Abstract
Unlabelled
RUNX2 expression in mesenchymal cells induces osteoblast differentiation and bone formation. BMP blocking agents were used to show that RUNX2-dependent osteoblast differentiation and transactivation activity both require BMP signaling and, further, that RUNX2 enhances the responsiveness of cells to BMPs.Introduction
BMPs and the RUNX2 transcription factor are both able to stimulate osteoblast differentiation and bone formation. BMPs function by activating SMAD proteins and other signal transduction pathways to stimulate expression of many target genes including RUNX2. In contrast, RUNX2 induces osteoblast-specific gene expression by directly binding to enhancer regions in target genes. In this study, we examine the interdependence of these two factors in controlling osteoblast differentiation in mesenchymal progenitor cells.Materials and methods
C3H10T1/2 mesenchymal cells and primary cultures of marrow stromal cells were transduced with a RUNX2 adenovirus and treated with BMP blocking antibodies or the natural antagonist, NOGGIN. Osteoblast differentiation was determined by assaying alkaline phosphatase and measuring osteoblast-related mRNA using quantitative RT/PCR. Activation of BMP-responsive signal transduction pathways (SMAD, extracellular signal-regulated kinase [ERK], p38, and c-jun-N-terminal kinase [JNK]) was assessed on Western blots.Results and conclusions
C3H10T1/2 cells constitutively synthesize BMP2 and 4 mRNA and protein, and this BMP activity is sufficient to activate basal levels of SMAD phosphorylation. Inhibition of BMP signaling was shown to disrupt the ability of RUNX2 to stimulate osteoblast differentiation and transactivate an osteocalcin gene promoter-luciferase reporter in C3H10T1/2 cells. BMP blocking antibodies also inhibited RUNX2-dependent osteoblast differentiation in primary cultures of murine marrow stromal cells. Conversely, RUNX2 expression synergistically stimulated BMP2 signaling in C3H10T1/2 cells. However, RUNX2 did not increase the ability of this BMP to activate SMAD, ERK, p38, and JNK pathways. This study shows that autocrine BMP production is necessary for the RUNX2 transcription factor to be active and that BMPs and RUNX2 cooperatively interact to stimulate osteoblast gene expression.Free full text

BMP Signaling Is Required for RUNX2-Dependent Induction of the Osteoblast Phenotype
Abstract
RUNX2 expression in mesenchymal cells induces osteoblast differentiation and bone formation. BMP blocking agents were used to show that RUNX2-dependent osteoblast differentiation and transactivation activity both require BMP signaling and, further, that RUNX2 enhances the responsiveness of cells to BMPs.
Introduction
BMPs and the RUNX2 transcription factor are both able to stimulate osteoblast differentiation and bone formation. BMPs function by activating SMAD proteins and other signal transduction pathways to stimulate expression of many target genes including RUNX2. In contrast, RUNX2 induces osteoblast-specific gene expression by directly binding to enhancer regions in target genes. In this study, we examine the interdependence of these two factors in controlling osteoblast differentiation in mesenchymal progenitor cells.
Materials and Methods
C3H10T1/2 mesenchymal cells and primary cultures of marrow stromal cells were transduced with a RUNX2 adenovirus and treated with BMP blocking antibodies or the natural antagonist, NOGGIN. Osteoblast differentiation was determined by assaying alkaline phosphatase and measuring osteoblast-related mRNA using quantitative RT/PCR. Activation of BMP-responsive signal transduction pathways (SMAD, extracellular signal-regulated kinase [ERK], p38, and c-jun-N-terminal kinase [JNK]) was assessed on Western blots.
Results and Conclusions
C3H10T1/2 cells constitutively synthesize BMP2 and 4 mRNA and protein, and this BMP activity is sufficient to activate basal levels of SMAD phosphorylation. Inhibition of BMP signaling was shown to disrupt the ability of RUNX2 to stimulate osteoblast differentiation and transactivate an osteocalcin gene promoter-luciferase reporter in C3H10T1/2 cells. BMP blocking antibodies also inhibited RUNX2-dependent osteoblast differentiation in primary cultures of murine marrow stromal cells. Conversely, RUNX2 expression synergistically stimulated BMP2 signaling in C3H10T1/2 cells. However, RUNX2 did not increase the ability of this BMP to activate SMAD, ERK, p38, and JNK pathways. This study shows that autocrine BMP production is necessary for the RUNX2 transcription factor to be active and that BMPs and RUNX2 cooperatively interact to stimulate osteoblast gene expression.
