Abstract
Free full text

Signal-Dependent Protein Synthesis by Activated Platelets New Pathways to Altered Phenotype and Function
Abstract
New biologic activities of platelets continue to be discovered, indicating that concepts of platelet function in hemostasis, thrombosis, and inflammation require reconsideration as new paradigms evolve. Studies done over 3 decades ago demonstrated that mature circulating platelets have protein synthetic capacity, but it was thought to be low level and inconsequential. In contrast, recent discoveries demonstrate that platelets synthesize protein products with important biologic activities in a rapid and sustained fashion in response to cellular activation. This process, termed signal-dependent translation, uses a constitutive transcriptome and specialized pathways, and can alter platelet phenotype and functions in a fashion that can have clinical relevance. Signal-dependent translation and consequent protein synthesis are examples of a diverse group of posttranscriptural mechanisms in activated platelets that are now being revealed.
Introduction: New Paradigms at the Vascular Interface
Platelets are unique cellular effectors of hemostasis and thrombosis that act rapidly and in concert with coagulation proteins and with other vascular cells. The acute hemostatic functions of platelets are well known, have dominated the attention of the field for decades, and have been the foundation for discoveries that generated new molecular therapies.1,2 Rapid, immediate activation responses mediate platelet-dependent thrombosis in a variety of pathologic conditions,3 and pharmacological antiplatelet strategies are largely aimed at these events.4 Nevertheless, the focus on adhesion, aggregation, and secretion, and the view that platelets have a repertoire of activities primarily restricted to these acute processes, have also generated a central dogma that may inappropriately limit our view of their actions at the vascular interface and in other settings in health and disease.5 Clearly, our understanding of the molecular mechanisms by which platelets influence hemostasis, thrombosis, regulated and dysregulated inflammation, and neoplasia remains incomplete and continues to evolve. New paradigms are emerging as previously unrecognized pathways in platelets are identified, and unanticipated activities are characterized. In this regard, the current state of the field of platelet biology may be akin to that of endothelial cells several decades ago, when endothelium was thought by most investigators and physicians to have a limited range of responses; on the contrary, however, when this dogma was reexamined using new approaches that included primary culture of human endothelium,6 active participation of these cells in interactions with leukocytes7,8 and a variety of other previously unrecognized functions were discovered.9 If the comparison is accurate, new paradigms relevant to activities of platelets at the vascular interface are likely to be reported with some frequency.
Alternative and traditional views of selected features of platelet biology are listed in the Table. There is already considerable evidence for some of the alternative themes, such as inflammatory and immune activities of platelets,10–16 whereas others are less well explored and more speculative. The remainder of this review summarizes evidence for one such functional capability not generally recognized in platelets until recent discoveries revealed it: synthesis of new protein products in response to cellular activation (reviewed in references5,17). General dogma in the field has assumed that platelets have limited or no capacity for protein synthesis because they are anucleate and do not have nuclear transcriptional capability. Nevertheless, observations from a number of laboratories now demonstrate that physiologically relevant activation signals induce translation of proteins with important functions from constitutive or posttranscriptionally processed messenger RNAs (mRNAs) in human and murine platelets, a process that we have termed signal-dependent translation. These and other studies indicate that the platelet has intricate posttranscriptional mechanisms that allow it to alter its proteome, phenotype, and functions by accomplishing new protein synthesis in response to cellular activation. This capacity may allow platelets to modify the complex milieu of the vascular interface in ways that were previously unrecognized.
