Abstract
Free full text

Somatic Hypermutation Introduces Insertions and Deletions into Immunoglobulin V Genes
Abstract
During a germinal center reaction, random mutations are introduced into immunoglobulin V genes to increase the affinity of antibody molecules and to further diversify the B cell repertoire. Antigen-directed selection of B cell clones that generate high affinity surface Ig results in the affinity maturation of the antibody response. The mutations of Ig genes are typically basepair substitutions, although DNA insertions and deletions have been reported to occur at a low frequency. In this study, we describe five insertion and four deletion events in otherwise somatically mutated VH gene cDNA molecules. Two of these insertions and all four deletions were obtained through the sequencing of 395 cDNA clones (~110,000 nucleotides) from CD38+IgD− germinal center, and CD38−IgD− memory B cell populations from a single human tonsil. No germline genes that could have encoded these six cDNA clones were found after an extensive characterization of the genomic VH4 repertoire of the tonsil donor. These six insertions or deletions and three additional insertion events isolated from other sources occurred as triplets or multiples thereof, leaving the transcripts in frame. Additionally, 8 of 9 of these events occurred in the CDR1 or CDR2, following a pattern consistent with selection, and making it unlikely that these events were artifacts of the experimental system. The lack of similar instances in unmutated IgD+CD38− follicular mantle cDNA clones statistically associates these events to the somatic hypermutation process (P = 0.014). Close scrutiny of the 9 insertion/deletion events reported here, and of 25 additional insertions or deletions collected from the literature, suggest that secondary structural elements in the DNA sequences capable of producing loop intermediates may be a prerequisite in most instances. Furthermore, these events most frequently involve sequence motifs resembling known intrinsic hotspots of somatic hypermutation. These insertion/deletion events are consistent with models of somatic hypermutation involving an unstable polymerase enzyme complex lacking proofreading capabilities, and suggest a downregulation or alteration of DNA repair at the V locus during the hypermutation process.
During the course of a T cell–dependent antibody response, B cells hone the specificity of their antibody molecules through a process of random somatic hypermutation of their V genes, followed by antigen driven selection. This is collectively referred to as affinity maturation. This process occurs within the germinal centers (GCs)1 of secondary follicles from peripheral lymphoid organs when antigen stimulated B cells receive proper signals from T and accessory cells. In the human system, GC B cells are characterized by the surface expression of CD38 and, in most cases, the loss of IgD (1–3). We have previously shown that the initiation of somatic hypermutation occurs within the CD77+ subset of these IgD−CD38+ B cells (4). Mutated V genes can be isolated from all subsequent stages of B cell differentiation and in cells from all IgD− and certain IgD+ B cell subsets (4, 5). The molecular process of somatic hypermutation remains elusive, primarily due to the lack of a good in vitro model until very recently (6). Much of what is known concerns: (a) localizing the somatic hypermutation process to particular B cell subsets and anatomical settings (4, 7–10); (b) delineating the limits and rates of mutational activity (11); (c) determining the minimal substrate through transgenic technology (12, 13); and (d) analyzing the mutations themselves in the context of the surrounding sequence to reveal tendencies such as strand polarity and “hotspots” of somatic hypermutation (for reviews see references 12 and 13).
Although somatic hypermutation is typically described as the generation of bp substitutions, insertions and deletions have been sporadically described. As with somatic point mutations, the analysis of these events can provide valuable information concerning somatic hypermutation itself. Analysis of human VH4 family genes generated from the amplification of cDNA from somatically mutated GC (IgD− CD38+) and memory (IgD−CD38−) B cell subpopulations led us to identify a number of cDNA clones from the mutated cell populations that contained insertions and deletions. We provide evidence that these events are linked to the somatic hypermutation process. Additionally, these events occur in a predictable fashion relative to the surrounding sequence, suggesting a model for their occurrence with implications for the molecular process of somatic hypermutation.
Materials and Methods
Isolation, Labeling, and Sorting of Tonsil B Cells.
Human tonsils were obtained during routine tonsillectomy. B cell isolation and sorting for CD38 and IgD expression were performed as previously described (4, 14). In brief, human tonsillar B cells were separated into IgD+CD38− follicular mantle (FM) B cells, IgD− CD38+ GC B cells, and IgD−CD38− memory B cells to 95–98% purity as predicted by FACS® analysis, as previously described (13). The mutation state of the VH gene cDNA clones from the various subpopulations was in agreement with our previous study (4). Clones were considered somatically mutated if they contained two or more bp substitutions, well beyond the expected error rates for the avian myeloblastosis virus reverse transcriptase (AMV-RT), Taq, and PFU polymerases used in these analyses (this mutation rate is based on our previous analyses; reference 4).
Sequencing the Ig VH Transcripts.
Total RNA was extracted from 1–5 × 105 B cells using guanidinium thiocyanate-phenol-chloroform in a single step using the Ultraspec RNA isolation system (BIOTECX Laboratories, Houston, TX), and was reverse transcribed using oligo-d(T) or specific V gene constant region oligonucleotides Cμ12 (5′-CTGGACTTTGCACACCACGTG-3′) for IgM transcripts or Cγ180 (5′-CTGCTGAGGGAGTAGAGTCC-3) for IgG transcripts and SuperScript II reverse transcriptase (GIBCO BRL, Gaithersburg, MD). First strand cDNA was used directly for second strand synthesis and amplification via PCR using internal primers corresponding to the Cμ or Cγ constant regions in combination with VH4 or VH6 family–specific leader oligonucleotides: Cγ140, 5′-GGCAAGGTGTGCACGCCGCTG-3′; Cμ10, 5′-TCTGTGCC CTGCATGACGTC-3′; L-4, 5′-ATGAAACACCTGTGGTTCTT-3′; L-6, 5′-ATGTCTGTCTCCTTCCTCAT-3′. The PCR products were purified using microconcentrators (Amicon, Beverly, MA), and then were kinased and blunt-end ligated into an EcoRV-digested and dephosphorylated pBluescript plasmid (Stratagene, La Jolla, CA; Polynucleotide Kinase, T4 DNA Ligase, and EcoRV were from Boehringer Mannheim, Amsterdam, Netherlands). After transformation by electroporation into electro-competent DH10α Escherichia coli (GIBCO BRL) and screening with consensus internal oligonucleotides as previously described (4, 15), positive colonies were picked, plasmid mini-preparations were made, and colonies were sequenced in both directions using an automated DNA sequencer and automated sequencer protocol (ABI-377; Advanced Biotechnologies Inc., Columbia, MD). All sequences were analyzed using DNAstar (DNAstar Inc., Madison, WI). In the first tonsil analyzed, 583 clones were picked, plasmid mini-preparations were made, and Southern blots were prepared by standard methods. These blots were screened with a set of oligonucleotides specific for the various VH4 family genes. Only those clones that screened positive with constant region probes but negative for the various VH4 complementarity-determining region (CDR)1–specific probes were sequenced (395 of 583 clones), thus enriching the somatically mutated populations analyzed, in that the CDR1 probes should anneal only to the sequences most similar to germline. The frequency of the occurrence of these events can therefore only be predicted to be between 6 out of 395 and 6 out of 583 clones (1–2%). Any sequence of interest was resequenced in both directions to ensure sequence fidelity.
