Abstract
Free full text

Osteoclast differentiation independent of the TRANCE–RANK–TRAF6 axis
Abstract
Osteoclasts are derived from myeloid lineage cells, and their differentiation is supported by various osteotropic factors, including the tumor necrosis factor (TNF) family member TNF-related activation-induced cytokine (TRANCE). Genetic deletion of TRANCE or its receptor, receptor activator of nuclear factor κB (RANK), results in severely osteopetrotic mice with no osteoclasts in their bones. TNF receptor-associated factor (TRAF) 6 is a key signaling adaptor for RANK, and its deficiency leads to similar osteopetrosis. Hence, the current paradigm holds that TRANCE–RANK interaction and subsequent signaling via TRAF6 are essential for the generation of functional osteoclasts. Surprisingly, we show that hematopoietic precursors from TRANCE-, RANK-, or TRAF6-null mice can become osteoclasts in vitro when they are stimulated with TNF-α in the presence of cofactors such as TGF-β. We provide direct evidence against the current paradigm that the TRANCE–RANK–TRAF6 pathway is essential for osteoclast differentiation and suggest the potential existence of alternative routes for osteoclast differentiation.
Osteoclasts are multinucleated cells responsible for bone resorption and are derived from hematopoietic precursor cells (HPCs) of myeloid lineage. It is currently thought that two critical factors supplied by osteoblasts, macrophage CSF (M-CSF) and TNF-related activation-induced cytokine (TRANCE, also called receptor activator of NF-κB [RANK] ligand, osteoclast differentiation factor, and osteoprotegerin ligand), are essential for the differentiation and maturation of osteoclast precursors in bones (1, 2). M-CSF–defective mice (op/op) show an osteopetrotic phenotype that can spontaneously reverse with age or be rescued by transgenic expression of Bcl-2. Mice deficient in either TRANCE or its receptor RANK also show an osteopetrotic phenotype that is caused by the complete lack of osteoclasts in their bones. TRANCE KO mice, however, cannot be rescued by transgenic expression of Bcl-2 (3), supporting the idea that TRANCE is a differentiation factor for osteoclasts. Ex vivo recombinant TRANCE, in combination with M-CSF, can induce osteoclast formation from mouse bone marrow cells, spleen cells, or human peripheral blood cells without stromal cells or osteoblasts. Moreover, recombinant TRANCE alone induces the differentiation of the monocytic cell line RAW264.7 into osteoclasts (1, 2).
Most TNF receptor family members, including RANK, interact with a family of adaptor proteins called TNF receptor-associated factors (TRAFs) (4). Among the known TRAF family members, TRAF2, TRAF5, and TRAF6 can activate transcription factors, such as NF-κB and AP-1, that are required for osteoclast differentiation (4). RANK interacts with most of the TRAF family members (4); however, TRAF6 seems to play a critical role in osteoclast differentiation mediated by RANK. Three independent TRAF6 KO mouse lines have been reported, and all exhibit osteopetrosis (5–7). Collectively, it has been proposed that TRANCE–RANK interaction and subsequent signaling via TRAF6 are essential for the differentiation of mature osteoclasts (1, 2, 4).
We show that osteoclasts can be generated from HPCs of TRANCE, RANK, or TRAF6 KO mice. These results provide direct evidence that osteoclast differentiation in vitro can occur independently of the TRANCE–RANK interaction and its signaling via TRAF6. Therefore, our data challenge the current paradigm for osteoclast differentiation and suggest the possibility that alternative pathways may exist for osteoclast differentiation that is independent of the TRANCE axis.
Results AND Discussion
Is the TRANCE–RANK interaction essential for osteoclast differentiation?
We revisited the issue of whether the TRANCE–RANK interaction is essential for osteoclast differentiation. Although this may be counterintuitive because it has been firmly established that TRANCE or RANK KO mice do not have any osteoclast in their bones, the following observations serve as the basis of this study. First, it was reported that TNF-α could support osteoclastogenesis in the presence of osteoprotegerin, an inhibitor of TRANCE–RANK interactions (8–10). These results suggest that TNF-α might induce osteoclast differentiation in the absence of TRANCE–RANK interactions. However, the issue of whether TNF-α is a direct inducer of osteoclast differentiation is still controversial. Lam et al. showed that TNF-α alone failed to induce osteoclast differentiation from murine precursors in similar culture conditions (11). Second, although no osteoclasts can be identified in the bones of TRANCE or RANK KO mice, this may not be simply because of the complete failure of osteoclastogenesis. For example, impaired osteoclast differentiation superimposed on a shortened lifespan may also explain the observed phenotype in TRANCE or RANK KO mice. Indeed, TRANCE is a survival factor for differentiated osteoclasts (12).
