Abstract
Free full text

Crystal engineering of HIV-1 reverse transcriptase for structure-based drug design
Associated Data
Abstract
HIV-1 reverse transcriptase (RT) is a primary target for anti-AIDS drugs. Structures of HIV-1 RT, usually determined at ~2.5–3.0 Å resolution, are important for understanding enzyme function and mechanisms of drug resistance in addition to being helpful in the design of RT inhibitors. Despite hundreds of attempts, it was not possible to obtain the structure of a complex of HIV-1 RT with TMC278, a nonnucleoside RT inhibitor (NNRTI) in advanced clinical trials. A systematic and iterative protein crystal engineering approach was developed to optimize RT for obtaining crystals in complexes with TMC278 and other NNRTIs that diffract X-rays to 1.8 Å resolution. Another form of engineered RT was optimized to produce a high-resolution apo-RT crystal form, reported here at 1.85 Å resolution, with a distinct RT conformation. Engineered RTs were mutagenized using a new, flexible and cost effective method called methylated overlap-extension ligation independent cloning. Our analysis suggests that reducing the solvent content, increasing lattice contacts, and stabilizing the internal low-energy conformations of RT are critical for the growth of crystals that diffract to high resolution. The new RTs enable rapid crystallization and yield high-resolution structures that are useful in designing/developing new anti-AIDS drugs.
INTRODUCTION
HIV-1 reverse transcriptase (RT) is the enzyme responsible for generating a double-stranded linear DNA from the single-stranded RNAs packaged in HIV-1 virions. Twelve of the 25 approved anti-AIDS drugs target RT (hivinsite.ucsf.edu, 2008) and are classified as either nucleoside/nucleotide RT inhibitors (NRTIs) or nonnucleoside RT inhibitors (NNRTIs). A high rate of viral replication combined with lack of efficient proofreading activities in both RT and human RNA polymerase II results in the rapid generation of mutant viruses (1). The generation of HIV-1 mutants in infected patients allows the virus to develop resistance to all of the available anti-AIDS drugs, sometimes within days to a few months of treatment (2). New anti-AIDS drugs should be designed to be effective against viruses that carry known resistance mutations.
Structural studies have been instrumental in developing the diarylpyrimidine (DAPY) class of NNRTIs, including TMC278/rilpivirine and TMC125/etravirine/Intelence, which effectively inhibit wild-type and drug-resistant HIV-1 viruses (3,4). The DAPY NNRTIs have strategic flexibility, allowing them to inhibit NNRTI-resistant viruses (5,6). Early attempts to crystallize the RT/TMC278 complex yielded crystals that failed to diffract beyond 6 Å resolution. The conformational flexibility of TMC278 may have introduced heterogeneity into the RT molecules in the crystal lattice (7), which might have been the primary cause of the persistently low resolution diffraction obtained in the many trials over a 5-year period. In an effort to restrict the conformations of RT in the crystal lattice and improve the diffraction quality, a systematic protein crystal engineering approach was taken to produce an RT that could give high-resolution crystal structures of the RT/TMC278 complex.
Three fundamental types of protein engineering approaches that are useful for crystallography include: (i) alterations that affect the suitability of the protein for biochemical study, including mutagenesis and the addition of tags for expression, solubility and purification; (ii) changes that increase the conformational homogeneity of the protein sample and (iii) modifications of the protein that directly alter interactions at crystal contact interfaces (8,9). Examples of these approaches include the addition and subsequent removal of purification tags; deletions of disordered regions including termini, loops and domains by recombinant techniques or limited proteolysis and replacement of highly entropic residues (e.g. lysines and glutamic acids) by the ‘surface entropy reduction’ method (10). Alterations of proteins to improve crystallization include the substitution of residues known to be involved in crystallization, systematic or random alteration of surface residues to create a library of potentially crystallizable proteins, and alteration of known crystal contacts that could potentially lead to new crystal forms.
HIV-1 RT is a heterodimer consisting of subunits with masses of 66 kDa (p66) and 51 kDa (p51). The two subunits, p66 and p51, consisting of 560 and 440 residues, respectively, are produced by cleavage of the Gag–Pol polyprotein precursor by HIV-1 protease. They share a common amino terminus. HIV-1 RT crystallizes with different space groups and unit cells, and the resulting crystals diffract X-rays to different resolution depending on the nature of the complex (e.g. ±nucleic acid, ±NNRTI, etc.) and of the HIV-1 RT itself. Three different versions of HIV-1 RT, varying in termini and HIV-1 strain sequence, have been used for crystallization of RT/NNRTI complexes. Each of the three versions crystallizes with a characteristic space group symmetry: P212121 (11); C2 (12,13) and C2221 (14).