INTRODUCTION
BMPs, MEMBERS OF TGF-β superfamily, are potent osteogenic agents that stimulate maturation of mesenchymal osteoprogenitor cells to osteoblast.(1–5) BMP signaling is initiated by binding of ligand to type I and II BMP receptors at the cell surface.(6) Ligand-induced receptor activation initiates two types of signal transduction pathways: (1) the canonical SMAD pathway in which receptor-specific SMAD1, 5, and 8 are activated, form complexes with the common partner, SMAD4, and translocate into the nucleus to regulate the transcription of target genes,(7–10) and (2) mitogen-activated protein kinase (MAPK) pathways including p38, c-jun-N-terminal kinase (JNK), and extracellular signal-regulated kinase (ERK) pathways.(9,11–14)
Among the downstream targets of BMPs are RUNX2 and other osteoblast-related transcription factors such as DLX5 and osterix.(15–18) RUNX2, the osteoblast-specific product of the Cbfa1 gene, is a runt domain-containing transcription factor that is essential for osteoblast differentiation and bone formation. Cbfa1 is also the locus of cleidocranial dysplasia (CCD), an autosomal dominant human disorder associated with defects in the cranial and appendicular skeleton.(19–21) RUNX2 overexpression in mesenchymal cell lines and primary cultures of marrow stromal cells induces osteoblast-specific gene expression and mineralization in vitro as well as bone formation after cell implantation into animal hosts.(22–30)
Because BMPs can upregulate RUNX2 under certain conditions, it has been proposed that RUNX2 is the principle mediator of downstream BMP actions and can function in place of BMPs in bone regeneration applications.(26) However, a number of lines of evidence suggest that certain BMP actions on bone are independent of RUNX2. For example, although BMPs are unable to induce mineralization in calvarial cells from Cbfa1−/− mice, they are still able to weakly induce osteocalcin and alkaline phosphatase gene expression as well as stimulate chondrocyte differentiation.(19,31) Also, several studies showed that pre-osteoblast cell lines such as MC3T3-E1 cells and marrow stromal cells produce BMPs, and this autocrine BMP signal is necessary for the differentiation of these cells even though they already express RUNX2.(32,33) Last, RUNX2 is known to physically interact with SMAD1 and 5 and functionally cooperate with these molecules to stimulate gene expression.(34–37)
There is also considerable evidence for the concept that members of the RUNX family of transcription factors are required for activity of TGF-β–related factors including BMPs. We previously reported that RUNX2 transduction dramatically increases the responsiveness of C3H10T1/2 cells to an adenovirus encoding BMP2.(25) By all criteria examined (induction of alkaline phosphatase, osteocalcin, or mineralization), combined transduction with AdRUNX2 and AdBMP2 vectors stimulated osteoblast differentiation to levels that were up to 10-fold greater than the sum of individual levels found in cells transduced with either AdRUNX2 or AdBMP2. Similarly, RUNX2 is required for TGF-β induction of collagenase-3 in osteoblast and breast cancer cells,(38,39) whereas RUNX3 is necessary for TGF-β to inhibit the growth of carcinoma cell lines.(40)
In view of these findings, in this study, we address the issue as to whether BMP signaling is necessary for RUNX2 transcriptional activity in mesenchymal osteoprogenitor cells and also further explore the basis for synergistic interactions between RUNX2 and BMPs. As will be shown, mesenchymal osteoprogenitor cells produce BMPs in an autocrine manner and inhibition of their activity using the natural antagonist, NOGGIN, or blocking antibodies prevents RUNX2 from stimulating osteoblast differentiation. Furthermore, the RUNX2-dependent stimulation of BMP responsiveness previously reported will be shown to occur in the absence of any changes in known BMP receptor–mediated signal transduction pathways.
MATERIALS AND METHODS
Reagents
Tissue culture medium and FBS were obtained from Invitrogen. L-ascorbic acid was purchased from Aldrich Chemicals. Recombinant human BMP2 (rhBMP2), monoclonal anti-human BMP2/4 antibody (BMP2/4 blocking antibody), and monoclonal anti-human BMP7 antibody (BMP7 blocking antibody) were purchased from R&D Bio-systems. NOGGIN was a generous gift from Dr Aris Economides (Regeneron Phamaceuticals). Anti-phospho-SMAD 1/5/8, anti-p38, anti-phospho-p38, anti-SAPK/JNK, anti-phospho-SAPK/JNK, anti-p42/44 ERK, and anti-phospho-p42/44 ERK were purchased from Cell Signaling Technologies. Anti-SMAD1/5 was obtained from Upstate Biotechnology. Anti- RUNX2 was purchased from Santa Cruz.
Cell cultures and transfections
The murine pluripotent mesenchymal cell line, C3H10T1/2,(41) was obtained from American Type Culture Collection. MC3T3-E1 subclone 4 (MC4) and subclone 42 (MC42) were previously described.(42) Both subclones express high levels of osteoblast-specific markers and mineralize after growth in ascorbic acid (AA)-containing medium for several days, although MC42 cells have higher levels of alkaline phosphatase (ALP). MC42 cells also contain stably transfected copies of a 1.3-kb murine osteocalcin gene 2 (mOG2) promoter driving a firefly luciferase reporter gene. Luciferase activity in these cells parallels endogenous levels of osteocalcin mRNA.(43) Both C3H10T1/2 cells and MC3T3-E1 cells were maintained in AA-free α-MEM containing 10% FBS and 1% penicillin/streptomycin. Primary marrow stromal cells were prepared from 4- to 6-week-old C57BL6 mice as previously described.(30) Unless indicated otherwise, cells were plated at a density of 50,000 cells/cm2 and treated every other day with α-MEM containing 10% FBS, 1% penicillin/streptomycin, and 50 μg/ml ascorbic acid (differentiating medium) and the indicated additions. Cell cultures were transfected with the indicated plasmids using LipofectAMINE reagent (Life Sciences Technologies) as previously described.(43)
Transduction
Control adenovirus (AdLacZ) was purchased from the University of Michigan vector core. Adenovirus expressing RUNX2 (AdRUNX2) was previously described.(25) Viruses were added to cells in serum-free medium. After 4 h, serum was added to a final concentration of 2%. Twenty-four hours later, cells were transferred to complete medium containing 50 μg/ml of ascorbic acid. BMP blocking antibodies (5 ηg/ml), NOGGIN (50 ηg/ml), or rhBMP2 at the desired dose were added to cells 24 h after transduction as indicated.