To test the hypothesis that the TRANCE–RANK interaction may not be essential for osteoclast differentiation, we examined whether TNF-α can substitute for TRANCE in inducing osteoclasts from HPCs. Osteoclast precursors were prepared with M-CSF from bone marrow cells and were further cultured with M-CSF and TNF-α. Similar to Lam et al., we could not find obvious multinucleated tartrate-resistant acid phosphatase (TRAP)–positive osteoclasts (TRAP+ mononuclear cells [MNCs]) after treatment with TNF-α, although TRAP+ MNCs were observed (Fig. 1, A and B). To test the possibility that our culture conditions might have insufficient cofactors, we examined the role of TGF-β in this process. We chose TGF-β for this study because it was shown to synergize with TRANCE in the induction of osteoclast differentiation and to be prevalent in the bone (13, 14) (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20050978/DC1). When TGF-β was added with M-CSF to prepare osteoclast precursors from bone marrow cells, subsequent TNF-α treatment resulted in a large number of TRAP+ MNCs (Fig. 1, A and B), and these formed actin rings (not depicted). Such results suggest that if proper cofactors are included to prepare osteoclast precursors, TNF-α can induce the formation of osteoclasts.
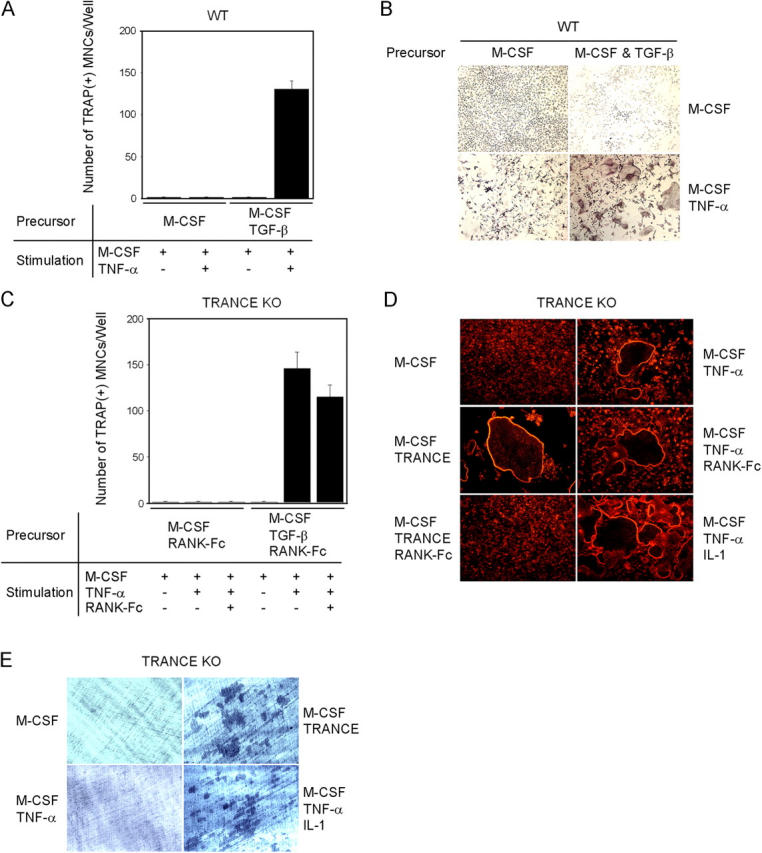
TNF-α–induced osteoclastogenesis in WT or TRANCE-deficient cells. (A–C) Osteoclast formation from WT or TRANCE-deficient osteoclast precursors. (A and B) BMMs were derived from bone marrow cells of WT mice by culturing them for 3 d with M-CSF alone or with M-CSF + TGF-β as indicated. BMMs were further cultured with M-CSF alone or with M-CSF + TNF-α as indicated. (C) Splenocytes isolated from TRANCE KO mice were cultured for 3 d with M-CSF alone or with M-CSF + TGF-β in the presence of 5 μg/ml RANK-Fc to generate BMMs, which were subsequently treated with M-CSF alone, M-CSF/TNF-α, or M-CSF/TNF-α/RANK-Fc (5 μg/ml), as indicated. Cultured cells were fixed and stained for TRAP. (D and E) TRANCE-deficient osteoclast precursors were prepared by culturing HPCs with M-CSF and TGF-β. F-actin ring staining (D) and a pit formation assay (E) on TRANCE-deficient cells that were subsequently cultured for 3 d with the indicated conditions are shown.
