Abstract
Free full text

Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA.
Abstract
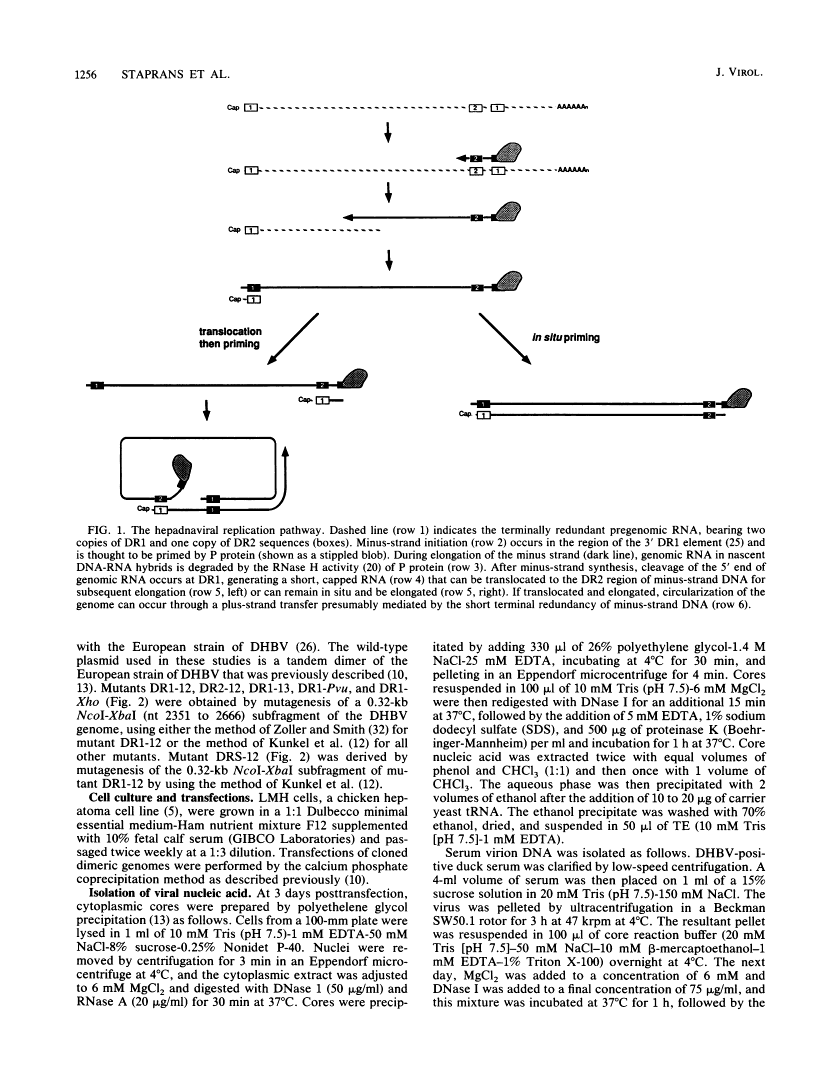
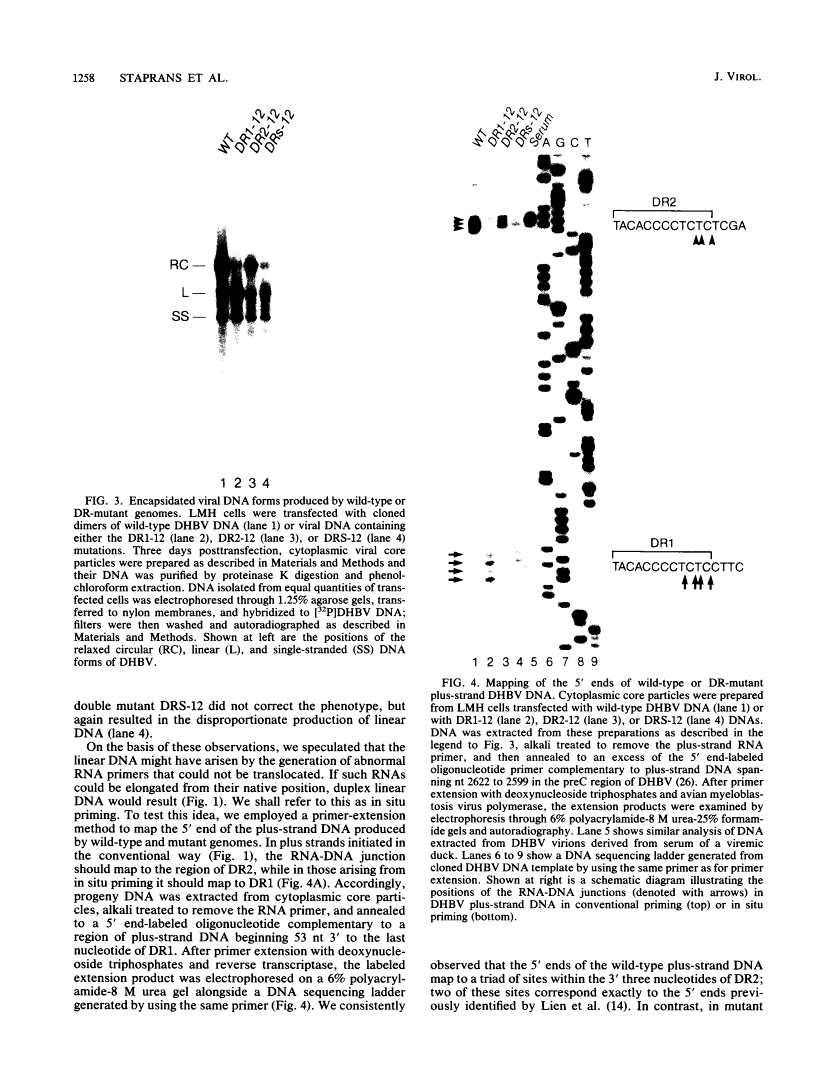
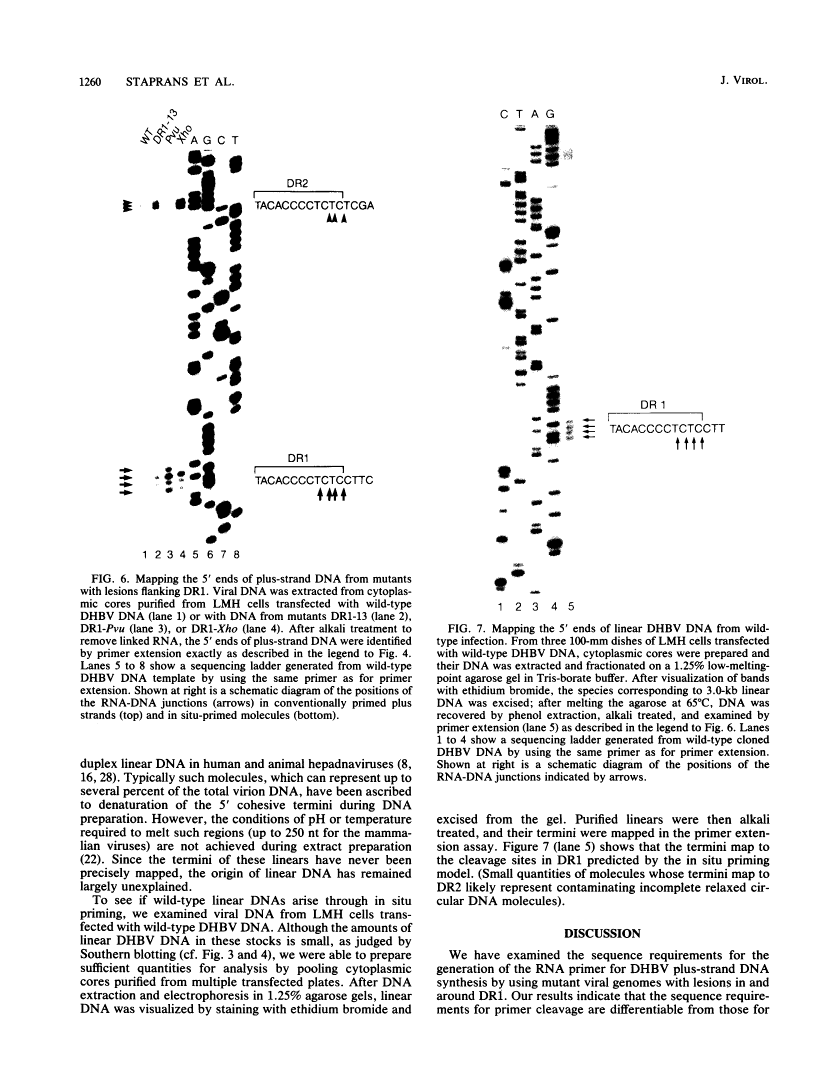
Hepadnaviruses replicate their circular DNA genomes via reverse transcription of an RNA intermediate. The initial product of reverse transcription, minus-strand DNA, contains two copies of a short direct repeat (DR) sequence, termed DR1 and DR2. Plus-strand DNA synthesis initiates at DR2 on minus-strand DNA, using as a primer a short, DR1-containing oligoribonucleotide derived by cleavage and translocation from the 5' end of pregenomic RNA. To clarify the sequence requirements for plus-strand primer cleavage and translocation, we have constructed mutants of the duck hepatitis B virus bearing base changes in or around the DR1 sequence in the primer. A point mutation at the terminal nucleotide of DR1 has a striking phenotype: normal levels of duplex viral DNA are produced, but nearly all of the DNA is linear rather than circular. Mapping of the 5' end of plus-strand DNA reveals that primer cleavage occurs with normal efficiency and accuracy, but the primer is not translocated to DR2; rather, it is extended in situ to generate duplex linear DNA. Other mutations just 3' to DR1 similarly affect primer translocation, although with differing efficiencies. Linear DNA found in wild-type virus preparations has the same fine structure as the mutant linears described above. These results indicate that (i) plus-strand primer cleavage and translocation are distinct steps that can be dissociated by mutation, (ii) lesions in sequences not included in the primer can severely inhibit primer translocation, and (iii) elongation of such untranslocated primers is responsible for the variable quantities of linear DNA that are found in all hepadnaviral stocks.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.7M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartenschlager R, Schaller H. The amino-terminal domain of the hepadnaviral P-gene encodes the terminal protein (genome-linked protein) believed to prime reverse transcription. EMBO J. 1988 Dec 20;7(13):4185–4192. [Europe PMC free article] [Abstract] [Google Scholar]
- Bosch V, Bartenschlager R, Radziwill G, Schaller H. The duck hepatitis B virus P-gene codes for protein strongly associated with the 5'-end of the viral DNA minus strand. Virology. 1988 Oct;166(2):475–485. [Abstract] [Google Scholar]