RNA analysis and real-time PCR
Total RNA was harvested from cells using TRIzol reagent (Invitrogen) and cleaned with RNeasy Mini Kit (Qiagen). Two micrograms of total RNA was reverse transcribed using Taqman reverse transcription reagent for first-strand synthesis (Applied Biosystems). All real-time PCR reactions contained the first-strand cDNA corresponding to 20 ηg of RNA, Taqman universal mastermix and Taqman assay reagents. Taqman predeveloped assay reagents were used for detection of mouse BSP (FAM/MGB probe; Applied Biosystems), mouse RUNX2 (FAM/MGB probe; Applied Biosystems), and mouse GAPDH (FAM/MGB probe; Applied Biosystems). Forward primer (5′-GACCTCACAGATGCCAAGCC-3′), backward primer (5′-CCCTCCTGCTTGGACGACATGA-3′), and Taqman TAMPA probe (5′-AGCGGCCCTGAGTCTGACAAAGCC-3′) were used to quantify osteocalcin expression. Universal mouse reference RNA (Stratagene) was used to generate a relative standard curve. Real-time detection of PCR products was performed using a ABI PRISM 7700 sequence detector (Applied Biosystems) in the Dental School Molecular Biology Core Facility. The real-time PCR product, mRNA expression, was calculated based on a relative standard curve and normalized to GAPDH. All real-time PCR analysis used triplicate independent samples.
Biochemical assays
ALP activity was measured in cell layers using p-nitrophenyl phosphate substrate as previously described.(44) ALP activity was normalized with DNA that was measured by PicoGreeen dsDNA quantitation kit (Molecular Probes). Luciferase activity was measured with reagents and protocols from Promega. The luciferase activity was normalized to renilla activity in C3H10T1/2 cells and DNA in MC42 cells.
Western blot analysis
Cells were lysed by incubation at 4°C for 30 minutes in buffer containing phosphatase inhibitor (20 mM sodium phosphate, pH 7, 250 mM NaCl, 30 mM sodium pyrophosphate, 0.1% NP-40, 5 mM EDTA, 10 mM NaF, 0.1 mM Na3VO4, and 1% protease inhibitor cocktail). Samples were fractioned by 10% SDS-PAGE gel and transferred to a nitrocellulose membrane (Schleicher & Schuel). Primary antibodies for all phospho-proteins were used at a dilution of 1:500; all other primary antibodies were used at 1:1000 dilution. Secondary antibody (horseradish peroxidase–conjugated goat anti-rabbit IgG) was used at 1:10,000 dilution. Immunoreactivity was detected by enhanced chemiluminescence (ECL; Amersham). BMP standards were prepared from conditioned medium of COS7 cells transduced with adenoviruses encoding BMP2, 4, or 7 as previously described.(45)
Statistical analysis
All experiments were repeated at least twice, and qualitatively identical results were obtained. Experimental results are reported as means ± SD based on triplicate independent samples.
RESULTS
Effects of RUNX2 on autocrine BMP synthesis
As discussed in the Introduction, osteoblasts and marrow stromal cells are known to produce BMPs that may stimulate the differentiation of these cells in an autocrine manner. The experiment shown in Figs. 1A and 1B was conducted to examine BMP expression in AdLacZ (control) and AdRUNX2-transduced C3H10T1/2 cells. The virus titer of 100 pfu/cell used in these studies was previously shown to maximally stimulate osteoblast differentiation.(25) BMP2, BMP4, and BMP7 mRNA was expressed in both cell populations (detected by quantitative RT/PCR; Fig. 1A). Similarly, BMP2 and 4 proteins were clearly seen when cell extracts from C3H10T1/2 cells were examined on Western blots. However, we were unable to detect BMP7 even though the BMP7 standard was clearly visualized (Fig. 1B). Figure 1A also shows that AdRUNX2 transduction stimulated BMP4 mRNA 2.2-fold. A similar increase in BMP4 protein levels was observed after densitometric analysis of Western blot data, whereas little or no change was observed for BMP2. Interestingly, BMPs were not detected in conditioned media from cells but were found exclusively associated with cell/extracellular matrix fractions.

Autocrine BMP synthesis in C3H10T1/2 cells. Cells were transduced with AdLacZ (open bars) or AdRUNX2 (gray bars) at 100 pfu/cell and harvested after 6 days for analysis of BMP (A) mRNA and (B) protein. Relative mRNA levels were measured by Q-RT/PCR and normalized to GAPDH mRNA. BMP protein in whole cell extracts was measured on Western blots. BMP-2, -4, and -7 standards (STD) were prepared from COS7 cell-conditioned medium.
BMP activity is required for induction of osteoblast differentiation by RUNX2
To determine whether autocrine BMP production in C3H10T1/2 cells is sufficient to stimulate BMP signaling, we examined basal SMAD phosphorylation in cell extracts from control and AdRUNX2-transduced cells in the absence of any added BMPs. Western blots were probed with an anti-phospho-SMAD1/5/8 antibody as well as antibodies against total SMAD1/5 and RUNX2 (Fig. 2A). Phosphorylated SMADs were detected in approximately equal amounts in control and AdRUNX2-transduced cells. Significantly, SMAD phosphorylation required autocrine BMP production in that it was almost completely eliminated if cells were treated with BMP blocking antibodies (combined treatment with monoclonal anti-human BMP2/4 antibody and monoclonal anti-human BMP7 antibody). In contrast, antibody treatment did not affect total SMAD1 and 5 or levels of virally expressed RUNX2 protein. This result shows that BMPs in the extracellular matrix (ECM) are accessible to antibodies that can penetrate the matrix to block BMP function.