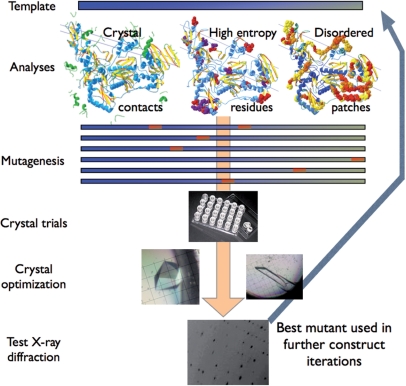
To produce crystals of HIV-1 RT/TMC278 complex that diffracted to high resolution, we used an iterative high-throughput approach involving multiple rounds of expression, purification and crystallization. In each round, the plasmid construct that produced crystals with the highest resolution of X-ray diffraction was used as the basis for the next round of mutagenesis. This iterative approach made it possible to develop HIV-1 RTs with better crystallographic characteristics (Figure 1).
MATERIAL AND METHODS
Expression vector and mutant construction
The HIV-1 RT-encoding DNA from the Q258C-RT construct (15) was ligation-independent cloned (LIC) into pCDF-2 Ek/LIC with the LIC Duet™ Minimal Adaptor (Novagen, San Diego, CA USA) according to manufacturer's recommendations. The HIV-1 RT-encoding dual expression vector is designated pRT1. Mutagenesis was completed using methylated overlap-extension ligation-independent cloning (MOE-LIC). See Figure 2A for the location and pairing of the primers on pRT1. The methylated and nonmethylated primers are listed in Supplementary Table 3.
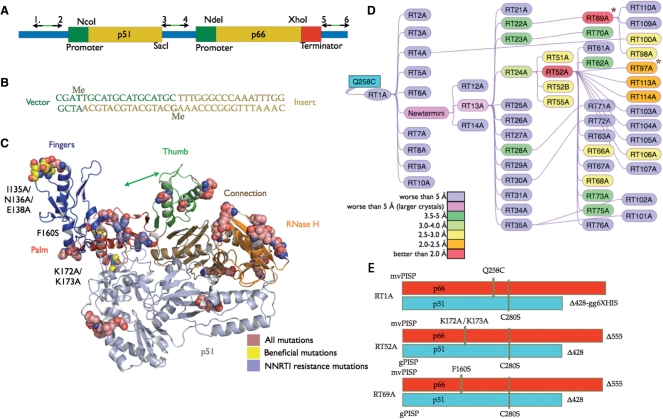
Mutagenesis of RT. (A) Schematic showing the three binding sites (arrows) of the 2′-O-methylated primers used in MOE-LIC. Locations of specific restriction enzyme cut sites are also indicated. (B) Annealed duplex of the primer terminated insert and vector with 2′-O-methyl nucleotides indicated by Me. (C) Cartoon of RT color-coded by the p66 subdomains. All mutations made in this study are indicated as spheres. The beneficial mutations are colored yellow and labeled. (D) Flowchart showing the generation of the mutants, which are color-coded to show X-ray diffraction resolution of the crystals. Stars denote those mutants for which the unliganded crystals show improved resolution. (E) Diagram of RT1A, RT52A and RT69A.
For mutagenizing ORF-2 (p66), mutagenesis overlap extension PCR was performed using mutated overlap segments with the 2′-O-methylated primers to amplify the full insert with PfuUltra™ II Fusion HS DNA Polymerase (Stratagene, La Jolla, CA USA). The vector, with p66 removed to minimize false positives, was amplified with complementary methylated primers in a separate reaction. The PCR products were then gel purified, and 0.04 pmols of vector and insert were mixed at a 1 : 1 molar ratio in a buffer containing 25 mM Tris pH 8.0, 5 mM MgCl2, 0.025 mg/ml BSA and 2.5 mM DTT in a 20 μl volume. The mixture was heated to 70°C and cooled slowly over 2 h in a water bath. Once cooled to ~40°C, 1 μl of 25 mM EDTA was added and the mixture incubated at room temperature for 5 min before being desalted using a Centri-Sep column (Princeton Separations, Adelphia, NJ USA) or by ethanol precipitation (16). Desalted annealed DNA of 5 μl was added to electrocompetent NovaBlue cells (Novagen) and electroporated according to manufacturer's recommendations.
Expression and purification of RT
pRT containing BL21-CodonPlus®-RIL cells were induced with 1 mM IPTG at an OD600 of 0.9 followed by expression at 37°C for 3 h. Ni-NTA purification was performed according to the manufacturer's recommendations (Qiagen, Valencia, CA USA) with the following modifications: no added lysozyme, 600 mM NaCl in each of the standard buffers, 0.1% Triton X-100 added to the lysate and wash buffers and a high-salt wash step performed with 1.2 M NaCl added to the standard wash buffer. After elution the HRV14 3C protease was added (1 : 100 ratio of protease : RT) and incubated at 4°C overnight. Mono Q was performed as described (17). The RT was buffer exchanged and concentrated to 20 mg/ml in 10 mM Tris pH 8.0 and 75 mM NaCl. The concentrated RT was aliquoted and stored at −80°C or placed at 4°C for immediate crystallization.