To rule out an indirect effect of TNF-α on osteoclast formation through up-regulation of TRANCE, we prepared osteoclast precursors by culturing splenocytes from TRANCE KO mice with M-CSF alone or with M-CSF + TGF-β for 3 d. To minimize the potential effect of TRANCE in the serum, we added RANK-Fc to the culture at a concentration 5 μg/ml, which completely inhibits osteoclast formation induced by recombinant soluble TRANCE (Fig. 1 C). Even in the presence of an excessive amount of RANK-Fc, TRAP+ MNCs were readily formed by M-CSF and TNF-α treatment when osteoclast precursors from TRANCE KO mice were prepared with M-CSF + TGF-β (Fig. 1 C). TRANCE-deficient TRAP+ MNCs induced by TNF-α successfully formed actin rings (Fig. 1 D); however, TNF-α–induced TRANCE-deficient osteoclasts failed to resorb bone (Fig. 1 E). TRAF6 activation has been reported to be important for osteoclast activation (15). Because TRAF6 is not activated by TNF-α, we next tested whether TRANCE-deficient osteoclasts can be further activated by IL-1, which activates TRAF6 (16). When TRANCE-deficient osteoclast precursors, prepared by M-CSF + TGF-β, were cultured with TNF-α and IL-1 in the presence of M-CSF, pits were clearly formed on dentine slices (Fig. 1 E).
Similar experiments were also performed with spleen cells from RANK KO mice and showed that TNF-α induced TRAP+ MNCs when osteoclast precursors cells were prepared in the presence of M-CSF + TGF-β (Fig. 2 A). These cells formed actin rings and expressed most of the molecular markers that characterize mature osteoclasts (Fig. 2, B and C, and Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20050978/DC1). However, these cells failed to form resorption pits (Fig. 2 D). Surprisingly, as with TRANCE-deficient cells, combined treatment with M-CSF, TNF-α, and IL-1 induced bone-resorbing osteoclasts from RANK-deficient osteoclast precursors that were prepared with M-CSF and TGF-β (Fig. 2 D).
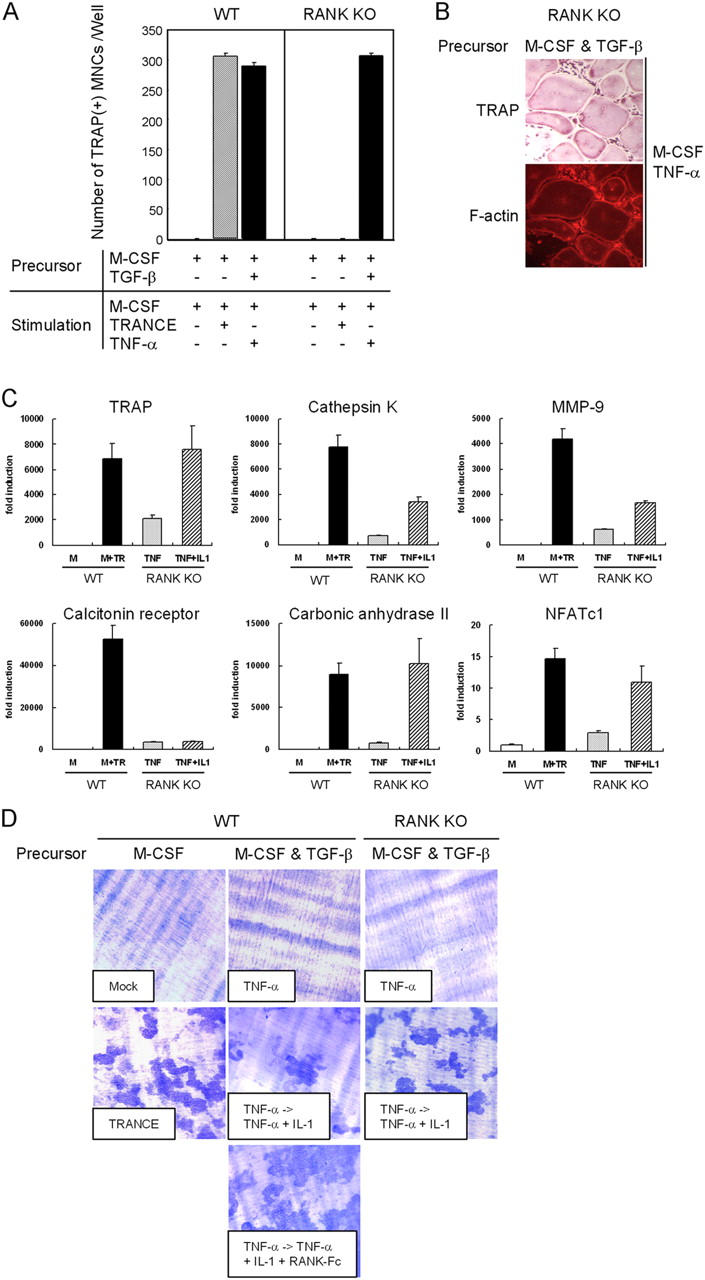
TNF-α–induced osteoclastogenesis from WT or RANK-deficient cells. (A) Osteoclast formation from WT or RANK-deficient osteoclast precursors. Spleen cells were incubated for 3 d with M-CSF alone or with M-CSF + TGF-β to generate osteoclast precursors as indicated. Precursors were further cultured with M-CSF alone, M-CSF/TRANCE, or M-CSF/TNF-α. Cultured cells were fixed and stained for TRAP. (B) Osteoclast precursors prepared with M-CSF and TGF-β were further cultured for 3 d with M-CSF/TNF-α and fixed and stained for F-actin rings and TRAP. (C) Osteoclast precursors were prepared by