Effect of BMP inhibition on SMAD activation and AdRUNX2-induced ALP activity. C3H10T1/2 cells or primary cultures of murine MSCs were transduced with AdLacZ or AdRUNX2 as in Fig. 1. (A) Inhibition of SMAD phosphorylation. Forty-eight hours after transduction, C3H10T1/2 cells were treated with preimmune serum or combined BMP2/4 and BMP7 blocking antibodies at 5 ηg/ml. After 4 h, whole cell extracts were prepared for Western blot detection of phospho-SMADs 1/5/8, total SMADs 1/5, and RUNX2 as indicated. (B) Inhibition of ALP induction in C3H10T1/2 cells. Cells were treated with NOGGIN (50 ηg/ml), BMP2/4 blocking antibody (5 ηg/ml), BMP7 blocking antibody (5 ηg/ml), or combined BMP2/4 and BMP7 blocking antibodies (5 ηg/ml each) as indicated for 5 days and harvested for measurement of ALP activity. (C) Inhibition of ALP induction in MSCs. Cells were treated with BMP blocking antibodies as indicated. AdLacZ, open bars; AdRUNX2, gray bars.
As shown in Fig. 2B, autocrine BMP production is also required for AdRUNX2-dependent osteoblast differentiation. BMP blocking antibodies and the natural BMP antagonist, NOGGIN, were used to inhibit BMP signals in AdRUNX2-transduced C3H10T1/2 cells. NOGGIN, an extracellular BMP-binding protein that forms inactive complexes with BMP2, 4, and to a lesser extent, BMP7,(46) inhibited AdRUNX2-induced ALP activity by 77.7%. Individual treatment with either BMP2/4 or BMP7 blocking antibody decreased AdRUNX2-induced ALP activity by 63.8% and 47.1%, respectively, whereas combined treatment with both antibodies further decreased AdRUNX2-induced ALP activity by 82.6%.
Because C3H10T1/2 cells are an immortalized mesenchymal cell line that may differ from normal mesenchymal cells, we considered it important to determine if BMP activity is also required for AdRUNX2-induced osteoblast differentiation of marrow stromal cells (MSCs). This cell population is known to contain pluripotent mesenchymal precursors having intrinsic osteogenic potential that can be greatly enhanced by transduction with RUNX2 expression vectors.(30) Furthermore, autocrine BMP production has previously been documented in these cells.(32) Marrow stromal cells were isolated from the marrow of C57BL6 mice and treated with AdRunx2 in the presence or absence of BMP2/4 and BMP7 blocking antibodies. As shown in Fig. 2C, antibody treatment inhibited ALP activity in marrow stromal cells by 65%.
To confirm that inhibition of BMP activity blocks overall osteoblast differentiation, we also examined the ability of BMP blocking antibodies to inhibit late osteoblast differentiation marker mRNA in C3H10T1/2 cells. As shown in Fig. 3, combined treatment with BMP2/4 and BMP7 blocking antibodies decreased AdRUNX2-induced osteocalcin (OCN) mRNA expression by 72.5% (Fig. 3A) and bone sialoprotein (BSP) mRNA expression by 98% in C3H10T1/2 cells (Fig. 3B).

Effect of BMP inhibition on AdRUNX2-induced late osteoblast differentiation markers. C3H10T1/2 cells were transduced with AdLacZ (open bars) or AdRUNX2 (gray bars) at 100 pfu/cells and treated with combined BMP2/4 and BMP7 blocking antibodies as indicated in Fig. 2. After 6 days, cells were harvested for mRNA analysis by Q-RT/PCR. (A) OCN mRNA. (B) BSP mRNA. Values are expressed as fold increase relative to Ad-LacZ-treated controls.
The studies described above all examined effects of BMP inhibition in virally transduced mesenchymal cells overexpressing RUNX2. To examine the role of BMP signaling on RUNX2 activity in a more physiological context, effects of BMP2/4 and BMP7 blocking antibodies were also examined in MC3T3-E1 pre-osteoblast cells. These cells constitutively express RUNX2, BMP2, BMP4, and BMP7.(33) Ascorbic acid (AA) induces differentiation of MC3T3-E1 cells by stimulating synthesis of a collagenous extracellular matrix.(47) This matrix interacts with cell surface integrins that subsequently activate the MAPK pathway, RUNX2 phosphorylation, and transcriptional activity.(43,48,49) Thus, much of the responsiveness of these cells to AA is mediated by RUNX2 and its associated enhancer sequences in the regulatory regions of target genes. As shown in Fig. 4, BMP blocking antibodies decreased AA-induced ALP activity by 67.2% (Fig. 4A), OCN expression by 60.7% (Fig. 4B), and BSP expression by 72% (Fig. 4C). AA also increased RUNX2 mRNA levels by 1.8- to 2-fold. However, this induction was not affected by antibody treatment (Fig. 4D). Related studies from our group previously showed that the BMP antagonist, NOGGIN, blocked AA-induced differentiation in these cells, further supporting the concept that BMP activity is essential for osteoblast differentiation.(33)

Effect of BMP inhibition on AA-induced osteoblast differentiation markers in MC3T3-E1 pre-osteoblast cells. (A) Clone 4 MC3T3-E1 cells were growth in the presence (gray bars) or absence of AA (open bars) and treated with combined BMP2/4 and BMP7 blocking antibodies (5 ηg/ml each) for 5 days and assayed for ALP activity. Total RNA was isolated from replicate samples and assayed for (B) OCN, (C) BSP, and (D) RUNX2 mRNA using Q-RT/PCR.
Taken together, these results clearly show that BMP activity is required for RUNX2 to exert its pro-differentiation effects on mesenchymal and pre-osteoblast cells.
