling complex has yet been identified. Furthermore, no cis-acting DNA elements have been identified that are required for targeting the TCR-γ locus. The inaccessibility of the chromatin at the TCR-γ locus in the absence of the IL-7 signal may serve as a model to understand the mechanism by which chromatin is altered to allow for recombination.
Acknowledgments
This work was funded in part by a grant from the National Institutes of Health to M.S. Schlissel (AI40227). M.S. Schlissel is a Scholar of the Leukemia Society of America. This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-C0-56000. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- Candeias S., Muegge K., Durum S.K. IL-7 receptor and VDJ recombinationtrophic versus mechanistic actions. Immunity. 1997;6:501–508. [Abstract] [Google Scholar]
- Haks M.C., Oosterwegel M.A., Blom B., Spits H.M., Kruisbeek A.M. Cell-fate decisions in early T cell developmentregulation by cytokine receptors and the pre-TCR. Semin. Immunol. 1999;11:23–37. [Abstract] [Google Scholar]
- Hofmeister R., Khaled A.R., Benbernou N., Rajnavolgyi E., Muegge K., Durum S.K. Interleukin-7physiological roles and mechanisms of action. Cytokine Growth Factor Rev. 1999;10:41–60. [Abstract] [Google Scholar]
- Puel A., Ziegler S.F., Buckley R.H., Leonard W.J. Defective IL7R expression in T−B+NK+ severe combined immunodeficiency. Nat. Genet. 1998;20:394–397. [Abstract] [Google Scholar]
- Noguchi M., Yi H., Rosenblatt H.M., Filipovich A.H., Adelstein S., Modi W.S., McBride O.W., Leonard W.J. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. [Abstract] [Google Scholar]
- Macci P., Villa A., Gillani S., Sacco M.G., Frattini A., Porta F., Ugazio A.G., Johnston J.A., Candotti F., O'Shea J.J. Mutations of Jak-3 gene in patients with autosomal severe combined immunodeficiency (SCID) Nature. 1995;377:65–68. [Abstract] [Google Scholar]
- Kim K., Lee C.-K., Sayers T.J., Muegge K., Durum S.K. The trophic action of IL-7 on pro-T cellsinhibition of apoptosis of pro-T1, T2 and T3 cells correlates with Bcl-2 and Bax levels and is independent of Fas and p53 pathways. J. Immunol. 1998;160:5735–5741. [Abstract] [Google Scholar]
- Akashi K., Kondo M., von Freeden-Jeffry U., Murray R., Weissman I.L. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041. [Abstract] [Google Scholar]
- Maraskovsky E., O'Reilly L.A., Teepe M., Corcoran L.M., Peschon J.J., Strasser A. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor deficient mice but not in mutant rag-1−/− mice. Cell. 1997;89:1011–1019. [Abstract] [Google Scholar]
- Maki K., Sunaga S., Ikuta K. The V–J recombination of T cell receptor γ genes is blocked in interleukin-7 receptor–deficient mice. J. Exp. Med. 1996;184:2423–2427. [Europe PMC free article] [Abstract] [Google Scholar]
- Candeias S., Peschon J.J., Muegge K., Durum S.K. Defective T-cell receptor γ gene rearrangement in interleukin 7 receptor knockout mice. Immunol. Lett. 1997;57:9–14. [Abstract] [Google Scholar]
- Durum S.K., Candeias S., Nakajima H., Leonard W.J., Baird A.M., Berg L.J., Muegge K. Interleukin 7 receptor control of T cell receptor gamma gene rearrangementrole of receptor-associated chains and locus accessibility. J. Exp. Med. 1998;188:2233–2241. [Europe PMC free article] [Abstract] [Google Scholar]
- Corcoran A.E., Riddell A., Krooshoop D., Venkitaraman A.R. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. [Abstract] [Google Scholar]
- Peschon J.J., Gliniak B.C., Morrissey P., Maraskovsky E. Lymphoid development and function in IL-7R deficient mice. In: Durum S.K., Muegge K., editors. Cytokine Knockouts. Humana Press; Totowa, NJ: 1998. pp. 37–52. [Google Scholar]
- Schlissel M., Constantinescu A., Morrow T., Baxter M., Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. [Abstract] [Google Scholar]
- Hempel W.M., Stanhope-Baker P., Mathieu N., Huang F., Schlissel M.S., Ferrier P. Enhancer control of V(D)J recombination at the TCRbeta locusdifferential effects on DNA cleavage and joining. Genes Dev. 1998;12:2305–2317. [Europe PMC free article] [Abstract] [Google Scholar]
- Stanhope-Baker P., Hudson K.M., Shaffer A.L., Constantinescu A., Schlissel M.S. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–897. [Abstract] [Google Scholar]
- Krangel M.S., Hernandez-Munain C., Lauzurica P., McMurry M., Roberts J.L., Zhong X.P. Developmental regulation of V(D)J recombination at the TCR alpha/delta locus. Immunol. Rev. 1998;165:131–147. [Abstract] [Google Scholar]
- Schlissel M.S., Stanhope-Baker P. Accessibility and the developmental regulation of V(D)J recombination. Semin. Immunol. 1997;9:161–170. [Abstract] [Google Scholar]
- Roth D.B., Craig N.L. VDJ recombinationa transposase goes to work. Cell. 1998;94:411–414. [Abstract] [Google Scholar]