culturing HPCs with M-CSF and TGF-β for 3 d. BMMs were incubated for an additional 3 d with M-CSF alone or with M-CSF/TRANCE for WT cells. RANK-deficient osteoclast precursors prepared with M-CSF + TGF-β were cultured for 3 d with M-CSF/TNF-α and an additional day with M-CSF/TNF-α in the presence or absence of IL-1 as indicated. Cells were subjected to real-time PCR analysis for TRAP, cathepsin K, MMP-9, calcitonin receptor, carbonic anhydrase II, and NFATc1. Values were normalized to 18S RNA expression. (D) Osteoclast precursors were prepared and cultured as described in C with the indicated stimuli, and dentine slices were stained with toluidine blue.
Collectively, these data provide rather surprising but direct evidence that osteoclast formation can be induced in vitro by a mechanism that is independent of TRANCE–RANK, if proper factors are included during osteoclast precursor preparation. In this study, these factors were M-CSF and TGF-β, followed by treatment with M-CSF, TNF-α, and IL-1 to induce differentiation and activation of osteoclasts.
How TNF-α induces osteoclast differentiation independent of TRANCE–RANK is unclear at this time. Although TRAF2 and TRAF5 are critical signaling adapters for TNF receptors, TRAF2- or TRAF5-deficient cells differentiate into mature osteoclasts upon TNF-α treatment if osteoclast precursors are prepared with M-CSF and TGF-β (unpublished data). TRAF6 is also dispensable for TNF-α–induced osteoclast differentiation (Fig. S3, A–C, available at http://www.jem.org/cgi/content/full/jem.20050978/DC1). However, TRAF6 appears to be critical for the complete activation of osteoclasts because TRAF6-deficient osteoclasts derived with TNF-α failed to resorb bone even in the presence of IL-1 (Fig. S3 D). It is of interest that, although TNF-α alone fails to activate nuclear family of activated T cells (NFAT) c1, it can strongly induce the transcription of NFATc1 to a level similar to that induced by TRANCE if osteoclast precursors are prepared with M-CSF and TGF-β (Fig. 3). It is unclear how TNF-induced early signaling events vary in osteoclast precursors prepared with M-CSF and TGF-β. Nevertheless, it appears that the ability of distinct stimuli to induce NFATc1 above a critical threshold is an important step in this process. This is consistent with the finding that overexpression of NFATc1 is sufficient to induce osteoclast differentiation (17) and is consistent with data demonstrating that high levels of CD40 stimulation can induce osteoclast differentiation independent of TRANCE–RANK activation (18).