RUNX2-dependent transcriptional activation of the osteocalcin gene also requires BMP activity
RUNX2 regulates gene expression by directly binding to specific enhancer sequences in regulatory regions of target genes. The murine osteocalcin gene 2 promoter (mOG2) contains two such sequences, designated OSE2a and OSE2b, at −131 and −602 bp upstream from the transcription start site that both participate in the transcriptional activation of this gene.(50) Because BMP blocking antibodies dramatically inhibited RUNX2-dependent osteoblast differentiation without altering RUNX2 levels, we hypothesized that BMPs may modify the transcriptional activity of RUNX2. To address this issue, we examined effects of BMP blocking antibodies on RUNX2-dependent activation of the mOG2 promoter (Fig. 5A). For this experiment, C3H10T1/2 cells were transfected with a 1.3-kb mOG2 promoter fragment driving firefly luciferase together with β-galactosidase or RUNX2 expression vectors. The 1.3-kb promoter fragment contains all known sequences necessary for the osteoblast-specific expression of Ocn.(50) Interestingly, BMP blocking antibodies totally prevented RUNX2 from stimulating luciferase activity in this system without affecting basal levels of promoter activity (Fig. 5A). To examine the role of BMPs on RUNX2 activity in pre-osteoblasts, blocking antibodies were also examined in MC42 cells, a highly differentiating MC3T3-E1 subclone that contains stably integrated copies of the 1.3-kb mOG2 promoter driving firefly luciferase.(43) Antibodies dramatically decreased AA-induced luciferase activity without affecting basal promoter activity (Fig. 5B).
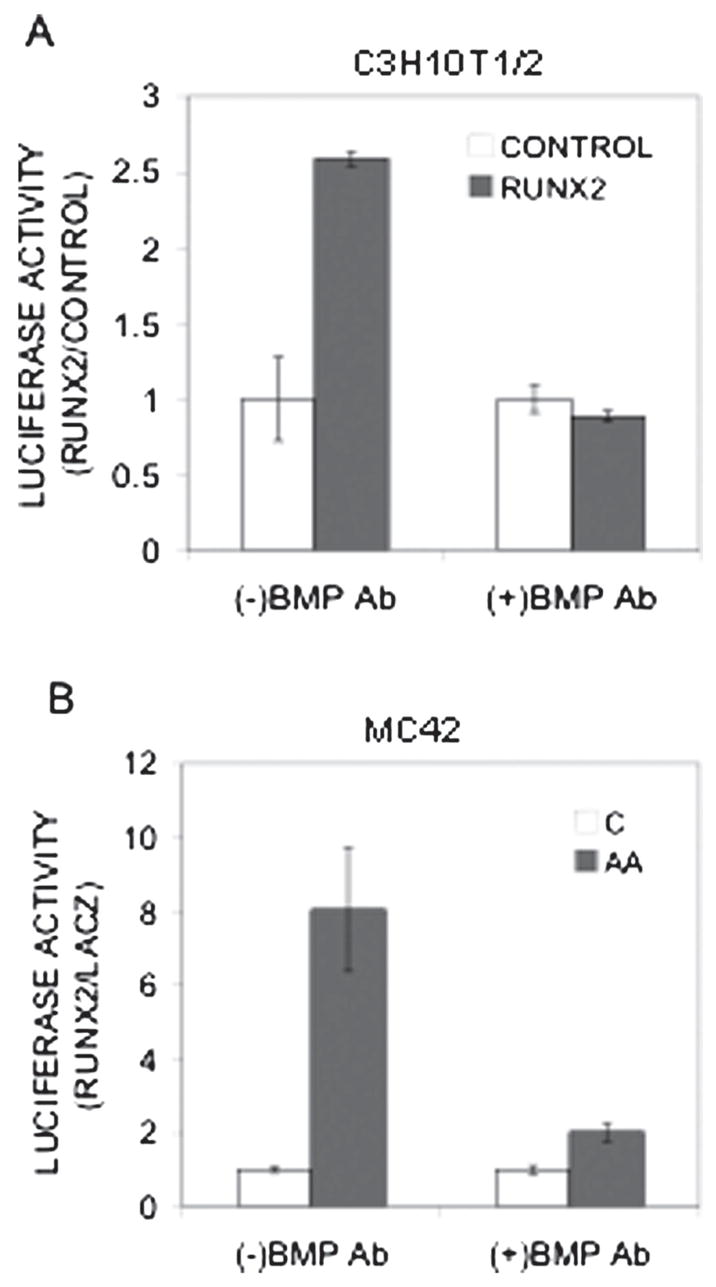
Effect of BMP inhibition on RUNX2 transcriptional activity. (A) Inhibition of RUNX2-activated OCN promoter activity. C3H10T1/2 cells were transfected with a 1.3-kb mOG2-luc reporter plasmid, renilla luciferase plasmid (to assess transfection efficiency), and either LacZ (open bars) or RUNX2 (gray bars) expression plasmids as indicated. Cells were treated with combined BMP2/4 and BMP7 blocking antibodies (5 ηg/ml each) and harvested 4 days later for measurement of luciferase activity. All data were normalized for transfection efficiency and expressed as fold increase relative to LacZ control samples. The result was reported as fold increase of luciferase activity normalized with Renilla activity. (B) Inhibition of AA-induced OCN promoter activity. MC42 cells were grown in control (open bars) or AA-containing medium (gray bars) and treated with combined BMP2/4 and BMP7 blocking antibodies (5 ηg/ml each) for 5 days before luciferase activity was measured. Results were normalized to DNA and reported as fold increase relative to untreated controls.