Table
New Biology of Platelets: Traditional Paradigms May Be Insufficient to Understand Platelet Activities at the Vascular Interface
| Traditional View | Alternate View |
|---|---|
| Platelets are biologically simple because they are anucleate and have a limited repertoire of responses. | Platelets have specializations and biologic activities that are novel and complex. Some activities are yet to be discovered. |
| Platelets do not express new gene products. | Platelets have diverse posttranscriptional mechanisms and use a transcriptome and specialized pathways to modify their proteome, phenotype, and functions. |
| Platelets are short-acting cells in clots and damaged tissue. | Platelets can be relatively long-lived and can mediate cell-cell interactions for many hours after initial adhesion, aggregation, and secretion. |
| Platelets operate exclusively in the intravascular compartment. | Platelets can influence critical events in the extravascular milieu in direct and indirect fashions. |
Mature Human Platelets Have Essential Components Required for Translation of mRNAs and Protein Synthesis
Human platelets freshly isolated from blood have ribosomes, polyribosome complexes (“polysomes”—the machinery on which mRNAs are translated and elongating polypeptide chains are assembled), and regulatory factors requisite for translational initiation and control.5,17 Ribosomes were identified in platelet preparations in early studies.18–21 Platelets from peripheral blood were also reported to contain polyribosomes in studies conducted 2 decades ago.21,22 More recently, modern approaches to profiling of mRNAs that are associated with polyribosomes and, therefore, are traditionally considered to be translationally competent23 have been applied to platelets to identify transcripts that are positioned for rapid, regulated translation in response to cellular activation.24 Furthermore, key ribosomal components, including ribosomal protein S6 and ribosomal P antigen, which are constituents of the 40S and 60S ribosomal subunits, respectively, are present in circulating platelets from normal subjects.17 These markers are consistent with ultrastructural evidence that ribosomes are present.
Using a cell-free system consisting of fractions from homogenates of resting human platelets, Booyse and Rafelson showed that platelet ribosomes are functional and that the lysate system mediated incorporation of radiolabeled amino acids into newly synthesized proteins.18 In parallel, these investigators and others reported that intact washed human platelets incorporate labeled amino acids into protein products.25–29 Rat platelets also incorporate exogenous amino acids into new protein.30 Puromycin, an inhibitor of translation—but not actinomycin D, an inhibitor of transcription—blocked new protein synthesis by isolated human platelets in these studies.25,27,31 This is a classic pattern indicative of posttranscriptional control. Essentially, all of the platelets isolated from normal subjects incorporated radiolabeled amino acids into new protein,28 demonstrating that this function is not a property of a subset of immature cells. Platelets from splenectomized subjects with idiopathic thrombocytopenic purpura had increased levels of amino acid incorporation into protein, indicating that the physiological state of the subject or the age and maturity of the platelets influence protein synthesis.21 Extracellular factors were reported to alter protein synthesis by human platelets under some conditions.31 This provided evidence suggesting that the synthetic mechanisms involved are regulated. Many of these early observations have been corroborated and extended in contemporary studies.5,17
The Platelet Transcriptome
Circulating human platelets have a substantial and diverse transcriptome, in addition to protein synthetic machinery. RNA-selective fluorescent dyes stain the entire population of platelets isolated from normal subjects, indicating the presence of RNA species transcribed by parent megakaryocytes.5,17 Messenger RNAs with 5′-methylguanosyl (m7G) caps and 3′ untranslated region polyadenylated tails are present, as are 18S and 28S ribosomal proteins, which are integral to the structure of ribosomes.17,32,33 Early experiments with intact platelets from normal subjects indicated that some of the mRNA transcripts are competent to serve as templates for proteins and have relatively long functional half lives that correlate with the lifespan of platelets in the circulation.26 This observation then lay fallow, for the most part, until the advent of reverse transcriptase polymerase chain reaction (RT-PCR) analysis and cDNA cloning methodologies.32,33 This infusion of new technology resulted in construction of cDNA libraries from platelet transcripts.33 Most recently, transcript profiling by microarray analysis and serial analysis of gene expression (SAGE) have been applied to platelets, identifying 1500 to 3000 unique transcripts in platelets from normal subjects, depending on the approach.24,34–39 Both cytoplasmic and mitochondrial transcripts are represented.35 There is substantial consistency between data generated by microarray analysis and SAGE,39 and in platelets isolated from different normal donors.17 As noted, transcriptional profiling has also been used to examine the complement of mRNAs associated with polyribosomes in platelets.24,34 Careful controls demonstrated that platelet transcripts were identified in profiling studies, and minimize or eliminate the question of confounding contributions from other blood cell types.40–42 Platelet transcripts have also been detected by Northern analysis and RNAse protection assays. These and other features of platelet mRNAs and their analytic assessment were recently reviewed.5 Integration of platelet genomic and proteomic information is now being accomplished.38
Remarkably, unspliced pre-mRNAs are included in the transcriptome of platelets from normal subjects.43,44 Equally unexpected, freshly isolated normal human platelets are invested by megakaryocytes with a functional spliceosome that can process pre-mRNAs to translationally competent mature transcripts.43,44 This provides a novel mechanism by which activated platelets can alter their transcriptome without new transcription, and contributes RNA templates for signal-dependent synthesis of specific proteins (see below).