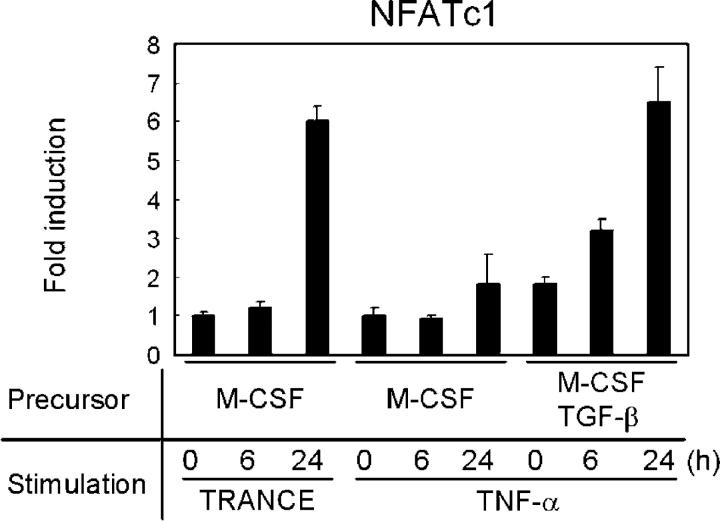
Induction of NFATc1. Osteoclast precursors were derived from bone marrow cells of WT mice by culture with M-CSF alone or with M-CSF/TGF-β and subsequently stimulated with TRANCE or TNF-α for the indicated times. M-CSF was present at all times. Samples were subjected to real-time PCR using NFATc1-specific primers. NFATc1 mRNA induction was normalized to HPRT expression.
Role of TRAF6 in osteoclast activation
TRAF6 KO mice were generated by three independent schemes, and all showed severe osteopetrosis (5–7) (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20050978/DC1). However, there were some discrepancies in the phenotypes of these mice. For example, though functionally defective, normal numbers of TRAP+ MNCs were observed in the long bone of TRAF6 KO mice reported by Lomaga et al. (6). However, Naito et al. reported no TRAP+ MNCs in their mice (5). In contrast, we found that the number of TRAP+ MNCs is considerably reduced, but still detectable, in the bones of TRAF6 KO mice (Fig. S4). One possible explanation for these discrepancies is that different regions of TRAF6 were deleted in the three lines and, thus, there may be variations in the leakiness of null mutations among the models. However, none of the TRAF6-null mice showed detectable levels of mutant or WT TRAF6 protein. Moreover, HPCs from all of the different TRAF6 KO lines failed to become osteoclasts in vitro when stimulated with M-CSF and TRANCE (19) (Boyle, W., personal communication; Fig. 4, A and B, left column). These results make it unlikely that there was residual TRAF6 protein function in any of the TRAF6 mutant lines. Thus, various experiments, both in vivo and in vitro, with three different lines of TRAF6 KO mice failed to resolve a critical question related to the role of TRAF6 in TRANCE-induced osteoclast differentiation: is TRAF6 essential for osteoclast differentiation induced by TRANCE–RANK interaction?

TRANCE-induced osteoclastogenesis in WT or TRAF6-deficient cells. (A and B) Osteoclast formation from WT or TRAF6-deficient cells. Spleen cells were incubated for 3 d with M-CSF alone or with M-CSF + TGF-β to generate osteoclast precursors, as indicated, and further cultured with M-CSF alone or M-CSF + TRANCE. Cultured cells were fixed and stained for TRAP. F-actin ring staining (C) and a pit formation assay (D) on WT or TRAF6-deficient TRAP+ MNCs derived from osteoclast precursors prepared with M-CSF and TGF-β are shown.