Characterizing the Genomic Repertoire.
Total genomic DNA was isolated from FM B cells (IgD+, CD38−) using the Puregene DNA isolation kit (Gentra Systems, Inc., Minneapolis, MN). VH4 genes were amplified using a VH4 leader-specific primer (L-4, as above) and a primer specific for all VH4 gene family heptamer–nonamer spacer regions as previously described (16). PCR products were agarose gel purified, then cloned into E. coli as described above for the cDNA clones. Clones identified in the cDNA analysis that contained insertions or deletions were used to design PCR primers to amplify both the exact sequence of clones with insertions/deletions as found and the predicted sequences based on the proposed germline counterparts. Oligonucleotides used in this analysis (Format, is as follows: clone: exact/predicted): g64:5′-GGACGGGTTGTACTTGGTTCC-3′/5′-GGACGGGTTGTAGGTCTCC-3′; g144:5′-TCTTGAGGGACGGGTTGGTGT-3′/5′-TCTTGAGGGACGGGTTGT-3′; g187:5′-CAGCTCCAGTAGTAAGCCCCG-3′/5′-CAGCTCCAGTAGTAACCACCG; g188: 5′-GAGGGATTGTAGTTGGAGCC-3′/5′-GAGGGGTTGTAGTTGGTCCC; g192:5′-CCAGCCCCAGTAGTAGTAACT-3′/(same); and g80:5′-GCGGATCCAATACCTCACACT-3′/ 5′-GCGGATCCAGTAGTAACC-3′.
Sequence Availability.
All cDNA sequences with insertions or deletions, and any genomic sequences unique to the literature as described in the results section are available from EMBL/Genbank/ DDBJ under accession numbers AF013615 through AF013626.
Assay for Screening VH Gene Lengths.
To facilitate the analysis of large numbers of VH gene transcripts for the presence of insertions or deletions, first strand cDNA produced as described above was PCR amplified using Expand high fidelity polymerase (Boehringer Mannheim) to reduce errors resulting from Taq polymerase alone. The products of this PCR amplification were cloned as described above and screened using 32P-labeled, gene-specific oligonucleotides (VH4-39:5′-ATTGGGAGTATCTATTATAGT-3′; L-6 as above). Positive colonies were picked and used to inoculate overnight cultures. A 1 μl aliquot from each 24-h culture was used to directly inoculate 25-μl PCR amplification mixtures in 96-well–format PCRs. The internal PCR reactions used 32P-labeled, gene-specific oligonucleotides to amplify a 230-base fragment including the VH4-39 CDR1 (L-4, as above, and VH4-39-3′: 5′-GCTCCCACTATAATAGATACT-3′) or for analysis of VH6 genes a 166-nucleotide fragment including the CDR1 and CDR2 of VH6 (VH6FW1: 5′-TGCCATCTCCGGGGACAGTGT-3′, VH6FW3: 5′-TGTGTCTGGGTTGATGGTTAT-3′). Aliquots of each clone were also used to inoculate amplifications of CDR3 regions using FW3-specific (ssFW3: 5′-CTGAA[C/G]CTGAGCTCTGTGAC[T/C]) and Cμ- or Cγ-specific oligonucleotides (CμD: 5′-GGAATTCTCACAGGAGACGA-3′, Cγ-140 as above) to analyze the diversity of the populations under study; the distribution of CDR3 size variations of several hundred VH sequences cloned in this analysis were used to produce an expected distribution of CDR3 sizes for comparison (see Fig. Fig.55 B). The amplification products were electrophoresed on 0.6X-TBE, 5% urea-acrylamide sequencing gels (Long Ranger; J.T. Baker, Phillipsburg, NJ) and analyzed with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA.) using the Image Quant software supplied by the manufacturer. Clones that differed from the expected size and those clones in lanes adjacent to aberrantly migrated bands were used to produce plasmid preparations from which the inserts were sequenced in either direction.
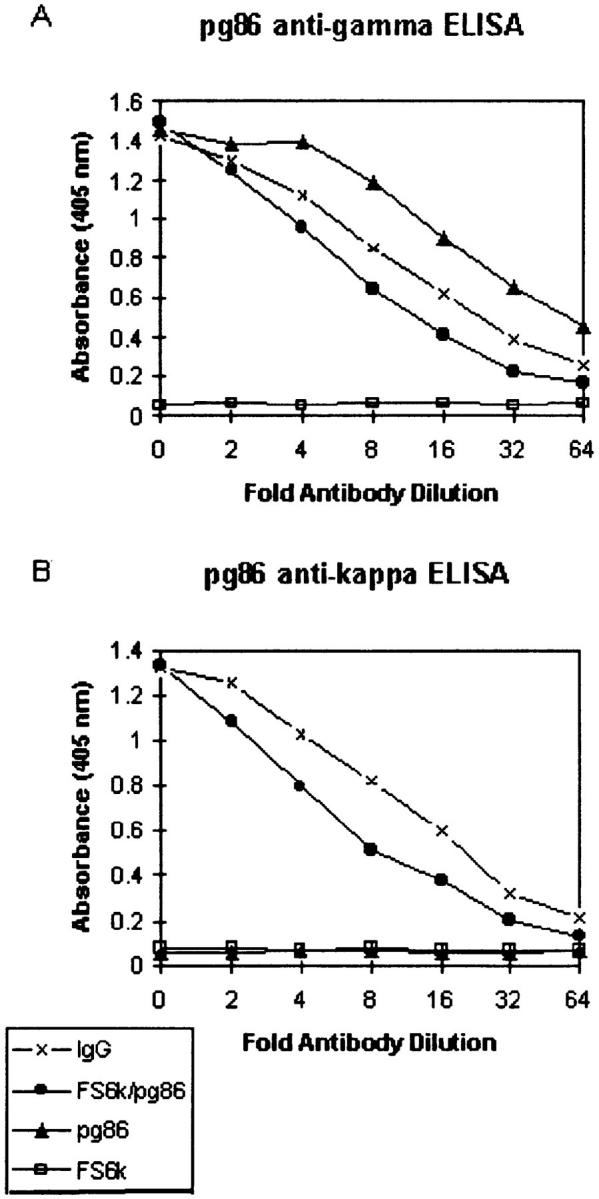
ELISA assays showing the expression of clone pg86 with a six amino acid insertion at the FW1/CDR1 junction. Clone pg86 (IgG heavy chain) was coexpressed with the κ light chain FS6κ in insect cells using the baculovirus expression system. Expression of pg86 and its ability to pair with the FS6κ light chain was tested using capture ELISAs. Wells were coated with goat anti–human IgG. Supernatants containing recombinant antibodies were added in serial twofold dilutions. Bound antibody was detected with phosphatase-conjugated goat anti-human IgG (A), and goat anti–human Cκ (B).