- Büscher M, Reiser W, Will H, Schaller H. Transcripts and the putative RNA pregenome of duck hepatitis B virus: implications for reverse transcription. Cell. 1985 Mar;40(3):717–724. [Abstract] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. [Europe PMC free article] [Abstract] [Google Scholar]
- Condreay LD, Aldrich CE, Coates L, Mason WS, Wu TT. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J Virol. 1990 Jul;64(7):3249–3258. [Europe PMC free article] [Abstract] [Google Scholar]
- Dressler D, Potter H. Molecular mechanisms in genetic recombination. Annu Rev Biochem. 1982;51:727–761. [Abstract] [Google Scholar]
- Enders GH, Ganem D, Varmus HE. 5'-terminal sequences influence the segregation of ground squirrel hepatitis virus RNAs into polyribosomes and viral core particles. J Virol. 1987 Jan;61(1):35–41. [Europe PMC free article] [Abstract] [Google Scholar]
- Ganem D, Greenbaum L, Varmus HE. Virion DNA of ground squirrel hepatitis virus: structural analysis and molecular cloning. J Virol. 1982 Oct;44(1):374–383. [Europe PMC free article] [Abstract] [Google Scholar]
- Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. [Abstract] [Google Scholar]
- Hirsch R, Colgrove R, Ganem D. Replication of duck hepatitis B virus in two differentiated human hepatoma cell lines after transfection with cloned viral DNA. Virology. 1988 Nov;167(1):136–142. [Abstract] [Google Scholar]
- Hirsch RC, Lavine JE, Chang LJ, Varmus HE, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature. 1990 Apr 5;344(6266):552–555. [Abstract] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. [Abstract] [Google Scholar]
- Lavine J, Hirsch R, Ganem D. A system for studying the selective encapsidation of hepadnavirus RNA. J Virol. 1989 Oct;63(10):4257–4263. [Europe PMC free article] [Abstract] [Google Scholar]
- Lien JM, Aldrich CE, Mason WS. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J Virol. 1986 Jan;57(1):229–236. [Europe PMC free article] [Abstract] [Google Scholar]
- Mason WS, Seal G, Summers J. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol. 1980 Dec;36(3):829–836. [Europe PMC free article] [Abstract] [Google Scholar]
- McGraw RA., 3rd Dideoxy DNA sequencing with end-labeled oligonucleotide primers. Anal Biochem. 1984 Dec;143(2):298–303. [Abstract] [Google Scholar]