RUNX2 regulation of BMP responsiveness
As developed in the Introduction, there is abundant evidence that the RUNX family of transcription factors is required for responsiveness of cells to BMPs and TGF-β. We previously reported that RUNX2 transduction dramatically increases the in vitro and in vivo responsiveness of C3H10T1/2 cells to an adenovirus encoding BMP2 and speculated that RUNX2 makes these cells more sensitive to the differentiating effects of BMPs.(25) The studies presented in Fig. 6 show this to be the case. C3H10T1/2 cells were transduced with control (LacZ) or RUNX2 virus, treated with rhBMP2, and examined for osteoblast differentiation. Time-course and BMP dose/response analyses for alkaline phosphatase induction (Figs. 6A and 6B) revealed that, in the absence of RUNX2, cells only exhibited a weak response to rhBMP2 beginning at day 6. In contrast, AdRUNX2 transduction dramatically increased the sensitivity of cells to BMP stimulation. Depending on the time-point examined, BMP-induced ALP activity in RUNX2-transduced cells was 2- to 10-fold greater than the sum of activities of cells separately treated with AdRUNX2 or rh-BMP2. The increase in BMP sensitivity with RUNX2 transduction was particularly striking when the dose dependence of the BMP response was examined at early times (day 5). At this time-point, rhBMP2 alone had minimal activity even at a saturating concentration of 100 ηg/ml. In contrast, rhBMP2 at concentrations as low as 25 ηg/ml stimulated ALP activity in RUNX2-expressing cells. Consistent with these results, the ability of rhBMP2 to induce OCN and BSP RNA was also greatly enhanced in AdRUNX2-transduced cells (Figs. 6C and 6D).
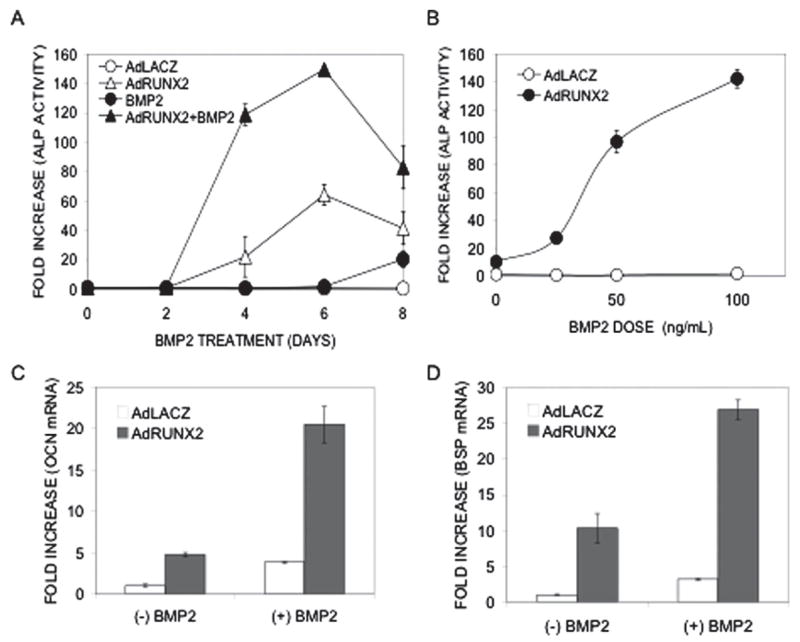
Effect of RUNX2 on cellular responsiveness to BMP2. (A) Synergistic induction of ALP activity. C3H10T1/2 cells were transduced with either AdLacZ or AdRUNX2 at 100 pfu/cells, treated with rh-BMP2 at 50 ηg/ml, and harvested for ALP activity at the indicated time. AdLacZ control (○), AdLacZ control plus rhBMP2 (●), AdRUNX2 (Δ), AdRUNX2 plus rhBMP2 (▲). (B) BMP dose-dependence. C3H10T1/2 cells were tranduced as in A, treated with the indicated concentration of rhBMP2 for 5 days, and harvested for ALP activity. Ad-LacZ control (○), AdLacZ control plus rh-BMP2 (●). (C and D) Induction of OCN and BSP mRNA. C3H10T1/2 cells were transduced as in A and B, treated with rhBMP2 at 100 ηg/ml for 5 days, and harvested for measurement of (C) OCN and (D) BSP mRNA levels by Q-RT/PCR. AdLacZ, open bars; AdRUNX2, gray bars.
Activation of BMP signal transduction pathways
There are a number of possible explanations for how RUNX2 might enhance BMP responsiveness of cells including effects on BMP receptors, SMAD protein intermediates, effects on other BMP-responsive signal transduction pathways, or related nuclear factors. As a first step in exploring this area, we examined the responsiveness of BMP-related signal transduction pathways in control and RUNX2-expressing C3H10T1/2 cells. For this experiment, cells were transduced with AdLacZ or AdRUNX2 and treated with rhBMP2 for 30 minutes before cell layers were harvested for analysis. Western blots were used to examine activation of SMAD, p38, JNK, and ERK pathways (Fig. 7). As expected, phospho-SMADs 1/5/8 were dramatically increased by rhBMP2 treatment. However, RUNX did not affect the extent of SMAD phosphorylation and only slightly increased total SMAD levels. Similarly, phospho-p38 was slightly increased by rhBMP2, but to the same extent in both control and AdRUNX2-transduced cells. Although total and phospho-JNK levels were strikingly increased in AdRUNX2-transduced cells, levels were not affected by rhBMP2 treatment. Last, phospho-ERK was not affected by either AdRUNX2 transduction and BMP2 treatment. In summary, effects of BMP2 on previously characterized BMP signal transduction pathways were not affected by RUNX2. Our results suggested that RUNX2 must affect events that are downstream from these membrane receptor–initiated signals.