Crystallization
Inhibitors were dissolved in dimethyl sulfoxide (DMSO) prior to addition to protein sample. The RT was screened unliganded, with a 2.5-fold molar excess of NNRTI, or with a 5-fold molar excess of RNase H inhibitor (RNHI) using the hanging-drop vapor diffusion method. The inhibitor–mutant complex was incubated at room temperature for 10 min prior to addition of the crystallization solution. Depending on the number of samples being screened, EasyXtal DG-Tools (Qiagen) or Linbro Plates (Hampton Research, Aliso Viejo, NJ USA) crystallization trays were used for screening. Based on visually identified crystal hits, further optimization was used to obtain diffraction quality crystals. RT52A and RT69A crystals were produced in a matrix of 24 conditions from 9% to 12% PEG 8000, 50 mM imidazole pH 6.0–6.8, 10 mM spermine, 15 mM MgSO4 and 100 mM ammonium sulfate. All successful crystallization experiments were performed at 4°C.
Data collection and structure determination
Crystals of RT52A were flash-cooled by immersion into liquid nitrogen after treating the crystals for 2–10 s in a cryoprotective solution containing crystallization well solution plus 27% ethylene glycol and the inhibitor at the same concentration as in the hanging drop. Best results, in terms of sharper diffraction spots and signal to noise ratio (I/σ), were obtained by using MicroMounts (MiTeGen, Ithaca, NY USA) for mounting the crystals. Crystals were screened and diffraction datasets were collected at the Cornell High Energy Synchrotron Source (CHESS) F1 and A1 beamlines, National Synchrotron Light Source (NSLS) beamlines X25 and X29, and Advanced Photon Source (APS) at Argonne National Laboratory (ANL), SER-CAT beamline 19ID. The diffraction data were indexed, integrated, scaled and merged using HKL2000 (18). The resolution of the data was estimated using the last resolution shell values for completeness, R-merge and the ratio of I to σ(I). X-ray diffraction data for apo-RT69A was collected at CHESS using the above protocol.
The crystal structure of apo-RT69A was solved by molecular replacement using apo-RT structure [PDB ID 1DLO (19)] as the starting model. Cycles of model building guided by high-resolution structures of RT/TMC278 complex and an RT/RNase H-inhibitor complex (Himmel et al., personal communication), solvent modeling and refinement generated the final model of apo-RT69A structure that is refined at 1.85 Å resolution to Rwork and Rfree of 0.238 and 0.252, respectively. The atomic coordinates and structure factors are deposited in Protein Data Bank (PDB) with accession code 3DLK.
RT activity assays
The processivity assay used a DNA/DNA template-primer (20). The RNase H activity assay was performed as described (21).
Additional methods
Other experimental methods, including CD spectroscopy, dynamic light scattering and details of RT enzymatic activity assays are given in Supplementary Methods.
RESULTS
Coexpression and mutant cloning
A coexpression system was used that makes it possible to specify the exact sequence and termini of the two subunits independently (Figure 2A). In the initial coexpression construct, the encoded p51 subunit consisted of 428 residues plus a hexahistidine purification tag at the C-terminus (15,22,23). This construct also encodes the p66 Q258C mutant, which is used to cross-link nucleic acid to the modified RT for X-ray crystallographic studies. In Escherichia coli the HIV-1 RT coexpression plasmid pRT1 produces large amounts of HIV-1 RT (~40 mg/l) under standard conditions.
RT mutants were generated using a rapid, high yield and inexpensive mutagenesis system. Donahue et al. (16) described a ligation-independent cloning technique in which terminator primers are used to create 12–15 nt complementary overhangs on the insert and vector. The insert and vector are annealed and transformed into bacteria, thereby avoiding any post-PCR enzymatic steps. The terminating residue in the primer is a 2′-O-methylated nucleotide, which causes the thermostable polymerases Taq or Pfu to terminate DNA synthesis (Figure 2B). There are two major limitations with this technique: (i) the 2′-O-methylated primers cost ~$100 per pair for each insert and (ii) the site of 2′-O-methylation has an ~20% mutation rate.