Scoring of Insertion/Deletion Events.
In the results section, insertions and deletions are scored as events per 104 nucleotides within the customary boundaries of CDR1 and CDR2. This unit was chosen because in the selected populations studied these events are generally only found in the CDR regions and therefore the comparison of events per total nucleotides would be misleading. In the PAGE analysis, each VH4-39 FM clone included only the CDR1 (21 nucleotides) within a total of 230 nucleotides/clone, whereas each VH6 FM clone was only 166 nucleotides but included both the CDR1 and CDR2 (75 CDR nucleotides). In the sequencing analysis, various B cell populations were analyzed involving a wide range of overall lengths. Comparisons of the frequency of insertions/deletions just within the CDRs allowed for a more standardized and quantitative analysis, and for more freedom in experimental design.
Baculovirus Expression System.
Cloning and coexpression of clone pg86 and κ light chain FS6κ in the baculovirus expression system was performed as previously described (17). Recombinant Autographa californica nuclear polyhedrosis virus (AcMNPV) was cloned using the pH360NX transfer vector and expressed in Sf9 cells.
Capture ELISA for γ Heavy Chain, and κ Light Chains.
Expression of recombinant antibodies of clone pg86 coexpressed with κ light chain FS6κ were measured by capture ELISA. Wells were coated with goat anti–human IgG and incubated with supernatant of recombinant pg86/FS6κ added in serial twofold dilutions. Bound antibody was detected using alkaline phosphatase–conjugated goat anti–human IgG, or goat anti–human Cκ. After 1-h incubation at 37°C, phosphatase substrate was added and absorbance was measured at 405 nm in an ELISA plate reader.
Results
Insertions and Deletions into Immunoglobulin VH Genes.
In a large scale analysis of VH genes from both the IgM and IgG compartments of B cell subpopulations separated from a single human tonsil, six clones that contained DNA insertions or deletions were isolated. These insertions and deletions were apparently selected in that they involved nucleotide triplets or multiples of nucleotide triplets, leaving the cDNAs (transcripts) in frame, and they were localized to the CDR1 and CDR2 (Fig. (Fig.1,1, A and B). The six clones with insertions or deletions were identified from the sequencing of 395 cDNA clones (~110,000 nucleotides) from GC and memory B cell subpopulations, resulting in a frequency of <2% of clones analyzed (~1 event/18,000 nucleotides). All six events were in IgG transcripts. Two events were obtained from IgD−CD38+ GC and four events from IgD−CD38− memory cell populations. None of the IgM VH cDNAs analyzed from this tonsil had insertions or deletions, although we have observed such events in IgM transcripts in the past and in subsequent analyses, as described below.

Predicted amino acid sequences of nine cDNA clones with insertions or deletions. Note that all of these clones are extensively mutated. In all but clone tm121, the insertions or deletions occur in CDR1 or CDR2. (A and B) Two clones with insertions (A) and four clones with deletions (B) from a single tonsil. (C) Three additional cDNA clones with insertions isolated from various sources. Sequence data available from GenBank/DDBJ under accession numbers AF013615 through AF013626.
The Insertions and Deletions Are Not Germline Encoded.
The analysis described above focused on the VH4 gene family, which consists of 10–14 members/genome, varying slightly between individuals (16, 18). As shown in Fig. Fig.2,2, the major difference between VH4 genes involves the length of CDR1. Because genomic diversity between VH4 family members resembles the events described in this paper we had to rule out possible alternative explanations for these events, such as: (a) different alleles of the detected genes; (b) rarely expressed or otherwise unknown VH4 gene family members; or (c) hybrids between known and detected VH genes and/or other artifacts of the experimental system. To address these issues, both the expressed and genomic repertoires from this tonsil were characterized. As indicated in Table Table1,1, 2 out of 118 VH4–39, 2 out of 49 VH4–31, 1 out of 87 VH4–34, and 1 out of 45 VH4–59 cDNA clones contained insertion/deletion events. cDNA clones were judged as unique isolates based on CDR3 analysis, and the few isolates that appeared to be clonally related differed in their patterns of somatic mutation beyond the level explainable by reverse transcription and PCR errors (maximum: >1 mutation/500 nucleotides of VH gene sequence as previously described [4]).
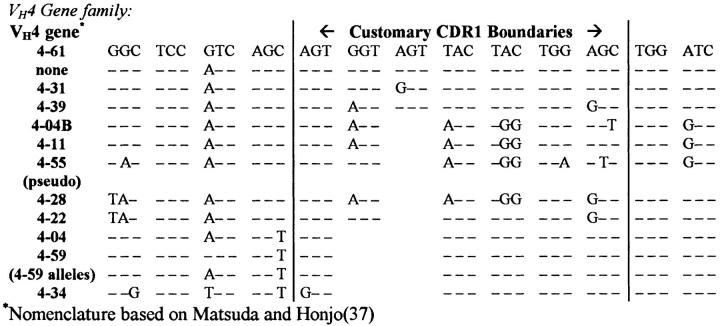
Comparison of the CDR1s of the human VH4 germline genes. The primary variability between VH4 family members is 3–6-bp size variances in the CDR1s which is similar to the short insertions and deletions that we attribute to somatic hypermutation in the selected B cell populations studied in this report.