Western blot analysis of BMP2-responsive signal transduction pathways. C3H10T1/2 cells were transduced with Ad-LacZ or AdRUNX2 at 100 pfu/cell. Forty-eight hours after transduction, cells were treated with rhBMP2 for 30 minutes, harvested, and assayed on Western blots for the following signaling intermediates: phospho-SMAD 1/5/8, total SMADs 1/5, phospho-JNK, total JNK, phospho-p38, total p38, phospho-ERK, total ERK, and total RUNX2.
DISCUSSION
This study examined interrelationships between BMPs and the RUNX2 transcription factor in stimulating osteoblast differentiation. Two general conclusions can be drawn from this work: (1) the transcriptional activity of RUNX2 in osteogenic precursor cells requires BMP signaling and (2) the sensitivity of cells to BMPs is enhanced in the presence of RUNX2. These findings extend previous work from this and other laboratories showing a requirement for autocrine BMP production in the differentiation of pre-osteoblasts and document the existence of synergistic interactions between RUNX2 and BMPs. Although RUNX2 is induced by BMPs in certain systems, our studies indicate that this transcription factor cannot strictly replace BMPs in the osteoblast differentiation pathway.
RUNX2 overexpression is known to induce osteoblast phenotypic markers in a number of cell types including fibroblasts, mesenchymal cell lines such as C3H10T1/2 cells, primary myoblasts, and marrow stromal cells.(22,25,26,28,30) In the case of C3H10T1/2 cells and MSCs, our group showed that RUNX2 induced in vitro osteoblast differentiation and mineralization as well as in vivo bone formation after implantation of cells at subcutaneous sites.(25,30) Because RUNX2 physically associates with enhancer sequences in target genes, it has generally been assumed that it directly stimulates gene expression in the absence of other signals. This study shows this not to be the case. Instead, we find that C3H10T1/2 cells constitutively express BMPs 2 and 4 at levels that can be detected on Western blots. This BMP activity was exclusively associated with cell layers, consistent with earlier studies showing sequestration of TGF-β and BMPs by the ECM. In the case of TGF-β, this is explained by the association of this growth factor with LTBP-1 (latent TGF-β –binding protein-1) that subsequently targets it to the ECM.(51) The basis for ECM sequestration of BMPs is less well understood. However, a recent report showed that BMP7, whereas not associating with LTBP-1, can form a complex with a homologous factor, fibrillin-1, that co-localizes with BMP7 in the ECM of several tissues.(52) Cell layer–associated BMPs are biologically active in that they can stimulate basal levels of SMAD 1/5/8 phosphorylation (Fig. 2). Thus, when RUNX2 is expressed in this system, it is in an environment where BMP signals (i.e., P-SMADS) are already present. These BMP signals are required for RUNX2 to be transcriptionally active because RUNX2-dependent stimulation of the mOG2 promoter and induction of osteoblast marker mRNA expression was completely inhibited by BMP blocking antibodies. Consistent with these findings, Lee et al.(15) previously examined the effects of RUNX2 induction in C2C12 pre-myoblast cells and concluded that RUNX2 could not induce osteoblast differentiation in the absence of BMP signals provided by either direct BMP treatment or SMAD5 overexpression.
As noted above, RUNX2 overexpression has been reported to stimulate osteoblast differentiation in a variety of cell types including fibroblasts, MSCs, and myoblasts. Because RUNX2 can be induced by BMPs in certain systems, it has been proposed that it is functionally downstream of the BMP pathway and can, therefore, substitute for BMPs as an osteogenic agent.(26) In light of these results, it is unlikely that RUNX2 can function in such an autonomous manner. In fact, BMPs are known to be constitutively expressed by most of the cell types that were used in RUNX2 overexpression experiments including fibroblasts, marrow stromal cells, and myocytes.(32,53,54) Therefore, it is likely that low levels of phospho-SMADs were present when RUNX2 was overexpressed.
We previously reported that adenoviruses expressing RUNX2 and BMP2 synergistically interact to stimulate osteoblast differentiation in C3H10T1/2 cells. This study extends these findings by showing that RUNX2 also dramatically increases the sensitivity of cells to rhBMP2. Although prolonged exposure of C3H10T1/2 cells to rBMPs has been reported to stimulate osteoblast differentiation,(5) these cells are relatively insensitive to BMPs, particularly after short treatment times. For example, in Fig. 6, rhBMP2 had little or no effect on alkaline phosphatase after 6 days and only weakly induced this differentiation marker after 8 days. In contrast, rhBMP2 was very active in RUNX2-expressing cells even after only 4 days of treatment and at rhBMP concentrations as low as 25 ηg/ml. This RUNX2 requirement is consistent with previous studies showing that calvarial cells from Cbfa1-deficient mice have a severely attenuated response to BMPs.(19)
There are several potential mechanisms to explain how RUNX2 might increase BMP responsiveness. Our studies clearly show that results cannot be explained by effects of RUNX2 on early components of the BMP signal transduction pathway including upregulation of BMP levels, SMAD levels/phosphorylation kinetics, or activation of other BMP-related pathways. Although RUNX2 did increase BMP4 levels ~2-fold, this change was not accompanied by increased SMAD1/5/8 phosphorylation. Similarly, the ability of rhBMP2 to stimulate non-SMAD pathways including p38, JNK, or ERK pathways was not affected by RUNX2 (Fig. 7). Interestingly, RUNX2 did increase total and phospho-JNK levels, although neither component was affected by rhBMP2 treatment. Taken together, these studies suggest that RUNX2 interacts with the BMP signaling pathway at stages after receptor activation.