The platelet transcriptome may vary in clinical conditions that involve enhanced thrombopoiesis, such as essential thrombocytosis.33 Young platelets from patients with increased thrombopoiesis secondary to immune thrombocytopenia, or recovery from stem cell transplantation, had increased expression of cyclooxygenase 2 (COX-2) protein, a marker of developing but not mature platelets, when compared with platelets from normal subjects.45 This may be explained by delivery of the COX-2 enzyme to juvenile and rapidly released platelets by parent megakaryocytes,45 but is also potentially attributable to sustained presence of COX-2 mRNA and its translation in circulating platelets under these conditions. There is additional evidence for variation in the platelet transcriptome under the influence of other diseases. Expression profiling was reported to reveal quantitative differences in several dozen mRNAs in platelets from subjects with ST-segment elevation myocardial infarction compared with mRNAs from subjects with stable coronary disease.46 As noted previously, translational capacity may also vary in platelet syndromes,21 in addition to the transcriptome.
Common and Specialized Elements in Platelet Translational Pathways and Transcripts
The initiation step in translation is rate-limiting and begins with interaction of the cap-binding complex of regulatory proteins termed eukaryotic initiation factor 4F to the 5′ cap structure of the mRNA transcript.47 This is a common and critical event that mediates linearization of the 5′ untranslated region (UTR), ribosomal association, and scanning of the leader sequence under basal and stimulated conditions of protein synthesis in cells of all lineages, although alternative mechanisms are used in a cell- and transcript-selective fashion in such conditions as stress and apoptosis.47 The cap-binding protein eIF4E, a central component of the eIF4F complex, is abundant in freshly isolated human platelets.17,34,48 Platelet activation redistributes eIF4E from the membrane skeleton and cytosol to the cytoskeletal core and facilitates its binding to mRNA transcripts, which are associated with the cytoskeleton and are physically separated from eIF4E under basal conditions. This spatial mechanism of translational control is regulated by engagement of platelet integrins.34 In platelets and other cells, activity of eIF4E is also regulated by phosphorylation via a p38 mitogen-activated protein (MAP) kinase-catalyzed pathway.17 Platelets, like other cell types, also have eIF2α,17,48 a critical regulator of binding of the initiator methionyl-tRNA complex to the 40S ribosomal subunit.47 Thus human platelets have key elements of a translation initiation pathway controlled by eukaryotic initiation factors that are common to a variety of cells. In addition, they have a specialized mechanism of regulation, involving redistribution of eIF4E mediated by cellular activation and integrins.