Table 1
cDNA and Germline Clones Isolated
| VH4 gene alleles isolated* | cDNA clones with ins/del | Total cDNA clones isolated | Germline clones isolated‡ | |||
|---|---|---|---|---|---|---|
| VH4–39 | 2 | 113 | 7 | |||
| VH4–31 | 2 | 49 | 8 | |||
| VH4–59 | 1 | 45 | 4 | |||
| VH4–34 | 1 | 87 | 11 | |||
| VH4–34 related | 0 | 0 | 4 | |||
| VH4–55 pseudogene§ | 0 | 0 | 12 | |||
| VH4–55-related pseudogene§ | 0 | 0 | 3 | |||
| VH4–04 | 0 | 17 | 7 | |||
| VH4–04-related pseudogene§ | 0 | 0 | 2 | |||
| VH4–61 | 0 | 25 | 7 | |||
New VH4 gene
| 0 | 33 | 3 | |||
| VH4–04B | 0 | 72 | 1 | |||
| VH4–28 | 0 | 0 | 1 |
 Nomenclature based on Matsuda and Honjo (37).
Nomenclature based on Matsuda and Honjo (37).  Nine unusual isolates were also cloned consisting of hybrids of two of the indicated genes, resumably do to PCR artifact. None of these artifacts were altered in size or resembled any of the insertion or deletion events observed.
Nine unusual isolates were also cloned consisting of hybrids of two of the indicated genes, resumably do to PCR artifact. None of these artifacts were altered in size or resembled any of the insertion or deletion events observed.  Pseudogenes contain stop codons or frameshift mutations and are not expressed.
Pseudogenes contain stop codons or frameshift mutations and are not expressed.  Newly identified VH4 gene is most closely related to VH4–04.
Newly identified VH4 gene is most closely related to VH4–04. To characterize the genomic repertoire of the initial tonsil, 80 germline VH4 gene clones were isolated and sequenced (Table (Table1),1), which encompassed all 14 VH4 family members or alternate alleles represented in the 446 cDNA clones analyzed from all of the tonsillar B cell subsets. In the course of this study, we isolated the germline counterpart of a novel VH4 gene segment for which transcripts had been found. In addition, germline genes corresponding to two apparently functional VH4 genes not found as cDNA clones in this analysis were isolated, as well as one nonfunctional VH4 gene and a divergent polymorphism of a known VH4 pseudogene. The proposed germline counterparts of each of the VH4 genes containing insertion/deletion events were isolated from 4 to 11 times (Table (Table1).1). 8 independent genomic isolates of VH4–31 and of VH4–39 were cloned. VH4–34 and VH4–59 were isolated 11 and 4 times, respectively. No germline genes were isolated that could have encoded the insertion/deletion events described.
To further be certain that the insertion/deletion events described herein were not germline encoded, two sets of PCR primers were designed to specifically recognize: (a) the exact sequence of the events; (b) the predicted, unmutated, germline sequence corresponding to the cDNAs containing insertion and deletion events. These primers were used to amplify genomic DNA from this individual, yielding negative results (data not shown). The unique nature of these events relative to both the expressed and genomic repertoire and our inability to amplify genomic counterparts for these events by PCR suggest that they are not germline encoded.
The Proposed Insertion/Deletion Events Are Not the Result of (VH/VH) Recombination.
As in most V gene repertoire analyses, we detected hybrid VH sequences that could be the result of either PCR splicing by overlap extension artifacts, or reciprocal homologous recombination between unrearranged V genes (19). However, none of these likely artifactual events were altered in size such that they resembled the insertion or deletion of DNA described above. A number of artifacts of this type had been isolated in the cDNA analysis as well; such artifacts are common to V gene analyses (20). The cDNA isolates with deletion and insertion events were stringently compared to all germline and cDNA isolates and were found to be unique relative to both the expressed and germline VH4 gene repertoires of this individual, supporting a somatic origin for their occurrence.
The Insertions and Deletions Are Associated with Somatic Hypermutation.
To determine whether or not these insertion/deletion events were associated with somatic hypermutation, we analyzed their occurrence in unmutated FM transcripts. This was done using either direct sequencing or PCR amplification of portions of the VH genes spanning the CDRs, followed by size comparisons on polyacrylamide gels (Fig. (Fig.3).3). Any clones that ran aberrantly, and the clones in adjacent lanes, were sequenced (75 out of the 485 clones). None of these 75 clones were related based on CDR3 homology. To ensure that the remaining 410 FM clones were polyclonal, the CDR3s were PCR amplified and loaded on the sequencing gels simultaneously to the VH gene amplification products for size comparisons (Fig. (Fig.33 A). The size distribution of these CDR3s was similar to that of ~500 VH gene sequences analyzed in this study (Fig. (Fig.33 B), providing evidence that our FM sample is polyclonal.

Polyacrylamide gel assay to identify insertions or deletions into VH genes. (A) Phosphorimage of a polyacrylamide gel: each lane contains the hot-PCR products (32P-labeled) of the VH gene and the CDR3 of an individual clone. (B) A comparison of the distribution of CDR3 sizes of the 485 CDR3s assayed to the distribution of 500 CDR3s observed in sequences from this report indicates that the clones assayed by electrophoresis were a polyclonal population. CDR3 sizes were measured from the most 3′ Tyr residue (common to all V genes analyzed) to the most 5′ Cμ or Cγ residue. CDR3 lengths for those assayed by electrophoresis were extrapolated based on sequencing of 75 out of the 485 clones assayed. The x-axis is the number of amino acids greater than the shortest CDR3 found.
The six events detected from a single tonsil were isolated from 395 mutated cDNA clones (25,482 CDR nucleotides), corresponding to a frequency of 2.35 events/104 CDR nucleotides. This is significantly different (p=0.014 by a one-sided χ2 test) from the analysis of unmutated FM-derived clones (25,515 CDR nucleotides) that yielded no insertions or deletions (Table (Table2).2).
Table 2
Analysis of Unmutated FM cDNA Clones for Insertion or Deletion Events
| Clone type | Clones assayed | CDR nucleotides‡ | Events observed | Frequency§ | Expected (events/104 CDR nucleotides)‖ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutated VH4 clones (GC and | 395 | 25,482 | 6 | 2.35 events/ | ||||||
 and memory B cells) and memory B cells) |  104 mn 104 mn | |||||||||
| Unmutated clones: | ||||||||||
 VH4–FM, CDR1* VH4–FM, CDR1*
| 265 | 5,565 | 0 | 0 | 1.31 | |||||
 VH6 IgM FM VH genes* VH6 IgM FM VH genes*
| 220 | 16,500 | 0 | 0 | 3.88 | |||||
 VH4 family FM sequences VH4 family FM sequences | 51 | 3,450 | 0 | 0 | 0.81 | |||||
| Total unmutated values | 25,515 | 0 | 0 | 2.35 events/104 CDR | ||||||
| (P = 0.014)¶ | nucleotides |
 Clones analyzed by hot-PCR/PAGE assay as described in the text.