- Molnar-Kimber KL, Summers JW, Mason WS. Mapping of the cohesive overlap of duck hepatitis B virus DNA and of the site of initiation of reverse transcription. J Virol. 1984 Jul;51(1):181–191. [Europe PMC free article] [Abstract] [Google Scholar]
- Nagaya T, Nakamura T, Tokino T, Tsurimoto T, Imai M, Mayumi T, Kamino K, Yamamura K, Matsubara K. The mode of hepatitis B virus DNA integration in chromosomes of human hepatocellular carcinoma. Genes Dev. 1987 Oct;1(8):773–782. [Abstract] [Google Scholar]
- Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990 Feb;64(2):613–620. [Europe PMC free article] [Abstract] [Google Scholar]
- Sattler F, Robinson WS. Hepatitis B viral DNA molecules have cohesive ends. J Virol. 1979 Oct;32(1):226–233. [Europe PMC free article] [Abstract] [Google Scholar]
- Seeger C, Ganem D, Varmus HE. Biochemical and genetic evidence for the hepatitis B virus replication strategy. Science. 1986 Apr 25;232(4749):477–484. [Abstract] [Google Scholar]
- Seeger C, Maragos J. Molecular analysis of the function of direct repeats and a polypurine tract for plus-strand DNA priming in woodchuck hepatitis virus. J Virol. 1989 May;63(5):1907–1915. [Europe PMC free article] [Abstract] [Google Scholar]
- Seeger C, Maragos J. Identification and characterization of the woodchuck hepatitis virus origin of DNA replication. J Virol. 1990 Jan;64(1):16–23. [Europe PMC free article] [Abstract] [Google Scholar]
- Sprengel R, Kuhn C, Will H, Schaller H. Comparative sequence analysis of duck and human hepatitis B virus genomes. J Med Virol. 1985 Apr;15(4):323–333. [Abstract] [Google Scholar]
- Summers J, Mason WS. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. [Abstract] [Google Scholar]
- Summers J, Smolec JM, Snyder R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4533–4537. [Europe PMC free article] [Abstract] [Google Scholar]
- Tuttleman JS, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986 Nov 7;47(3):451–460. [Abstract] [Google Scholar]
- Will H, Reiser W, Weimer T, Pfaff E, Büscher M, Sprengel R, Cattaneo R, Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987 Mar;61(3):904–911. [Europe PMC free article] [Abstract] [Google Scholar]
- Zoller MJ, Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. Methods Enzymol. 1987;154:329–350. [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.65.3.1255-1262.1991
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/65/3/1255.full.pdf