We combined the terminator primer technique with overlap-extension mutagenesis (24) to develop a rapid mutagenesis protocol for HIV-1 RT called MOE-LIC (see Supplementary Movie). In the MOE-LIC approach, the terminators lie outside the open reading frame (ORF), which avoids problems that could arise from unwanted mutagenesis in the coding or regulatory regions. Overlap-extension PCR makes it possible to generate either novel or mutagenized inserts that can be cloned into the expression plasmid (24,25). For the coexpression system a total of three terminator primer pairs (Figure 2A) was required which cost approximately $300. The terminator primer pairs are complementary; there is no complementarity between pairs, which allows for efficient cloning and directional specificity. The technique is cost effective because the same terminator primers were used in all of the cloning. Error rates were found to be extremely low; we found one unintended mutation per 30 mutants produced (1 error in ~50 000 nt sequenced).
Mutagenesis and crystallization
A protein engineering strategy designed to improve the crystallization of HIV-1 RT was developed as follows: (i) disrupt or enhance known common crystal contacts in the existing crystal forms of HIV-1 RT; (ii) remove high B-factor patches, primarily the disordered termini seen in the parent C2 RT/NNRTI crystal form; (iii) reduce surface entropy by converting lysine and glutamic acid patches to alanine (10); (iv) choose amino acids to mutate based on the available information about multiple crystal forms of HIV-1 RT (e.g. sequence variations, different sets of crystal contacts, ordered/disordered regions, etc.); (v) avoid mutating conserved residues and (vi) use iterative rounds of mutagenesis/crystallization to improve the quality of X-ray diffraction (Figure 1). Figure 2C shows the location of the mutations that were made for the crystallization trials (see Supplementary Table 1 for a complete list of the 59 HIV-1 RT variants and the diffraction resolution of the crystals). For the initial crystal screening of each HIV-1 RT variant (Supplementary Table 2), 18 crystallization conditions were chosen from previously reported crystallographic studies of HIV-1 RT (14,17,26,27, Himmel, D.M). Crystallization of individual HIV-1 RT samples was attempted in parallel experiments with unliganded RT, RT complexed with TMC278, and in complexes with other NNRTIs or RNHIs [e.g. β-thujaplicinol (28)].
The first round of mutagenesis/crystallization produced RT1–RT10 and crystals of the resulting modified RT/TMC278 complexes that diffracted to worse than 10 Å resolution (Figure 2D). However, one HIV-1 RT mutant, in which p66 was terminated at residue 555, produced larger crystals than those terminated at residue 560. In the second round, we attempted to optimize the termini for both the p66 and p51 subunits. To avoid possible interference with optimal packing in the crystal lattice as well as to allow for additional packing arrangements, the disordered residues at the termini of both subunits, including purification tags, were removed prior to crystallization. The C-termini were truncated at residue 555 for p66 and 428 for p51 based on the knowledge of less-ordered regions at the termini in published RT crystal structures. Of the three constructs generated in round two, RT13A, which had an N-terminal HRV14 3C cleavable (His)6-tag, gave the highest yield of monodisperse protein [post-(His)6-tag cleavage], as measured by dynamic light scattering (data not shown), and larger crystals (but with no improvement in X-ray diffraction quality), suggesting this version as the best candidate to be used as the template for the next round of mutants.
RT13A was the template for the third round of mutagenesis, resulting in constructs RT21–RT35. The crystals of RT24A/TMC278 complex diffracted X-rays to 3.3 Å resolution, which was better resolution than had been obtained with any previous version of HIV-1 RT complexed with TMC278. The 3.3 Å diffraction dataset was anisotropic and produced multiple lattices in the diffraction patterns; we did not obtain a dataset suitable for structure determination. Up to this point, all HIV-1 RT versions we tested had the p66 Q258C mutation that was used for cross-linking HIV-1 RT to nucleic acid (15,22,23). In the fourth round of mutagenesis, we reverted residue 258 to glutamine to remove any unwanted chemical reactivity that might result from having a surface cysteine residue not cross-linked to nucleic acid.
New crystal form and high-resolution diffraction from RT52A/NNRTI crystals
RT52A (Figure 2E), which is the same as RT24A with the original glutamine at position 258, produced crystals within 1–3 days when complexed with TMC278 and other NNRTIs. The crystals of the RT52A/NNRTI complexes diffracted X-rays to high resolution (often better then 2.0 Å). The quality of the 1.8 Å RT52A/TMC278 structure (6) is evident from the electron density map of the inhibitor shown in Figure 3A. The RT52A/NNRTI complexes represent a new crystal form of HIV-1 RT. This new crystal form has preserved the symmetry of its parent crystal space group C2 but has distinctly different unit cell parameters and crystal contacts (Figure 3B–D). Tighter packing of RT52A molecules in the crystal is evident from a 14% decrease in solvent content and a 19% decrease in unit cell volume compared to NNRTI (Janssen-R129385) complexed with the form of HIV-1 RT we used previously (expression construct designated 1B1) (17). There are nearly twice as many residues involved in crystal packing (within 4.5 Å of each other), 194 residues in RT52A/TMC278 structure compared with 97 in the 1B1 RT/R129385 structure. The surface area involved in crystal contacts is increased from 1556 Å2 in the IB1 RT/R129385 structure to 2707 Å2 in the RT52A/TMC278 structure, calculated using the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html).