Clones analyzed by hot-PCR/PAGE assay as described in the text.  CDR nucleotides are those within the customary bounds of the CDR1 and CDR2. (See Materials and Methods for a more detailed explanation of this unit).
CDR nucleotides are those within the customary bounds of the CDR1 and CDR2. (See Materials and Methods for a more detailed explanation of this unit).  Events per 104 CDR nucleotides.
Events per 104 CDR nucleotides.  Expected frequency (events/104 CDR nucleotides) derived from sequencing data: 6 events in 25,482 CDR nucleotides; 6/(25,482 CDR nucleotides/104) = 2.35.
Expected frequency (events/104 CDR nucleotides) derived from sequencing data: 6 events in 25,482 CDR nucleotides; 6/(25,482 CDR nucleotides/104) = 2.35.  Statistical analysis: χ2 test for independence.
Statistical analysis: χ2 test for independence. In the course of the analysis described above, we isolated one IgM clone containing a 6-nucleotide insertion into framework (FW)3 (see below). We believe that this clone is part of the mutated GC or memory repertoire because it contained 4 bp substitutions in addition to the insertion. In this study, the B cell populations analyzed were 95–98% pure, and the FM B cell subpopulation could therefore include between 2 and 5% contaminating clones, that is, IgM-expressing cells not from the naïve population that can therefore be somatically mutated. However, none of the unmutated FM clones analyzed had insertions or deletions.
Other Insertions and Deletions into VH Genes.
We have observed similar instances of insertions and deletions into the coding regions of apparently functional immunoglobulin V genes, including: (a) a VH6 IgM isolate containing a triplet duplication/insertion into the CDR1 in addition to several bp substitutions (Figs. (Figs.11 C and and44 A), which was derived from a human hybridoma secreting high affinity mAb against Bordetella pertussis (21, 22); (b) a 6-nucleotide insertion into the FW3 region of a mutated IgM VH6 gene, representing the only insertion or deletion observed outside of the CDRs (Figs. (Figs.11 C and and44 A, clone tm121); and (c) an 18-nucleotide duplication/insertion into a human plasma cell cDNA transcript at the boundary between the FW1 and CDR1 (Figs. (Figs.11 C and and44 C), doubling the length of this hypervariable loop. The viability of clone pg86 was tested by expressing it in the baculovirus system in association with a κ light chain encoding construct (FS-6κ; Fig. Fig.5).5). The efficient expression, secretion, and pairing with light chain in the baculovirus system suggest that the product of clone pg86 is a functional heavy chain despite the large duplication/insertion.

The insertions and deletions are related to the surrounding DNA sequence. (A) The insertions involve repetitions of the immediately adjacent sequence. (B) The deletions are deletions of tandem repeats. (C) The 18-base insertion in clone pg86 is a duplication of the adjacent sequence. Nucleotides that mutated before the duplication/insertion are indicated. Sequence data available from EMBL/GenBank/DDBJ under accession numbers AF013615 through AF013626.
The Insertions and Deletions Are Related to the Surrounding Sequence.
As shown in Fig. Fig.4,4, the insertions reported are duplications of the immediately adjacent sequence, and the deletions involve elements of repetitive tracts. In addition, a higher incidence of these events involve sequence motifs that resemble intrinsic hotspots of somatic hypermutation (12, 23–27): (a) four of eight events involved the serine codon AGC that has been reported as the “hottest” of hotspots (24–27) (Fig. (Fig.4,4, sequences HBp2, g187, g188, and g86); (b) two events involved TAC motifs (Fig. (Fig.4,4, g192 and g64); and (c) two events involved the motif AAC (Fig. (Fig.1,1, g144, and tm121). In general all of the clones found to contain insertions and deletions were highly mutated (Fig. (Fig.1).1). Several of these clones had bp substitutions clustered with the insertions or deletions (Figs. (Figs.11 and and4).4). The plasma cell transcript depicted in Fig. Fig.44 C contained an 18-nucleotide insertion that duplicated the 5′ adjacent sequence. The central nine nucleotides of the duplicated sequence form a partial palindrome (..GGtGaCtCC..). This clone was mutated (G to A at position 80 and an A to T at position 85) before the duplication/insertion event, as these mutations were perpetuated in the inserted sequence.
Discussion
Somatic modification of V genes encoding immunoglobulin and T cell receptors recapitulates most mechanisms observed in the evolutionary diversification of DNA: (a) V gene recombination, including imprecise junctions, P nucleotides, and untemplated N nucleotide addition; (b) gene conversion; and (c) bp substitutions in Ig somatic hypermutation. The insertion and deletion of nucleotides is another means for the evolutionary diversification of DNA, and has been proposed as an explanation for unusual V gene sequences in the past (Table (Table3).3). In this study, we show that insertions and deletions are associated with the somatic hypermutation process.