Unexpectedly, platelets also have a specialized pathway that controls translation of a subset of mRNAs with specific features in their 5′ UTRs. This signaling cascade is controlled by mammalian target of rapamycin (mTOR), a serine/threo-nine kinase of the phosphoinositide kinase–related kinase family that is an ortholog of TOR I and II, which have “master switch” roles in nutrient balance in yeast.49 mTOR was identified and characterized in mammalian cell types using the probe and exquisitely selective inhibitor rapamycin.49–51 Discovery of mTOR in platelets52 was unanticipated because its best-known activities in mammalian cells are as a regulator of cell cycle progression and cell growth: activities that are not features of the biology of mature, terminally differentiated platelets. Nevertheless, platelets isolated from the blood of normal subjects have mTOR and the distal components of the mTOR pathway, and the cascade is activated by outside-in signals delivered by receptors at the platelet surface.52,53 The components of the pathway downstream from mTOR are S6K1 (p70S6K) and eIF4E-binding protein 1 (4E-BP1) and related isoforms.49,50 S6K1 phosphorylates ribosomal protein S6, as do p85 and p90 ribosomal S6 kinases; each isoform is present in human platelets.5,53,54 4E-BP1 regulates binding of eIF4E to capped transcripts and is present in human platelets.52 mTOR and the downstream elements of the pathway are delivered to proplatelet tips in model megakaryocytes cultured from human CD34+ hematopoietic stem cells, consistent with their presence in mature platelets from blood.53 Phosphorylation of S6K1 and 4E-BP1 is a marker of mTOR activation.49,50,55 Phosphorylation of both proteins is induced by activation of human platelets with thrombin. These activation responses are selectively inhibited by rapamycin, indicating regulation by mTOR in this specialized cell type.52,53 In addition, the subcellular distribution of mTOR is spatially regulated in aggregated platelets.53
As in other cell types, rapamycin inhibits synthesis of a subset of protein products—not all—in activated human platelets,52 indicating that mTOR is a specialized, rather than a general, translational control pathway.49 Discrimination and differential translation of these mTOR-regulated mRNAs lie in sequence features of the 5′ UTR of the transcripts, which include complex secondary structure that requires significant free energy to unwind, as well as other sequence variables.49,56
In addition to specialized mechanisms depending on 5′ UTR sequence features, the 3′ UTR is also a key regulatory element in many mRNAs that code for proteins with critical biologic activity and can dramatically influence transcript half-life and translational efficiency.57,58 Recent SAGE analysis indicates that platelets are enriched in mRNAs with long 3′ UTRs, and that platelet transcripts have the longest 3′ UTRs of any cell or tissue analyzed in a large number that were chosen for examination.39 Human platelets contain RNA-binding proteins that recognize AU-rich mRNA decay elements (AREs) in the 3′ UTR of specific mRNAs and alter transcript stability and translational efficiency,59,60 including T-cell internal antigen-1 (TIA-I), TIA-I-related protein (TIA-R), and Hu protein R (HuR).17 Activated platelets also express an inducible form of cytoplasmic poly (A) binding protein,61 which, in other cell types, recognizes poly A sequences in the 3′ UTR and alters translation.17 Taken together, these observations indicate that the 3′ UTRs of platelet transcripts and specific RNA-binding proteins are likely to be key elements in the control of expression of specific gene products.