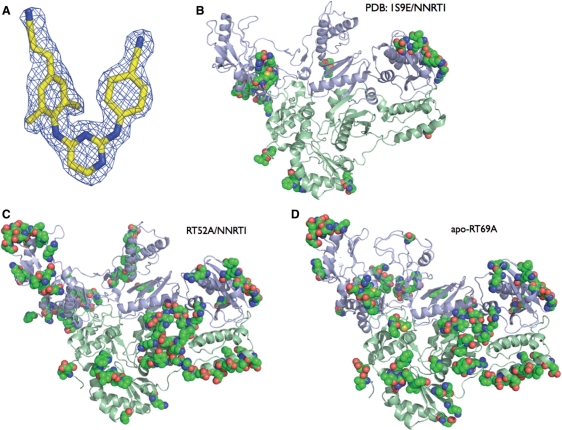
Crystal Structure of RT52A with TMC278 at 1.8 Å resolution. (A) Simulated annealed Fo–Fc omit map (3σ contours) for TMC278. (B) Typical 1B1-RT/NNRTI residues involved in crystal packing (pdb code: 1S9E). Residues involved in crystal contacts of HIV-1 RT are shown as space filled (residues within 4.5 Å of the asymmetric unit). (C) RT52A/TMC278 complex residues involved in crystal contacts. (D) Unliganded RT69A residues involved in crystal contacts.
Fragment screening with RT52A/TMC278 crystals
Drug fragment cocktail screening (29,30) is a potentially powerful technique for finding new inhibitors and new sites for inhibitors to bind, but this approach was difficult with the earlier, moderate resolution crystals of HIV-1 RT. Drug fragment cocktails are usually dissolved in DMSO and soaked into preformed crystals of the target protein in the crystallization solution plus DMSO. To determine if RT52A/TMC278 crystals could be used for fragment cocktail screening, crystals were soaked in 10–20% DMSO before and during cryoprotection. No loss in diffraction quality was found with 10% DMSO, and there was only a moderate decline in diffraction quality when 20% DMSO was used (2.0 versus 1.8 Å, data not shown). The DMSO-soaked crystals were also isomorphous to the original RT52A/TMC278 crystals. These results indicate that RT52A/TMC278 crystals are suitable for screening for binding of drug-like molecules and small chemical fragments and for lead optimization at both existing and novel-binding sites. The high-resolution HIV-1 RT crystals enable the acquisition of fast and reliable structures and will be critical in structure-based lead optimization.
Validation of RT52A and its derivatives
Comparison of the RT52A/TMC278 structure with 1B1 RT/NNRTI structures showed that the overall RT fold, distribution of secondary structure elements, and mode of NNRTI binding (6) are very similar, suggesting that the crystal engineering mutations had no significant impact on the structure of RT. To test for possible functional effects, proteins RT35A (RT52A without the K172A/K173A mutation), RT51A (RT52A + L100I/K103N), RT52A and RT55A (RT52A + K103N/Y181C) were assayed for DNA-dependent DNA polymerase activity and processivity and for RNase H activity (20,21). Supplementary Figure 2A shows that RT52A has processivity that is similar to wild-type HIV-1 RT (in this assay the wild-type HIV-1 RT was produced by coexpressing p66 with HIV-1 protease). RT51A has diminished processivity and RT55A an apparent increase in processivity. Each of the mutants has similar RNase H activity and specificity (Supplementary Figure 2B).
For 1B1-RT/NNRTI crystallization studies, only the p66 form of HIV-1 RT was expressed. A p51-like chain was produced via cleavage at residue 447 by a copurifying bacterial protease (Boyer, P.L.), ultimately yielding p66/p51 heterodimer (17). Altering RT52A at the C-terminus of p51 to produce a version that terminates at 447, instead of 428, changed the crystal unit cell to that seen with 1B1-RT complexed with NNRTIs, but with a significant reduction in X-ray diffraction resolution to 2.7 Å (Supplementary Table 1). Additional mutants were constructed to test the contribution of each of the changes in RT52A (Figure 2E), and each of the changes was found to be required for X-ray diffraction at high resolution (Supplementary Table 1).