To investigate the importance of TRAF6 in TRANCE-induced osteoclast differentiation in the presence of cofactors, splenocytes from TRAF6 KO mice were collected and cultured for 3 d in the presence of M-CSF and were then used as osteoclast precursors. As expected, no TRAP+ MNCs were detected by subsequent treatment with M-CSF and TRANCE (Fig. 4, A and B), which is consistent with previous experiments (19) (Boyle, W., personal communication). However, when TRAF6-deficient osteoclast precursors were prepared by cotreatment with M-CSF and TGF-β, TRAF6-deficient TRAP+ MNCs were generated by subsequent treatment with M-CSF and TRANCE (Fig. 4, A and B). These cells expressed high levels of TRAP or osteoclast-associated receptor (OSCAR), markers that are specific for authentic osteoclasts (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20050978/DC1). To determine whether TRAF6-deficient TRAP+ MNCs were functional, F-actin ring formation and bone resorption on dentine slices were assessed. Unlike WT osteoclasts, TRANCE-induced TRAF6-deficient TRAP+ MNCs failed to form actin rings or resorb bone (Fig. 4, C and D).
These results have several implications. First, TRAF6-deficient HPCs can be induced to differentiate into osteoclasts by TRANCE if the proper cofactors, such as TGF-β, are provided in the culture. This result implies that TRAF6 is not likely to be essential for osteoclast differentiation, in contrast to what has been previously proposed (19) (Boyle, W., personal communication). Thus, the varying degree of osteoclast development observed in the three different lines of TRAF6 KO mice may be caused by different cofactors that vary with genetic background, age, or housing environment. Nevertheless, TRANCE-induced osteoclastogenesis is still considerably reduced in the absence of TRAF6 (Fig. 4 A), implying that TRAF6 is a major signaling adaptor for RANK during osteoclast differentiation. Second, TRAF6 seems to be critical for complete activation of osteoclasts, even in the presence of excessive cofactors. Consistent with our data, RANK–TRAF6 interactions have been implicated for osteoclast cytoskeletal organization and resorptive function in vitro (15). Thus, in conjunction with our data that IL-1/TRAF6 signals can induce osteoclast resorption (Fig. 1 and Fig. 2), the most critical role of TRAF6 is likely to be its ability to activate mature osteoclasts to resorb bone.
Conclusion
Our study provides direct evidence that functional bone-resorbing osteoclasts can be generated in vitro in the absence of TRANCE–RANK if cultures are supplemented with the proper factors. This is clearly against the current paradigm of how an osteoclast is generated, which states that TRANCE–RANK interactions are absolutely required. Our data do not, however, refute the role of TRANCE–RANK as a major regulator of osteoclast differentiation. Rather, our findings suggest the possibility that an alternative pathway of osteoclast differentiation may exist in vivo.
Based on the limited published studies using TRANCE or RANK KO mice, such alternative pathways have not thus far been identified, even in a model of experimental arthritis (20). Nonetheless, the attempt to elucidate alternative pathways for osteoclast differentiation and the responsible factors in vivo should be justifiable based on our study. Of note, although we have used TNF-α and TGF-β to induce osteoclasts in the absence of TRANCE–RANK in vitro, the same factors may not be responsible for a putative alternative pathway of osteoclast differentiation in vivo. The identification of distinct osteoclast differentiation pathways will be important to design better therapeutical approaches to treat chronic inflammatory bone diseases.
MATERIALS AND METHODS
Reagents.
All cell culture media and supplements were obtained from Life Technologies. Soluble recombinant mouse TRANCE and RANK-Fc were purified from insect cells as described previously (21), and recombinant human M-CSF was a gift from D. Fremont (Washington University, St. Louis, MO). TNF-α, IL-1, and TGF-β were purchased from R&D Systems. Texas red-X phalloidin was obtained from Molecular Probes.
Mice.
C57BL/6 male mice (4–6 wk of age) were obtained from the Jackson Laboratory. The breeding and genotyping of mice deficient in TRANCE, RANK, TRAF6, TRAF5, or TRAF2 was performed as previously described (7, 21–24).
Osteoclast formation.