Multiple Proteins Are Synthesized by Activated Human Platelets
Although early studies indicated that platelets have protein synthetic capacity, the general concept in the field has been that it is low level, vestigial, and likely inconsequential. Several texts of hemostasis and platelet biology do not mention this function, and some commentaries conclude that platelets are simply incapable of any new protein synthesis.62 Consistent with the notion that platelets have low basal protein synthesis, little incorporation of the radiolabeled amino acid is detectable when freshly isolated human platelets are incubated with [35S] methionine under resting conditions in the absence of activation. However, when an activating signal is delivered to platelets incubated in parallel, multiple labeled proteins are synthesized when lysates and soluble fractions are analyzed by 1-dimensional or 2-dimensional gel electrophoresis5,52 (Lindemann S, Weyrich AS, Zimmerman GA, 2001). Some of these newly synthesized proteins have been identified and mechanisms of their signal-dependent translation determined24,43,44,52 (Lindemann S, Weyrich AS, Zimmerman GA, 2001). Extensive controls demonstrate that these proteins are products of activated platelets and not contaminating cells. Among these control and validation approaches, single-cell assays provide unambiguous evidence for new synthesis of key proteins by activated platelets using immunocytochemical analysis and confocal microscopy alone or in combination with in situ detection of the corresponding mRNAs.43,44,52,63
Signal-Dependent Translation of Bcl-3: An Index Example of Activation-Dependent Protein Synthesis by Activated Platelets
In studies of platelet signaling of altered gene expression in adherent human monocytes—a process by which platelets can influence new synthesis of proteins in inflammation and thrombosis60,64–66—control experiments demonstrated that isolated platelets, incubated in the absence of the leukocytes, synthesize B cell lymphoma-3 (Bcl-3) when activated by thrombin. A protooncogene member of the IκB family of transcriptional regulators, Bcl-3 had not previously been found in platelets. Subsequent analysis demonstrated that little or no Bcl-3 protein is present in freshly isolated human platelets, but, in contrast, that the Bcl-3 mRNA is present52 (Figure 1A). This pattern—in which there is a dramatic difference in abundance of mRNA and the corresponding protein product—indicates posttranscriptional regulation. In vitro translation of total platelet mRNA demonstrated that the Bcl-3 transcript is functional, and the incubations yielded a protein of expected size by immunoprecipitation.52 In parallel, activation of platelets with nanomolar concentrations of thrombin induced synthesis of Bcl-3 protein as detected by Western analysis (Figure 1A) and by metabolic labeling with radiolabeled methionine followed by immunoprecipitation of the protein product.52 In addition, immunocytochemical analysis demonstrated differential expression of Bcl-3 protein in activated, but not resting, platelets (Figure 1B and 1C) and that this is a response of virtually all platelets isolated from normal volunteers rather than a subset of circulating cells.52,63 New synthesis of Bcl-3 was inhibited by puromycin and cycloheximide but not by actinomycin D, indicating that translation—but not new transcription—is required. Together, these findings provided clear evidence for signal-dependent (that is, induced by activating signals) translation of Bcl-3 from mRNA that is transcribed in parent megakaryocytes but is repressed, or “silenced,” in circulating platelets under resting, basal conditions.52 Immunocytochemical detection of Bcl-3 in platelets in inflamed and thrombosed human vessels in surgical specimens (Figure 1D) provided in situ evidence that the experimental observations have physiological and clinical relevance.63 We subsequently found that collagen, platelet-activating factor, ADP, and epinephrine are also agonists for signal-dependent translation in platelets.5,24,63 Collagen was recently reported to induce Bcl-3 synthesis by platelets in experiments by other investigators.67 The time course of Bcl-3 synthesis in response to thrombin yielded additional important insights: newly synthesized Bcl-3 could be detected in activated platelets within 15 to 30 minutes in some experiments, consistent with translation of constitutively present mRNA without a requirement for new transcription.52 This feature is also consistent with the biology of platelets as rapid response cells. Nevertheless, synthesis of Bcl-3 is also prolonged over many hours,5,17,52 indicating that platelets may have important functions in thrombi and injured vessels well beyond the first few minutes of acute activation.