Engineering of high-resolution apo-RT crystals
Multiple conformations of proteins in a crystal can limit the ability of the crystals to diffract X-rays to a high resolution, a problem that is particularly acute for a flexible protein like HIV-1 RT. Complexes of HIV-1 RT bound to inhibitors, antibodies, or substrates that may favor a single conformation or a subset of conformations have been used to reduce the flexibility of HIV-1 RT. While RT52A successfully produced crystals of RT/NNRTI complexes that diffracted to high resolution, the unliganded RT52A crystals diffracted to only ~3 Å resolution (Supplementary Table 1). The apo-form of 1B1 RT (19) crystallizes with different unit cell parameters than the 1B1 RT/NNRTI complexes (Table 1). The difference in the unit cell between unliganded 1B1 RT and the 1B1 RT/NNRTI crystals is a consequence of packing of two structurally distinct (thumb up versus down) conformations of RT. This may explain why RT52A, which was optimized to produce HIV-1 RT/NNRTI crystals diffracting to high resolution, failed to produce crystals that diffract to high-resolution when crystallized without an NNRTI. A different set of mutations may therefore be necessary to obtain a high-resolution apo-RT crystal form.
Table 1.
Comparison of engineered and nonengineered crystal forms
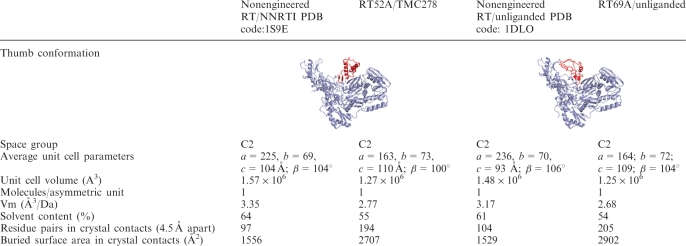 |
Subsequent rounds of mutagenesis focused on obtaining high-resolution crystals of apo-RT and HIV-1 RT complexes with RNHI-bound or DNA-bound RT. RT69A, which contains the mutation F160S, produced crystals of apo and RNHI-bound RT that diffracted X-rays to 1.8 Å resolution (Table 2). Crystals of unliganded RT69A contain a unit cell similar to the unit cell of NNRTI bound RT52A but quite distinct from other unliganded structures (Table 1). F160S is located adjacent to the binding cleft for nucleic acid and causes a 1.5 Å shift in Y115, which interacts directly with incoming nucleotides during polymerization. RT69A has wild-type levels of RNase H activity, indicating that the enzyme binds nucleic acid efficiently and that the RNase H active site is unaffected; however, RT69A has reduced processivity, indicating that there may be a reduction in polymerase activity or reduced ability to remain bound to the nucleic acid during DNA synthesis (Supplementary Figure 2C–D). Consequently, RT69A may not be the optimal form of HIV-1 RT for studies that involve nucleic acid or studies in which the region near the polymerase active site is important; however, it is suitable for structural studies of RT in complexes with RNHIs (Figure 4). RT97A, which contains the mutations P468T/N471D in addition to the mutations present in RT52A, also produced improved RNHI-containing crystals. RT97A forms apo-crystals that diffract X-rays to 2.1 Å resolution.
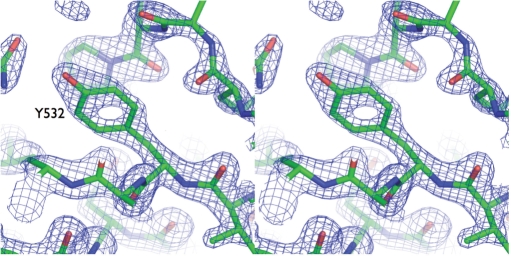
Stereo view of electron density in the RNase H domain of apo-RT69A. Stereo view of the 3Fo–2Fc map (calculated at 1.85 Å resolution and contoured at 2.5σ) surrounding Tyr532 in the RNase H domain of RT69A.
Table 2.