- Villey I., Caillol D., Selz F., Ferrier P., de Villartay J.P. Defect in rearrangement of the most 5′ TCR-J alpha following targeted deletion of T early alpha (TEA)implications for TCR alpha locus accessibility. Immunity. 1996;5:331–342. [Abstract] [Google Scholar]
- Nutt S.L, Urbanek P., Rolink A., Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell developmentdifference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–491. [Abstract] [Google Scholar]
- Kappes D.J., Browne C.P., Tonegawa S. Identification of a T-cell-specific enhancer at the locus encoding T-cell antigen receptor gamma chain. Proc. Natl. Acad. Sci. USA. 1991;88:2204–2208. [Europe PMC free article] [Abstract] [Google Scholar]
- Spencer D.M., Hsiang Y., Goldman J.P., Raulet D.H. Identification of a T-cell-specific transcriptional enhancer located 3′ of Cγ1 in the murine T-cell receptor γ locus. Proc. Natl. Acad. Sci. USA. 1991;88:800–804. [Europe PMC free article] [Abstract] [Google Scholar]
- Bouvier G., Watrin F., Naspetti M., Verthuy C., Naquet P., Ferrier P. Deletion of the mouse T-cell receptor β gene enhancer blocks αβ T-cell development. Proc. Natl. Acad. Sci. USA. 1996;93:7877–7881. [Europe PMC free article] [Abstract] [Google Scholar]
- Tourigny M.R., Mazel S., Burtrum D.B., Petrie H.T. T cell receptor (TCR)-β gene recombinationdissociation from cell cycle regulation and developmental progression during T cell ontogeny. J. Exp. Med. 1997;185:1549–1556. [Europe PMC free article] [Abstract] [Google Scholar]
- Ismaili J., Antica M., Wu L. CD4 and CD8 expression and T cell antigen receptor gene rearrangement in early intrathymic precursor cells. Eur. J. Immunol. 1996;26:731–737. [Abstract] [Google Scholar]
- Ye S.-K., Maki K., Kitamura T., Sunaga S., Akashi K., Domen J., Weissman I.L., Honjo T., Ikuta K. Induction of germline transcription in the TCRγ locus by Stat5implications for the accessibility control by the IL-7 receptor. Immunity. 1999;11:213–223. [Abstract] [Google Scholar]
- Schlissel M.S., Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of kappa gene transcription. Cell. 1989;58:1001–1007. [Abstract] [Google Scholar]
- Golding A., Chandler S., Ballestar E., Wolffe A.P., Schlissel M.S. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:3712–3723. [Europe PMC free article] [Abstract] [Google Scholar]
- Kwon J., Imbalzano A.N., Matthews A., Oettinger M.A. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol. Cell. 1998;2:829–839. [Abstract] [Google Scholar]
- Gascoigne N.R.J. Genomic organization of the T cell receptor genes in the mouse. In: Bell J.I., Owen M.J., Simpson E., editors. T Cell Receptors. Oxford University Press; Oxford: 1995. pp. 288–302. [Google Scholar]
Articles from The Journal of Experimental Medicine are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1084/jem.191.6.1045
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jem/article-pdf/191/6/1045/1126607/991727.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1084/jem.191.6.1045
Article citations
Advances in IL-7 Research on Tumour Therapy.
Pharmaceuticals (Basel), 17(4):415, 25 Mar 2024
Cited by: 0 articles | PMID: 38675377 | PMCID: PMC11054630
Review Free full text in Europe PMC
Harnessing the Power of IL-7 to Boost T Cell Immunity in Experimental and Clinical Immunotherapies.
Immune Netw, 24(1):e9, 15 Feb 2024
Cited by: 2 articles | PMID: 38455462
Review
Interleukin-7 receptor signaling is crucial for enhancer-dependent TCRδ germline transcription mediated through STAT5 recruitment.
Front Immunol, 13:943510, 19 Aug 2022
Cited by: 0 articles | PMID: 36059467 | PMCID: PMC9437428
αβ/γδ T cell lineage outcome is regulated by intrathymic cell localization and environmental signals.
Sci Adv, 7(29):eabg3613, 14 Jul 2021
Cited by: 2 articles | PMID: 34261656 | PMCID: PMC8279519
From thymus to periphery: Molecular basis of effector γδ-T cell differentiation.
Immunol Rev, 298(1):47-60, 15 Nov 2020
Cited by: 30 articles | PMID: 33191519 | PMCID: PMC7756812
Review Free full text in Europe PMC
Go to all (65) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Cutting edge: histone acetylation and recombination at the TCR gamma locus follows IL-7 induction.
J Immunol, 167(11):6073-6077, 01 Dec 2001
Cited by: 30 articles | PMID: 11714763
Interleukin 7 receptor control of T cell receptor gamma gene rearrangement: role of receptor-associated chains and locus accessibility.
J Exp Med, 188(12):2233-2241, 01 Dec 1998
Cited by: 82 articles | PMID: 9858510 | PMCID: PMC2212428
Defective T-cell receptor gamma gene rearrangement in interleukin-7 receptor knockout mice.
Immunol Lett, 57(1-3):9-14, 01 Jun 1997
Cited by: 31 articles | PMID: 9232418
Control of chromatin accessibility for V(D)J recombination by interleukin-7.
J Leukoc Biol, 69(6):907-911, 01 Jun 2001
Cited by: 16 articles | PMID: 11404375
Review
Funding
Funders who supported this work.
NIAID NIH HHS (3)
Grant ID: R37 AI040227
Grant ID: AI40227
Grant ID: R01 AI040227