Table 3
Insertions and Deletions into Somatically mutated V Genes Reported in the Literature
| Name | Source | Ins/Del (position) | Relation to surrounding sequence | References | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Selected populations or coding regions: | ||||||||||
 L4-le L4-le | Human VH4–34 (4.21) | ACC insert (within CDR2) | 4-34: | AGC ACC AAC (RT) | 38 | |||||
| L4-1e: | AGg ACC ACC AAC (RT) | |||||||||
 3B62 3B62 | Murine VH 186.2 | GTT deletion (CDR2) | VH186.2: | AGT GGT GGT ACT (RT) | 39 | |||||
| 3B62: | AGT GGT ACT | |||||||||
| Unselected populations or untranslated regions: | ||||||||||
 3B62 3B62 | Murine VH 186.2/D/JH2 to JH4 | ACT deletion (3′ untranslated) | GL: | GTG ACT ACT TTG (RT) | 40 | |||||
| 3B62: | GTG ACT TTG | |||||||||
 3B44 3B44 | Murine VH 186.2 | 4 single-base deletions (leader intron) | VH186.2: | GGC, GGT (RT) | 39, 40 | |||||
| 3B62: | G C, G T | |||||||||
| Other 2 events unrelated | ||||||||||
 M167 M167 | Murine VH107/DFL16.1/JH1 | 2 single-base insertions (leader intron) | GL: | ATAG AAGATTAGTAG (RT) | 41 | |||||
| M167 | ATAGTAAGATTAGTAG | |||||||||
| (3′ untranslated) | GL: | TTTC AGGTCATGAAGGA (RT) | 29, 41* | |||||||
| M167: | TTTgAAGGTCATGAAGGA | |||||||||
| 5 single-base deletions | GL: | GCTTTTC TGTA...CCCAGGAAAAGA | ||||||||
| M167 | GCTTTTC TGTA...CCCAG AAAA GA (IR) | |||||||||
| (all in 3′ untranslated region) | GL: | CTTTTTTCTT (RT) | 41 | |||||||
| M167 | CTTTT TCTT | |||||||||
| GL: | AGATTTTAC (RT) | |||||||||
| M167 | AGAccT AC | |||||||||
| GL: | TCATTGG (RT) | |||||||||
| M167 | TCAT GG | |||||||||
| GL: | GTGACTACTTTGACTACTG (RT) | |||||||||
| M167 | GT ACTACTTTGACTACTG | |||||||||
| TA deletion (3′) | None found (possible hotspot) | |||||||||
| GTGT deletion (leader intron) | GL: | TCTGTGTGTGTGTAT (RT) | ||||||||
| M167: | TCTGTGT GTAT | |||||||||
 M603 M603 | Murine VHS10 7/DFL16.1/JH1 | TC Deletion (leader intron) | GL: | TTTTCCTTGTCCTTGTTTT (RT) | 42, 41 | |||||
| M603: | TTTTCCTTG CTTGTTTT | |||||||||
| AAAT deletion (3′ Ei/MAR) | GL: | GCATTTCTAAAATAAGTTGAGGA (IR) | ||||||||
| M603 | GCATTTCTA AAGTTGAGGA | |||||||||
 MC101 MC101 | Murine VHQ52/D/JH3 | GG deletion (3′ untranslated) | GL: | AAACGGGAATC (RT) | 42, 43 | |||||
| MC101: | AAAC GAATC | |||||||||
 M511 M511 | Murine Vκ167/Jκ5 | GAA deletion (3′ untranslated) | GL: | TTTGAAGATAAA (RT) | 42, 44 | |||||
| m511: | TTT GATAAA | |||||||||
 H37-65 H37-65 | Murine Vκ Vκ21E/Jκ1-Jκ2 | 11 base deletion (Jκ1/Jκ2 intron) | GL: | AGGGACACCAGTGTGTGTACAC (IL) | 45 | |||||
| H37: | AGGGc GTACAC | |||||||||
 296.4C11, 253.12D3 296.4C11, 253.12D3 | Murine JκC intron | 7 base deletion and a 154 base deletion | No good relationship to surrounding sequence | 46 | ||||||
 2G7 2G7 | Murine transgene | single base deletion, and a 49 | CTTTGAAGAT . . (N30) . . CAGATCAAAG | 47 | ||||||
 nucleotide deletion nucleotide deletion | (Repeats form ends of deleted “loop”) (IL) | |||||||||
 85κ 85κ | Human myeloma Vκ genes | single-base (T) insertions into the | No relation, however, event followed the proposed | 48 | ||||||
 CDR1/FW2 junction rendering CDR1/FW2 junction rendering  genes out-of-frame genes out-of-frame |  hotspot motif TAC hotspot motif TAC | |||||||||
 HF-1‡ clone A6: HF-1‡ clone A6: | Human lymphoma (JH untranslated) | AG insertion into VH3′ | Consensus‡: | GGGGCAG GGC (RT) | 49 | |||||
 several several |  untranslated region untranslated region | clone A6: | GGGGCAGAGGGC | |||||||
 | 30 base deletion | No association | ||||||||
Continued RT, repetitive tract; IR, inverted repeat (loop with local DNA); IL, internal loop.
 Secondary structure reported by Golding et al. (29).
Secondary structure reported by Golding et al. (29).  This study is difficult to interpret in the context of the current report as the genomic JH locus was not available. The 10 clones were only 80% homologous to the closest JH locus reported in the literature with most alterations being similar between all of the isolates. Therefore, only 2 of the 26 proposed insertions/deletions can be attributed to somatic mutation with certainty, as they were unique to the consensus of the individual clones.
This study is difficult to interpret in the context of the current report as the genomic JH locus was not available. The 10 clones were only 80% homologous to the closest JH locus reported in the literature with most alterations being similar between all of the isolates. Therefore, only 2 of the 26 proposed insertions/deletions can be attributed to somatic mutation with certainty, as they were unique to the consensus of the individual clones. Complexities of the Analysis of Insertions and Deletions into V Genes.
The formal characterization of these events has been a daunting task because of their low frequency, and the complexity of the germline VH repertoire. According to our study, these events occur in <2% of somatically mutated clones. As shown in Fig. Fig.2,2, the primary variability between VH4 family members is 3–6-bp size variances in the CDR1s, which is comparable to the short insertions and deletions that we attribute to somatic hypermutation (in selected B cell populations). The similarity between evolutionary diversity and somatic diversification was expected, as the molecules are likely subject to the same functional and structural constraints. This has made it difficult to determine whether these events were generated somatically, versus germline encoded, or if they were artifacts of the experimental system: they could result from homologous recombination between alternate alleles or imperfect recombination between identical alleles, or they could have occurred during B cell replication independent of somatic hypermutation. In fact, VH genes may exhibit particularly unstable sequence characteristics evolved to help support both germline diversity and the generation of somatic mutations, as suggested by the identification of intrinsic hotspots of somatic hypermutation within the CDRs of V genes (25, 26). Perhaps the area of greatest contention in this complex system remains the possibility that these low frequency events are artifacts of the experimental manipulations performed, the AMV-RT, Taq, or PFU polymerases, and/or the cloning in E. coli.
The Insertion/Deletion Events Are The Result of the Somatic Hypermutation Process.