Murine osteoclasts were prepared from bone marrow cells or splenocytes. In brief, cells from bone marrow or spleens were cultured in α-MEM containing 10% FBS with 5 ng/ml M-CSF for 16 h to separate adherent cells from nonadherent cells. Nonadherent cells were harvested and cultured with 30 ng/ml M-CSF alone or with 30 ng/ml M-CSF plus 1 ng/ml TGF-β, as indicated in the figures. After 3 d of culture, floating cells were removed, and the attached cells were used as bone marrow–derived monocyte/macrophage precursors (BMMs). To generate osteoclasts, BMMs were further cultured with various combinations of 30 ng/ml M-CSF, 100 ng/ml TRANCE, and 20 ng/ml TNF-α, as indicated in the figures. After an additional 3 d of culture, cells were fixed and stained for TRAP. TRAP-positive multinucleated cells containing more than three nuclei were considered multinucleated TRAP+ osteoclasts. M-CSF was included at all times. In some experiments, 5 μg/ml RANK-Fc was added to the cultures. All cells were cultured at 37°C and 5% CO2.
Pit formation assay and F-actin ring staining.
Bone marrow cells or splenocytes were incubated for 3 d with 30 ng/ml M-CSF alone or with 30 ng/ml M-CSF plus 1 ng/ml TGF-β to generate BMMs. BMMs were placed on dentine slices and were cultured for an additional 3 d with various combinations of 30 ng/ml M-CSF, 100 ng/ml TRANCE, 20 ng/ml TNF-α, and 10 ng/ml IL-1, as indicated in the figures. In Fig. 2, however, 10 ng/ml IL-1 was added only for one additional day after RANK-deficient cells were cultured for 3 d with M-CSF and TNF-α. The slices were then recovered, cleaned by ultrasonication in 0.5 M NH4OH to remove adherent cells, and stained with toluidine blue to visualize resorption pits. For F-actin ring staining, BMMs prepared with M-CSF and TGF-β were further cultured for 3 d with various combinations of 30 ng/ml M-CSF, 100 ng/ml TRANCE, 20 ng/ml TNF-α, 10 ng/ml IL-1, and 5 μg/ml RANK-Fc, as indicated in the figures. Cultured cells were fixed and stained with Texas red-X phalloidin.
RT-PCR.
RT-PCR analysis was performed using cDNA from WT or TRAF6-deficient osteoclasts. Spleen cells were incubated for 3 d with 30 ng/ml M-CSF alone or 30 ng/ml M-CSF plus 1 ng/ml TGF-β to generate BMMs. BMMs were cultured for an additional 3 d with 30 ng/ml M-CSF alone or with 30 ng/ml M-CSF plus 100 ng/ml TRANCE. Total RNA was extracted from cultured cells using TRIZOL (Life Technologies). First-strand cDNA was transcribed from 1 μg of RNA using Superscript RT (Life Technologies) according to the protocol provided by the supplier. The PCR was performed at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for a total of 25 (for TRAP and hypoxanthine ribosyltransferase [HPRT]) or 30 (for OSCAR) cycles. The following primers were used: 5′-OSCAR, 5′-CTGCTGGTAACGGATCAGCTCCCCAGA-3′; 3′-OSCAR, 5′-CCAAGGAGCCAGAACCTTCGAAACT-3′; 5′-TRAP, 5′-CAGTTGGCAGCAGCCAAGGAGGAC-3′; 3′-TRAP, 5′-GTCCCTCAGGAGTCTAGGTATCAC-3′; 5′-HPRT, 5′-GTAATGATCAGTCAACGGGGGAC-3′; 3′-HPRT, 5′-CCAGCAAGCTTGCAACCTTAACCA-3′.
Real-time PCR.
Quantitative real-time PCR was performed using the TaqMan universal PCR master mix. TaqMan primers and probes were purchased from Applied Biosystems. To make first-strand cDNA, 4 μg RNA was reverse transcribed in a 20-μl reaction. The cDNA was diluted to fivefold, and 4 μl was used as a template in each PCR with PCR TaqMan universal master mix containing 1 × PCR buffer and the appropriate concentrations of gene-specific primers, the TaqMan probe, 5 mM MgCl2, 0.025 U AmpliTaq Gold, and 0.2 mM dNTPs combined in a total volume of 25 μl. A negative cDNA control lacking reverse trancriptase was also included in each assay. The PCR was performed at 95°C for 10 min (denaturation), 95°C for 15 s, and 60°C for 1 min for a total of 40 cycles. The cycle threshold (Ct) values, corresponding to the PCR cycle number at which fluorescence emission in real time reaches a threshold above the baseline emission, were determined. The Ct value assigned to a particular well thus reflects the point during the reaction at which a sufficient number of amplicons have accumulated in that well to be at a point well above the baseline.