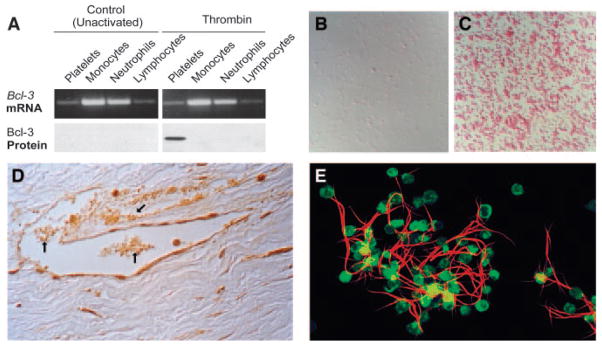
Bcl-3 is synthesized by activated human platelets and mediates fibrin clot retraction. A, Bcl-3 mRNA was present in resting unactivated platelets and in leukocyte subtypes when assayed by PCR (upper panels), but the protein was not detected by Western analysis (lower panel, left). When the cells in each suspension were activated with thrombin, Bcl-3 protein was synthesized by platelets but not by the other cell types (lower panel, right). B and C, Bcl-3 protein was absent from resting, unactivated platelets by immunocytochemical analysis (B) but was easily detected in platelets stimulated with thrombin (C). D, Bcl-3 was detected in aggregated and adherent platelets in inflamed and thrombosed vessels in human tissues (arrows), similar to its expression in activated platelets in vitro (C). E, Activated human platelets condense fluorescently-labeled fibrin strands in an in vitro model of clot retraction. Functional studies demonstrate that this is dependent on synthesis of Bcl-3 via activation of mTOR. A, B, and C are from reference 52 with permission of the publisher; panel D is from reference 63 with permission; panel E is from reference 53 with permission.
In initial studies, thrombin-stimulated expression of Bcl-3 appeared to be enhanced in aggregated platelets, compared with single cells in suspension that were not adherent to other platelets.52 Therefore, we examined the role of integrin αIIbβ3 and found that it regulates this synthetic response. Engagement of integrin αIIbβ3 by immobilized fibrinogen or an activating antibody induced synthesis of Bc1–3 in the absence of thrombin. Conversely, antibodies or peptides that block αIIbβ3 engagement and platelet aggregation, including small molecules and therapeutic antibodies in clinical use, blocked or attenuated Bcl-3 synthesis in response to thrombin.63 Bcl-3 synthesis was dramatically reduced in activated platelets from a patient with Glanzmann Thrombasthenia deficient in αIIbβ3. These studies indicated that intregin αIIbβ3 delivers outside-in signals to the pathway that mediates Bcl-3 synthesis, and provide direct evidence that signaling to translational control pathways and checkpoints is a mechanism of regulation of gene expression by integrin signaling.63 Integrins were previously thought to primarily, or exclusively, influence gene expression by signaling to transcriptional pathways.68 We also found that integrin engagement regulates spatial distribution of eIF4E and mRNA transcripts in activated platelets.34
The characteristics of signal-dependent synthesis of Bcl-3 in activated platelets suggest the possibility of a specialized pathway of translational control. We found that the 5′ UTR of the Bcl-3 transcript has complicated secondary structure (with predicted free energy required to “unwind” and linearize the 5′ UTR of approximately −76 kcal/mol) and other sequence features that made it a candidate for translational regulation by mTOR.52,56 We then examined the effect of rapamycin and found that it completely and selectively inhibited Bcl-3 synthesis in thrombin-stimulated platelets, and also inhibited phosphorylation of 4E-BP1 assayed as a marker of mTOR activation in parallel.52 Pharmacological inhibition of phosphatidylinositol-3-kinase, which lies upstream from mTOR in signaling cascades linking surface receptors to mTOR activation,49 also blocked both 4E-BP1 phosphorylation and Bcl-3 synthesis.52 Together, these studies demonstrated that synthesis of Bcl-3 is controlled by mTOR and provided evidence for a new and previously unrecognized activity of mTOR as a regulator of expression of specific protein products and phenotypic changes in terminally differentiated cells in response to signals delivered via G protein–coupled receptors and integrins.52 This observation in platelets contributed to a parallel set of discoveries demonstrating that mTOR has similar roles in myeloid leukocytes.69–71 The findings also suggest that inhibition of mTOR by rapamycin may have novel therapeutic effects on gene expression by platelets and leukocytes independent of inhibition of proliferation of other cell types when this agent is applied in antiangiogenic strategies and in “drug-eluting” vascular stents in the clinic.72