Our system addresses several key issues that associate the occurrence of insertions and deletions to the somatic hypermutation process. (a) Six of the nine insertions/deletions were identified within the VH4 gene repertoire of a single tonsil, providing an experimental system that could be characterized extensively as described below. (b) All of the insertion/deletion events reported involved triplets or multiples of triplets, leaving the transcripts in frame and therefore functional, and eight of nine events reported were localized to the CDRs. As with somatic point mutations, no insertions or deletions were observed in the 80 to 120 nucleotides of constant region (Cμ or Cγ) DNA sequenced with each cDNA clone. These hallmarks of somatic hypermutation and selection argue strongly that these events are not artifacts. (c) The B cells analyzed were processed and separated into highly pure, mutated B cell populations including GC (IgD− CD38+) and memory (IgD−CD38−) B cells, and an unmutated FM B cell population (IgD−CD38−), making it possible to focus our analysis on the mutated populations and use the unmutated population as a negative control, which in turn allows the statistical association of the observed insertion and deletions to the somatic hypermutation process (P = 0.014). In addition, the isolation of four of the insertion/deletion events from memory B cells provides evidence that these events did not result from artifacts related to contamination from endonucleolytically cleaved DNA from the apoptotic GC cells. (d) Seven of nine events reported in this study involved γ heavy chains that contain nearly twice the mutations of μ heavy chains (4), further correlating the events described here to somatic hypermutation. (e) As discussed below, the insertion/deletion events described tended to involve sequence motifs resembling previously described hotspots of somatic hypermutation, providing evidence that these events occur by the same process. (f) Finally, we extensively analyzed the VH4 gene family of the tonsil donor at both the expressed and genomic levels, facilitating the assignment of the insertions/deletions as somatic rather than germline encoded. 6 of the clones with insertions and deletions were unique among 395 VH4 cDNA clones sequenced from a single tonsil, including many independent isolates of each of the VH4 genes expressed (Table (Table1).1). In addition, we were unable to isolate genomic templates for any of the insertion or deletion events either by PCR or through the extensive characterization of the genomic VH4 repertoire of the tonsil donor (Table (Table1).1). Templating of these events from any other VH gene family can also be ruled out as members of the seven human VH gene families differ significantly in the CDR sequences where the events described had occurred.
Structural and Functional Considerations of Insertions and Deletions into VH Genes.
The events involving the insertion or deletion of a single amino acid from the CDR1 or CDR2 would not be expected to profoundly alter the backbone structure of these molecules, as the CDRs are the most malleable portions of antibodies. The clone g80 has two of the five amino acids that are customarily considered its CDR1 deleted, leaving only three amino acids to form this hypervariable loop (Fig. (Fig.11 B). Thus, this is one of the shortest CDR1s reported to date. The clone tm121 has two amino acids inserted into the FW3 region. The portion of the FW3 where this insertion occurred is believed to be solvent exposed and corresponds to the region where the B cell superantigen staphylococcal protein A binds to most VH3-encoded Ig molecules (28); therefore, it is likely that the insertion into this VH6 clone can be tolerated as a loop or bulge on the molecule's surface. The most complex structural change observed in our study involved clone pg86, with a six amino acid insertion at the FW1/CDR1 junction that would presumably double the length of this hypervariable loop and require dramatic structural accommodation. However, we were able to express this heavy chain and found it paired with light chain, indicating that it is likely functional (Fig. (Fig.5).5). The clone HBp2, containing a triplet insert into its CDR1, is particularly interesting because it has a known specificity. This VH6 gene was isolated from a human B cell hybridoma with anti–Bordetella pertussis specificity (21, 22). Clone HBp2 has also been expressed in the baculovirus system and is fully functional. We are currently performing mutational analysis of this heavy chain molecule to determine if the additional inserted amino acid plays a role in the affinity and/or specificity of this antibody.
Analysis of Insertions and Deletions Reported in the Literature.
Various groups have reported a number of insertion and deletion events (Table (Table3).3). Virtually all of the insertions and deletions reported from somatically mutated V genes involved the untranslated regions or occurred in silent passenger transgenes. 19 out of 25 insertions or deletions into somatically mutated genes involved predominantly repetitive elements, or in several cases other sequence patterns associated with secondary structures such as internal homologies or inverted repeats (Table (Table3).3). With the inclusion of the 9 events described in this work, 28 out of 34 insertions and deletions involved such elements. Thus, the proximity of sequence elements that can be predicted to cause secondary structural changes in the DNA seems to be a hallmark of insertions and deletions into somatically mutated VH genes.
A Model for the Occurrence of Insertions and Deletions during Somatic Hypermutation.
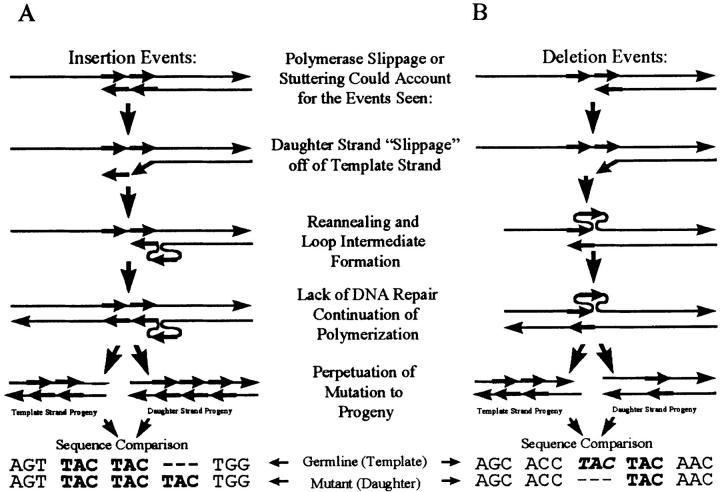
The evidence for the involvement of DNA secondary structure in the production of insertion or deletion mutations during somatic hypermutation, as suggested in 1986 by Golding et al. (29), now seems unequivocal. The insertions and deletions described in our study, and those illustrated in Table Table3,3, occur in a predictable fashion, involving sequence motifs that could form loop intermediates reminiscent of the replication slippage model of Streisinger et al. (30) and Ripley and Glickman (for review see 31) as presented in Fig. Fig.6.6. Such mutations are postulated to occur when DNA polymerase slips or stutters and the newly synthesized strand shifts on the template and reanneals to an adjacent repetitive element, producing unpaired loop intermediates localized to one or the other strands. If this unpaired loop intermediate is not repaired then it will be perpetuated as an insertion of an instance of the repetitive element if in the daughter strand, or a deletion if in the template strand.
A Possible Correlation to Intrinsic Hotspots.
A higher frequency of somatic hypermutation has been reported to occur at sequence motifs referred to as intrinsic hotspots (for review see reference 12). Interestingly, every insertion/deletion event reported in our study resembled one of these hotspots (AGC, TAC, and AAC; references 12 and 27; Fig. Fig.4).4). The analysis of selected populations may have influenced this tendency because seven out of eight of these events occurred in the CDRs where it has been shown that hotspot motifs are preferentially found (25, 26). Furthermore, only a weak correlation to hotspots could be found for the previously reported insertions/deletions involving unselected regions of V loci (Table (Table3).3). However, the single event found in this analysis that occurred outside of the CDRs in FW3 (clone tm121, Figs. Figs.11 C and and44 A), also involved a tandem of possible hotspots (AAG, AAC). A more extensive and directed analysis is required to fully address this issue.