Diffraction data and refinement statistics
| Unliganded RT69A | |
|---|---|
| PDB ID | 3DLK |
| X-ray source | CHESS F1 |
| Wavelength (Å) | 0.9176 |
| Space group | C2 |
| Cell constants (a, b, c in Å; β in degrees) | 164.01, 72.04, 109.33; 104.38 |
| Resolution range (Å) (last shell) | 50–1.85 (1.88–1.85) |
| Number of unique reflections (number of observations) | 99 493 (257 025) |
| Completeness (%) (in last shell) | 94.5 (84.8) |
| R-merge (in last shell) | 0.074 (0.588) |
| Average I/σ(I) (in last shell) | 14.4 (1.9) |
| Sigma cut-off | |I| < −0.5σ(I) |
| Refinement statistics | |
    Total number of atoms (solvent atoms) Total number of atoms (solvent atoms) | 8051 (188) |
    Resolution (Å) Resolution (Å) | 40.0–1.85 |
    Number of reflections (Rfree set) Number of reflections (Rfree set) | 99 441 (2991) |
    Completeness (%) (minus Rfree set) Completeness (%) (minus Rfree set) | 94.3 (91.4) |
    Cutoff criteria Cutoff criteria | |F| < 0 |
    Rwork Rwork | 0.238 |
    Rfree Rfree | 0.252 |
| Root mean square deviations | |
    Bond lengths (Å) Bond lengths (Å) | 0.006 |
    Bond angles (degrees) Bond angles (degrees) | 1.313 |
An 1.85 Å structure of apo-RT
Crystal structure of apo-RT69A is refined at 1.85 Å resolution to an Rwork and Rfree of 0.238 and 0.252, respectively. Like the previously determined apo-RT structures [PDB ID: 1DLO (19), 1HNV (27)], apo-RT69A contains no NNRTI-binding pocket and has the thumb and fingers subdomains in a closed conformation. Superposition of the p51 subunit and the connection-RNase H domains of p66 for apo-RT69A, 1HNV and 1DLO structures results in a RMSD of 1.83 and 1.24 Å, respectively, with the major conformational differences in all three structures between the fingers and thumb subdomains (Supplementary Figure 4). Most of the apo-RT69A structure is well defined by high-resolution electron density (Figure 4). Like the high-resolution RT/NNRTI (TMC278) structure, the high-resolution apo-RT69A structure provides another distinct functional state of RT that can be used in designing new classes of inhibitors.
DISCUSSION
There is no generalized blueprint for determining the best way to crystallize a protein, and there is no simple protocol for improving the quality of diffraction of protein crystals. We were able to use protein engineering to improve the diffraction resolution of a very important HIV-1 drug complex from ~6 to 1.8 Å; this result has implications for the design of anti-AIDS drugs and also provides support for the idea that rational approaches can be used to enhance the diffraction quality of macromolecular crystals. Further mutagenesis showed that the terminal truncations primarily define the unit cell of the RT52A/NNRTI complexes and that the other mutations increase the resolution of the X-ray diffraction by stabilizing the crystallized conformation of RT (Figure 5 and Supplementary Table 1). The stabilization of a particular crystallized conformation within the confines of tighter crystal packing appears to be responsible for the improved diffraction (Figure 5). However, the improved resolution is not apparent from the thermal stability measurements by circular dichroism (Supplementary Figure 3).
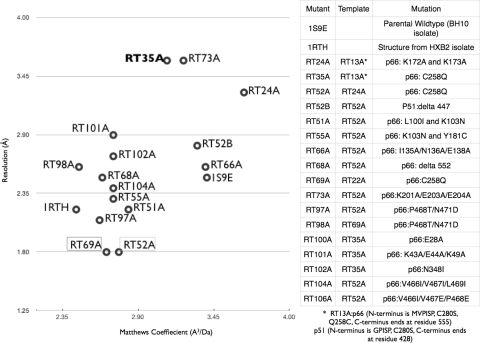
Comparison of unit cell and X-ray diffraction resolution of mutants. Plot of unit cell (Matthews coefficient) and X-ray diffraction resolution (Å) of the mutants that produced crystals that diffracted X-rays to better than 4 Å resolution. The legend of the table indicates the mutations and the parental template for each of the mutants. RT69A and RT97A are plotted based on crystals with RNHIs bound; all others were complexed with NNRTIs. RT35A is highlighted in bold and RT52A and RT69A are boxed for emphasis.
Comparison of X-ray diffraction resolution versus Matthews coefficients (A3/Da) of crystal structures of various HIV-1 RT forms indicates that there is an increase in resolution as solvent content (Matthews coefficients) decreases (Figure 5). The highest resolution diffraction previously seen was the 2.2 Å structure of RT/nevirapine (11), which was obtained by dehydrating the crystals (31). Apparently, dehydrating the crystals reduced the solvent content without fully optimizing the internal stability of the protein molecules in the dehydrated crystal lattice. We introduced mutations on the surface of HIV-1 RT that enhance the stability of RT molecules in the new crystal lattice. The addition of NNRTI-resistance mutations to RT52A/TMC78 crystals caused a decrease in X-ray diffraction resolution (2.9 Å for L100I/K103N and 2.1 Å for K103N/Y181C). The L100I/K103N double mutant increases the EC50 of TMC278 from 0.4 to ~8.0 nM while the K103N/Y181C double mutant's EC50 is 1.0 nM (3). The structures of RT/TMC278 complexes have revealed differences in the mode of binding for TMC278 with the NNRTI-resistance wild-type and mutant RTs (6). We have shown that the change in pocket conformation is accompanied by changes in the overall conformation of RT (5). Apparently, the L100I/K103N mutated RT52A, which has a large change in the pocket conformation and in the mode of TMC278 binding, is not as optimized in the crystal lattice as the RT52A/TMC278 structure, leading to significant drop in resolution. Because apo-RT and RT in a complex with NNRTI crystallize differently and make distinct crystal contacts, two distinct sets of mutations are required to optimize two distinct conformations of RT in the two different crystal forms (NNRTI-bound and apo).