Although Bcl-3 provided an index example of specialized, signal-dependent translation of a protein product in activated platelets (Figure 2),5,17,34,52,63 the functional relevance of this event was not immediately obvious and was initially perplexing because the activity assigned to Bcl-3 at that time was as a transcriptional regulator.73 A clue lay in the domain structure of Bcl-3, which includes ankyrin repeats and proline-rich N and C termini, suggesting the possibility of multiple protein-protein interactions. Based on this information, we designed experiments to determine whether newly synthesized Bcl-3 interacts with other intracellular proteins. We found that Bcl-3 specifically binds to the tyrosine kinase Fyn via the Fyn SH2 domain in activated platelets and transfected COS cells.52 Bcl-3 also associates with the actin cytoskeleton in platelets.53
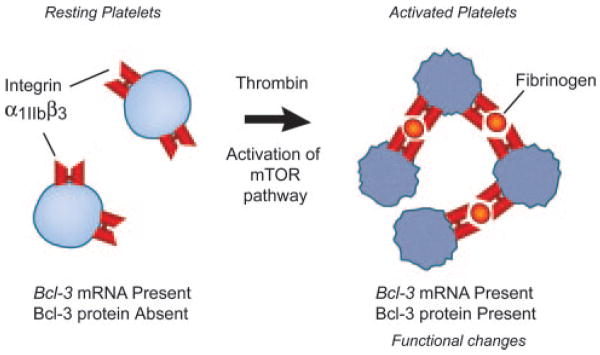
Synthesis of Bcl-3 is an index example of signal-dependent translation of key mRNAs and consequent synthesis of the corresponding proteins by activated human and murine platelets.
Because Fyn and related intracellular tyrosine kinases influence contractile responses of activated platelets,53,74 we examined the contributions of Bcl-3 and mTOR to fibrin clot retraction.53 Clot retraction is proposed to stabilize thrombi and to modify thrombus remodeling and resolution.53,74 It can be modeled in vitro, where activated platelets retract and condense fibrin strands in a fashion that can be examined macroscopically and microscopically (Figure 1E).53 In parallel loss-of-function and gain-of-function strategies, inhibition of mTOR activity in human platelets using rapamycin under conditions that block Bcl-3 synthesis52 inhibited clot retraction, whereas overexpression of Bcl-3 in a surrogate cell line that also expressed integrin αIIbβ3 enhanced fibrin retraction in a similar assay.53 These experiments provided evidence that signal-dependent translation of Bcl-3 under control of mTOR in activated human platelets alters fibrin retraction, a physiologically and clinically relevant functional response.53,75
To further examine its roles in clot retraction, we used platelets from mice with targeted deletion of Bcl-373 in an additional loss-of-function strategy. Initially, we found that little or no Bcl-3 is present in resting, unstimulated platelets from wild-type mice, but that its expression is induced by cellular activation with thrombin,53 recapitulating the pattern in human platelets. The protein was absent in thrombin-stimulated platelets from Bcl-3−/− animals, as expected, although platelet number was unaffected, and aggregation, upregulation of αIibβ3, and translocation of P-selectin were intact. In a clot retraction assay using exogenous fluorescently labeled fibrinogen similar to the assay used with human platelets (Figure 1E), Bcl-3−/− platelets exhibited a defect in fibrin retraction and stabilization that reproduced the one observed when mTOR activity was knocked out in human platelets with rapamycin.53 Thus, studies with platelets from genetically altered mice provided additional evidence that new synthesis of Bcl-3 by activated platelets mediates retraction and remodeling of fibrin clots, and that signal-dependent translation of protein products by activated platelets imparts physiologically relevant changes in cellular function and phenotype. Bcl-3–mediated fibrin retraction may alter the evolution and resolution of intravascular thrombi, and may also be important when platelets interact with deposited fibrin in the complex milieu of a wound, or at the surfaces of stents, shunts, or other intravascular devices.76,77
Biologic Advantages of Signal-Dependent Translation, and Potential Roles in Disease