Implications for the Molecular Mechanism of Somatic Hypermutation.
The instability of repetitive tracts during DNA replication is a hallmark of defects in postreplicative mismatch repair (33), and the locus-specific downregulation of DNA mismatch repair in response to UV irradiation has recently been reported for immunoglobulin VH genes in freshly sorted GC B cells (CD38+IgD−) compared to mantle zone B cells (CD38−IgD+; reference 34). In a recent study by Tran et al. (35), it was shown that tract instability of homonucleotide runs associated with mismatch repair defects occur more frequently in long than in short runs. These authors suggested that if loop intermediates occur in long repetitive tracts (>8 bp for a homonucleotide run) they could involve a distal repetitive element out of reach of the polymerase proofreading activity and only be subjected to mismatch repair. However, for short repetitive tracts, as for the events reported in this analysis, loop intermediates can only occur proximal to the polymerase complex and are therefore subjected to both polymerase proofreading and mismatch repair mechanisms.
All 9 events in this analysis, and 19 out of 25 events from the literature (28 out of 34 insertions and deletions reported), appeared to result from secondary structural intermediates. Loop intermediates proximal to the polymerase complex during DNA polymerization should be repaired by the polymerase proofreading mechanisms immediately, or by the postreplicative DNA repair systems. This analysis suggests the following characteristics for the polymerization process during somatic hypermutation. (a) The polymerase interacts with the V locus in a particularly unstable or “loose” fashion, especially when hotspot motifs or elements capable of forming secondary structures are encountered, allowing bp substitutions in most instances, and insertions or deletions via polymerase slippage at a much lower frequency; (b) it has limited proofreading capabilities; and (c) there is a downregulation of postreplicative mismatch repair. An efficient means to downregulate mismatch repair during somatic hypermutation could be through the lack of differentiation of the template and progeny strands for the mismatch repair system; lack of strand differentiation has been shown to increase the rate of mutations introduced (36). Such a system would be advantageous for the locus-specific V gene somatic hypermutation in that it could involve alterations of a single enzymatic complex (polymerase complex) rather than multiple systems (proofreading and mismatch repair). Another system, which would have the same advantage, i.e., the alteration of a single complex, would be the alteration of a DNA repair system such as transcription-coupled repair to be the somatic mutator, as suggested in recent studies (13). Alternatively, the insertions and deletions might result solely from a downregulation of postreplicative mismatch repair at the V locus in the rapidly proliferating centroblasts that are undergoing somatic hypermutation or due to a polymerase enzyme with such a high fault rate as to overwhelm any repair.
All currently accepted models of somatic hypermutation, whether related to DNA excision-repair–like systems or transcription-repair, or to DNA polymerization or reverse transcription, involve transcriptional activation involving cis-factors in the V locus (enhancers, etc.) followed by the activity of unknown polymerase enzymes of some type. This analysis does not refute or corroborate any of these models directly, but it does provide further characterization of the polymerization system involved, based on the types of mutations observed and on the molecular biology that is known to cause such mutations. This analysis and the model presented here provide further information or criteria to be contemplated as the various possible polymerase systems involved are considered.
Conclusions.
Insertions and deletions into immunoglobulin VH genes during somatic hypermutation are additional means by which the immunoglobulin repertoire can be diversified. These events display characteristics supporting models of somatic hypermutation involving a particularly unstable or error-prone polymerase to allow the introduction of mutations, and involving the downregulation of DNA repair to allow the perpetuation of these mutations. Additionally, we show that these events tend to involve sequence motifs resembling intrinsic hotspots of somatic hypermutation, suggesting that the polymerase complex is destabilized in a sequence-specific manner to allow preferential mutation at these sequence elements.
Acknowledgments
We are grateful to Yucheng Li, Fang Zhao, Steve Scholl, Carol Williams, Shirley Hall, and Robin Wray, all of whom provided unprecedented technical assistance for various aspects of this work. We also thank Kimble Frazer for excellent discussions.
These studies were supported by a grant from the National Institutes of Health (AI-12127). J. Donald Capra holds the Edwin L. Cox Distinguished Chair in Immunology and Genetics.
Footnotes
1 Abbreviations used in this paper: FM, follicular mantle; FW, framework; GC, germinal center.
References
Articles from The Journal of Experimental Medicine are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1084/jem.187.1.59
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jem/article-pdf/187/1/59/1113232/97-1247.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1084/jem.187.1.59
Article citations
Molecular mechanisms of DNA lesion and repair during antibody somatic hypermutation.
Sci China Life Sci, 67(11):2344-2353, 23 Jul 2024
Cited by: 0 articles | PMID: 39048716
Review
Whence and wherefore IgE?
Immunol Rev, 326(1):48-65, 23 Jul 2024
Cited by: 0 articles | PMID: 39041740
Review
MICA-specific nanobodies for diagnosis and immunotherapy of MICA+ tumors.
Front Immunol, 15:1368586, 14 Mar 2024
Cited by: 2 articles | PMID: 38550583 | PMCID: PMC10973119
Identification of B cell subsets based on antigen receptor sequences using deep learning.
Front Immunol, 15:1342285, 21 Mar 2024
Cited by: 0 articles | PMID: 38576618 | PMCID: PMC10991714
B-Cell Receptor Repertoire: Recent Advances in Autoimmune Diseases.
Clin Rev Allergy Immunol, 66(1):76-98, 01 Feb 2024
Cited by: 2 articles | PMID: 38459209
Review
Go to all (167) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (3 citations) ENA - AF013615
- (3 citations) ENA - AF013626
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Somatic diversification and selection of immunoglobulin heavy and light chain variable region genes in IgG+ CD5+ chronic lymphocytic leukemia B cells.
J Exp Med, 181(4):1507-1517, 01 Apr 1995
Cited by: 81 articles | PMID: 7535340 | PMCID: PMC2191964
Amino acid insertions and deletions contribute to diversify the human Ig repertoire.
Immunol Rev, 162:143-151, 01 Apr 1998
Cited by: 26 articles | PMID: 9602360
Review
Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+ IgD+ B cell line in vitro: definition of the requirements and modalities of hypermutation.
J Immunol, 162(6):3437-3447, 01 Mar 1999
Cited by: 62 articles | PMID: 10092799 | PMCID: PMC4623562