As noted in the Introduction, RUNX2 is known to physically and functionally interact with Smads.(35,37) A CBFA1 mutant containing a nonsense mutation at A376 of the human sequence was previously identified in a cleidocranial dysplasia family. The resulting truncated Runx2 was unable to interact with Smads or induce an osteoblast phenotype in C2C12 mesenchymal cells after BMP stimulation.(37) Recently, the Smad binding site of Runx2 was more precisely localized to the region between amino acids 432–391 and shown to be embedded in a previously defined nuclear matrix targeting region.(55) Based on these studies, the increased BMP responsiveness of RUNX2-expressing cells may be at least partially explained by RUNX2–SMAD interactions.
However, time-course studies of BMP induction of osteoblast differentiation in the presence of RUNX2 indicate that additional secondary mediators of the BMP response may also be required. Specifically, we find that osteoblast marker induction (i.e., induction of ALP activity, BSP and OCN mRNA) is not seen until 48–96 h after BMP addition to RUNX2-expressing cells (Fig. 6 and unpublished studies). This is not the result that would be expected if a RUNX2–SMAD complex directly regulated osteoblast marker genes, because this complex would be expected to form within a few minutes after BMP treatment (recall that Smad phosphorylation is stimulated within 30 minutes of BMP treatment; Fig. 7). Possible candidates for downstream BMP-inducible signals that might mediate this response include DLX5 and OSX transcription factors.(17,18,56) These factors may function alone or together with RUNX2 to induce gene expression. Ongoing studies in the project laboratory are exploring the role of these and related factors in the RUNX2/BMP response.
Our work suggests that a BMP/RUNX2 axis controls osteoblast differentiation, with BMPs providing signals that enhance RUNX2-dependent transcription and RUNX2 providing information necessary for BMP activity. Viewing BMPs and RUNX2 as working together may explain widely observed discrepancies between the rapid early effects of BMPs on SMAD phosphorylation and long-term effects on osteoblast-specific gene expression and differentiation of mesenchymal cells that normally require many hours to days. Cooperativity between BMPs and RUNX2 might explain this delay between early and late BMP responses as follows: BMP treatment would initially upregulate RUNX2 making cells more responsive to BMPs that would in turn further upregulate RUNX2 and other downstream mediators to induce the osteoblast phenotype.
Acknowledgments
The authors thank Dr Aris Economides for kindly providing NOGGIN. This work was supported by National Institutes of Health Grants DE13386 and DE11723 to RTF.
References
Full text links
Read article at publisher's site: https://doi.org/10.1359/jbmr.060109
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1359/jbmr.060109
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1359/jbmr.060109
Article citations
Selective inhibition of interleukin 6 receptor decreased inflammatory cytokines and increased proteases in an experimental model of critical calvarial defect.
Braz J Med Biol Res, 57:e13913, 19 Aug 2024
Cited by: 0 articles | PMID: 39166608 | PMCID: PMC11338548
Impact of Strontium, Magnesium, and Zinc Ions on the In Vitro Osteogenesis of Maxillary Sinus Membrane Stem Cells.
Biol Trace Elem Res, 16 Aug 2024
Cited by: 0 articles | PMID: 39150638
Anti-Microbial Drug Metronidazole Promotes Fracture Healing: Enhancement in the Bone Regenerative Efficacy of the Drug by a Biodegradable Sustained-Release In Situ Gel Formulation.
Biomedicines, 12(7):1603, 18 Jul 2024
Cited by: 0 articles | PMID: 39062176 | PMCID: PMC11274654
USP36 regulates the proliferation, survival, and differentiation of hFOB1.19 osteoblast.
J Orthop Surg Res, 19(1):483, 17 Aug 2024
Cited by: 0 articles | PMID: 39152465 | PMCID: PMC11330066
Nanotopography Influences Host-Pathogen Quorum Sensing and Facilitates Selection of Bioactive Metabolites in Mesenchymal Stromal Cells and Pseudomonas aeruginosa Co-Cultures.
ACS Appl Mater Interfaces, 16(33):43374-43386, 08 Aug 2024
Cited by: 0 articles | PMID: 39113638 | PMCID: PMC11345723
Go to all (243) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Synergistic effects of Nell-1 and BMP-2 on the osteogenic differentiation of myoblasts.
J Bone Miner Res, 22(6):918-930, 01 Jun 2007
Cited by: 51 articles | PMID: 17352654 | PMCID: PMC2866074
Reconstitution of Runx2/Cbfa1-null cells identifies a requirement for BMP2 signaling through a Runx2 functional domain during osteoblast differentiation.
J Cell Biochem, 100(2):434-449, 01 Feb 2007
Cited by: 55 articles | PMID: 16927309
Harmine promotes osteoblast differentiation through bone morphogenetic protein signaling.
Biochem Biophys Res Commun, 409(2):260-265, 06 May 2011
Cited by: 38 articles | PMID: 21570953
Smad-Runx interactions during chondrocyte maturation.
J Bone Joint Surg Am, 83-A Suppl 1(pt 1):S15-22, 01 Jan 2001
Cited by: 53 articles | PMID: 11263661
Review
Funding
Funders who supported this work.
NIDCR NIH HHS (6)
Grant ID: R01 DE011723-10
Grant ID: DE11723
Grant ID: DE13386
Grant ID: R56 DE011723
Grant ID: R01 DE011723
Grant ID: R01 DE013386