Free to read at jvi.asm.org
http://jvi.asm.org/cgi/content/abstract/65/3/1255
Free after 4 months at jvi.asm.org
http://jvi.asm.org/cgi/reprint/65/3/1255
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.65.3.1255-1262.1991
Article citations
The impact of integrated hepatitis B virus DNA on oncogenesis and antiviral therapy.
Biomark Res, 12(1):84, 15 Aug 2024
Cited by: 0 articles | PMID: 39148134 | PMCID: PMC11328401
Review Free full text in Europe PMC
Insights into Immune Exhaustion in Chronic Hepatitis B: A Review of Checkpoint Receptor Expression.
Pharmaceuticals (Basel), 17(7):964, 21 Jul 2024
Cited by: 1 article | PMID: 39065812 | PMCID: PMC11279883
Review Free full text in Europe PMC
Mechanisms of Hepatitis B Virus cccDNA and Minichromosome Formation and HBV Gene Transcription.
Viruses, 16(4):609, 15 Apr 2024
Cited by: 1 article | PMID: 38675950 | PMCID: PMC11054251
Review Free full text in Europe PMC
Heat Shock Protein Family A Member 1 Promotes Intracellular Amplification of Hepatitis B Virus Covalently Closed Circular DNA.
J Virol, 97(1):e0126122, 15 Dec 2022
Cited by: 1 article | PMID: 36519896 | PMCID: PMC9888207
Pregenomic RNA Launch Hepatitis B Virus Replication System Facilitates the Mechanistic Study of Antiviral Agents and Drug-Resistant Variants on Covalently Closed Circular DNA Synthesis.
J Virol, 96(24):e0115022, 30 Nov 2022
Cited by: 5 articles | PMID: 36448800 | PMCID: PMC9769369
Go to all (118) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sequence identity of the direct repeats, DR1 and DR2, contributes to the discrimination between primer translocation and in situ priming during replication of the duck hepatitis B virus.
J Mol Biol, 364(1):32-43, 07 Sep 2006
Cited by: 5 articles | PMID: 17005197 | PMCID: PMC1803024
Mutations within DR2 independently reduce the amount of both minus- and plus-strand DNA synthesized during duck hepatitis B virus replication.
J Virol, 70(12):8684-8690, 01 Dec 1996
Cited by: 13 articles | PMID: 8970995 | PMCID: PMC190963
The sequence of the RNA primer and the DNA template influence the initiation of plus-strand DNA synthesis in hepatitis B virus.
J Mol Biol, 370(3):471-480, 04 May 2007
Cited by: 26 articles | PMID: 17531265 | PMCID: PMC1991300
Hepatitis B virus reverse transcriptase and its many roles in hepadnaviral genomic replication.
Infect Agents Dis, 3(2-3):85-93, 01 Apr 1994
Cited by: 23 articles | PMID: 7529120
Review