Iterative protein engineering for crystallization can be applied to other proteins of interest, including other drug targets. Based on our results, the protein of interest should be modified to remove unstructured residues based on crystallographic and partial proteolysis results. Further mutagenesis, based on principles outlined in this article, can be used to improve crystal contacts and reduce the conformational flexibility of the protein. Although the effects of any one set of mutations are difficult to predict, taking a parallel approach that involves iterative steps can be used to improve the X-ray diffraction quality of the crystals.
We were successful in our initial goal of finding an HIV-1 RT mutant that gave diffraction quality crystals in a complex with TMC278 (6). This success, and in particular, the considerable decrease in the time it takes to grow good crystals, demonstrates the feasibility of high-throughput crystallization of HIV-1 RT in complexes with NNRTIs. In addition to improving the opportunities to develop more effective NNRTIs and RNHIs, the ability to produce high-resolution HIV-1 RT crystals quickly and easily should make it possible to use HIV-1 RT in fragment screening assays (29).
ACKNOWLEDGEMENTS
We acknowledge personnel at the Cornell High Energy Synchrotron Source (CHESS), Brookhaven National Laboratory (BNL), Advanced Photon Source Argonne National Laboratory (APS) and Liang Tong of Columbia University for support of data collection. Members of our laboratories provided valuable discussions and assistance, including Stefan Sarafianos, Chhaya Dharia, Chun Chu, Rajiv Bandwar, Thomas Acton, Sergio Martinez and Jason Schifano. E.A. is grateful to the National Institutes of Health (Grants AI 27690 MERIT Award and P01 GM 066671) for support of RT structural studies. S.H.H. was supported by the Intramural Research Program of National Institutes of Health, National Cancer Institute, Center for Cancer Research and National Institute of General Medical Sciences. Funding to pay Open Access publication charges for this article was provided by the NIH.
Conflict of Interest statement. None declared.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkn464
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2528191?pdf=render
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/abstract/36/15/5083
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/full/36/15/5083
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/reprint/36/15/5083.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/nar/gkn464
Article citations
Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J Med Chem, 66(9):6013-6024, 28 Apr 2023
Cited by: 2 articles | PMID: 37115705 | PMCID: PMC10184123
Targeting HIV-1 Reverse Transcriptase Using a Fragment-Based Approach.
Molecules, 28(7):3103, 30 Mar 2023
Cited by: 3 articles | PMID: 37049868 | PMCID: PMC10095864
Finding a chink in the armor: Update, limitations, and challenges toward successful antivirals against flaviviruses.
PLoS Negl Trop Dis, 16(4):e0010291, 28 Apr 2022
Cited by: 7 articles | PMID: 35482672 | PMCID: PMC9049358
Review Free full text in Europe PMC
HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J Med Chem, 64(22):16530-16540, 04 Nov 2021
Cited by: 2 articles | PMID: 34735153 | PMCID: PMC10655131
Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12(1):7127, 08 Dec 2021
Cited by: 5 articles | PMID: 34880240 | PMCID: PMC8654897
Go to all (60) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (4)
-
(3 citations)
PDBe - 1DLOView structure
-
(2 citations)
PDBe - 3DLKView structure
-
(2 citations)
PDBe - 1HNVView structure
-
(1 citation)
PDBe - 1S9EView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations.
Proc Natl Acad Sci U S A, 105(5):1466-1471, 29 Jan 2008
Cited by: 203 articles | PMID: 18230722 | PMCID: PMC2234167
Crystal structures of HIV-1 reverse transcriptase with etravirine (TMC125) and rilpivirine (TMC278): implications for drug design.
J Med Chem, 53(10):4295-4299, 01 May 2010
Cited by: 108 articles | PMID: 20438081
A comparison of the ability of rilpivirine (TMC278) and selected analogues to inhibit clinically relevant HIV-1 reverse transcriptase mutants.
Retrovirology, 9:99, 05 Dec 2012
Cited by: 19 articles | PMID: 23217210 | PMCID: PMC3549755
Looking for an active conformation of the future HIV type-1 non-nucleoside reverse transcriptase inhibitors.
Antivir Chem Chemother, 20(6):213-237, 11 Aug 2010
Cited by: 14 articles | PMID: 20710063
Review
Funding
Funders who supported this work.
Intramural NIH HHS
NIAID NIH HHS (2)
Grant ID: R37 AI027690
Grant ID: AI 27690
NIGMS NIH HHS (2)
Grant ID: P01 GM 066671
Grant ID: P01 GM066671