| PMC full text: | Published online 2008 May 12. doi: 10.1093/nar/gkn249
|
Figure 7.
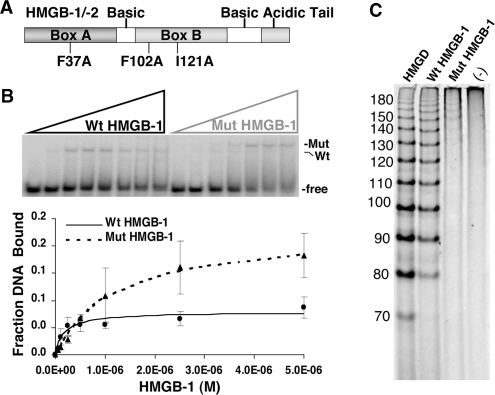
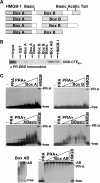
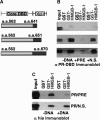
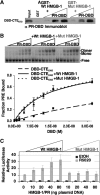
Functional effect of mutations in the DNA intercalating residues of HMGB-1. (A) Schematic of HMGB-1 domains. Mut-HMGB-1 contains alanine substitutions for the residues that intercalate in the DNA (Phe37, Phe102 and Ile121) as described previously (45,62). (B) Binding of wild-type (Wt) and mutant (Mut) HMGB-1 with PRE DNA as detected by EMSA. Increasing concentrations of WT HMGB-1 or Mut HMGB-1 proteins (0–5 µM) were incubated with a single concentration of [32P]-labeled PRE (0.6 nM) under conditions in the absence of competitor DNA that permit the detection of nonsequence-specific DNA binding. The distinct mobilities of Wt-HMGB-1 and Mut HGMB-1 complexes are indicated by the arrows. Free and shifted DNA bands were quantitated and graphed as a fraction of bound DNA. (C) DNA-bending properties of HMGB-1 Wt and Mut proteins as analyzed by circularization assay. HMGD, HMGB-1 Wt, HMGB-1 Mut (2 μM) or no protein was incubated with 10-mer linear duplex DNA in the presence of DNA ligase. Reactions were digested with exocnuclease III to eliminate linear DNA fragments. HMGD as a positive control was used to generate a ladder of known DNA circle sizes. Formation of DNA circles was detected by EMSA and cyber green staining (Invitrogen) as described in Materials and methods section.