Histology.
Osteoclasts were visualized by histochemical staining for TRAP. In brief, tibiae were dissected, fixed in gluatraldehyde, demineralized with EDTA, and embedded in glycol methacrylate. 3-μm sections were cut and mounted on glass slides without heating. Osteoclasts were stained by incubation in chromogenic acid phosphatase substrate in the presence of tartaric acid.
Online supplemental material.
Fig. S1 shows the effect of TGF-β on osteoclast differentiation. Fig. S2 depicts a real-time PCR analysis of mRNA from WT or RANK-deficient osteoclasts. Fig. S3 shows TNF-α–induced osteoclastogenesis of WT and TRAF6-deficient cells. Fig. S4 depicts an osteopetrotic phenotype of TRAF6 KO mice. Fig. S5 shows an RT-PCR analysis of mRNA from WT or TRAF6-deficient osteoclasts. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20050978/DC1.
Acknowledgments
We thank D. Fremont for reagents and C. Ware for helpful comments.
This work was supported in part by National Institutes of Health grants AR49078 (to Y. Choi); AR48714, AR46026, and AR38933 (to J.A. Lorenzo); and AR48231 (to S.-K. Lee). Additional support was provided by grant R13-2002-013-03001-0 (to N. Kim) from the Korea Science & Engineering Foundation through the Medical Research Center for Gene Regulation at Chonnam National University.
The authors have no conflicting financial interests.
References
Articles from The Journal of Experimental Medicine are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1084/jem.20050978
Read article for free, from open access legal sources, via Unpaywall:
http://jem.rupress.org/content/jem/202/5/589.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Gingival fibroblasts produce paracrine signals that affect osteoclastogenesis in vitro.
Bone Rep, 22:101798, 20 Aug 2024
Cited by: 0 articles | PMID: 39252697 | PMCID: PMC11381831
Comparative Transcriptomic Analyses of Peripheral Blood Mononuclear Cells of COVID-19 Patients without Pneumonia and with Severe Pneumonia in the First Year of Follow-Up.
Viruses, 16(8):1211, 28 Jul 2024
Cited by: 0 articles | PMID: 39205185 | PMCID: PMC11358892
Osteogenic Differentiation of Human Gingival Fibroblasts Inhibits Osteoclast Formation.
Cells, 13(13):1090, 24 Jun 2024
Cited by: 0 articles | PMID: 38994943 | PMCID: PMC11240541
The miR-29-3p family suppresses inflammatory osteolysis.
J Cell Physiol, 239(8):e31299, 19 May 2024
Cited by: 0 articles | PMID: 38764231
Osteogenesis imperfecta type 10 and the cellular scaffolds underlying common immunological diseases.
Genes Immun, 25(4):265-276, 29 May 2024
Cited by: 0 articles | PMID: 38811682
Review
Go to all (230) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A new member of tumor necrosis factor ligand family, ODF/OPGL/TRANCE/RANKL, regulates osteoclast differentiation and function.
Biochem Biophys Res Commun, 256(3):449-455, 01 Mar 1999
Cited by: 269 articles | PMID: 10080918
Review
Transforming growth factor beta affects osteoclast differentiation via direct and indirect actions.
J Bone Miner Res, 16(10):1787-1794, 01 Oct 2001
Cited by: 150 articles | PMID: 11585342
The TRAF family of signal transducers mediates NF-kappaB activation by the TRANCE receptor.
J Biol Chem, 273(43):28355-28359, 01 Oct 1998
Cited by: 279 articles | PMID: 9774460
Funding
Funders who supported this work.
NIAMS NIH HHS (10)
Grant ID: R01 AR049078
Grant ID: AR48714
Grant ID: R01 AR048714
Grant ID: AR48231
Grant ID: AR46026
Grant ID: AR49078
Grant ID: P01 AR038933
Grant ID: P30 AR046026
Grant ID: AR38933
Grant ID: R03 AR048231