Posttranscriptional regulation and, specifically, control at translational checkpoints provide biologic advantages in expression of key gene products. Translational regulation allows cells to rapidly synthesize proteins without new mRNA transcription, processing, or nuclear export. In addition, translational regulation provides mechanisms for precise control and “editing” of timing and magnitude of expression of proteins with potent biologic activities, and for spacial control of local protein synthesis in precise subcellular regions.58,78 Each of these variables is important in host defense including hemostasis, homeostatic inflammation, and wound repair.52,53,66,69–71 Therefore, specialized translational control offers functional advantages to key effector cells—such as platelets and leukocytes—that are critical for these processes. Nevertheless, there is evidence that dysregulation of posttranscriptional pathways and checkpoints is a mechanism of disease59 and thus may mediate thrombotic and inflammatory vascular syndromes.66
Activated Platelets Synthesize Additional Proteins Under Signal-Dependent Control
We have identified additional proteins of physiological and pathophysiologic relevance that are synthesized by platelets activated by outside-in signals. One is the pleiotropic proinflammatory factor interleukin-1β (IL-1β), providing a mechanism for rapid and sustained generation of this cell-associated and locally released cytokine in hemostatic and thrombotic responses.24 In earlier studies, several laboratories reported that activated platelets use IL-1 in intercellular signaling interactions,79–82 but the assumption was that it was preformed and stored in subcellular reservoirs in the resting state. A recent report concluded that IL-1β synthesis by platelet suspensions is attributable to contaminating leukocytes,83 but may not have fully appreciated controls in previous studies or single-cell assays that demonstrated IL-1β mRNA and the precursor pro–IL-1β protein in adherent, anucleate human platelets.43 The IL-1β transcript was also identified in platelets in an independent analysis.39 As a second example, tissue factor (TF), a critical procoagulant protein, is also synthesized by activated human platelets in a regulated fashion.44 The original observation has recently been independently confirmed.67,84 Of interest, IL-1β, TF, and Bcl-3 each have activities that can initiate, propagate, or stabilize a clot, or modify fibrin clot retraction. Thus their synthesis by activated platelets in a signal-dependent fashion may have evolved as a way to provide fine control of local hemostasis and thrombus remodeling at the vascular interface. Synthesis of IL-1β and TF by activated platelets is initiated by a novel pre-mRNA splicing mechanism (see next section).
Other proteins are also reported to be synthesized by activated human platelets, although in some cases the control mechanisms have not been explored. These include plasminogen activator inhibitor (PAI) 1, tetrahydrobiopterin, cyclo-oxygenase-1, and the SVCT2 ascorbate transporter.85–88 There is also evidence that the inducible isoform of nitric oxide synthase is produced in activated platelets, and that apoptotic proteins may be synthesized as platelets age (reviewed in reference5). As noted, experiments using radiolabeled amino acid incorporation and electrophoretic separation of newly synthesized proteins indicate that there are many other protein products, with preliminary identification of some of these in progress.5
Novel Pathways to Signal-Dependent Translation in Activated Human Platelets
Discovery of synthesis of Bcl-3 by activated platelets sparked a search for the identities of other protein products, yielding IL-1β and TF. It also led to the unexpected discovery that their synthesis is preceded by signal-dependent cytoplasmic splicing of IL-1β and TF pre-mRNAs, yielding mature transcripts that are translated into precursor (IL-1β) and active (TF) proteins.24,43,44 This identified a novel mechanism not previously recognized in activated mammalian cells.89 The splicing capacities of activated platelets are intricate and will be reviewed separately. Signal-dependent splicing, together with the mTOR-dependent translational control mechanism and other regulatory pathways discussed here, indicate that platelets have unexpected diversity in posttranscriptional control. Previous and ongoing studies add to this conclusion and suggest that platelets may also use ribosomal “stalling” or polypeptide termination,24 participation of micro RNAs (Denis MM, Trask B, Schwertz H, Weyrich AS, Zimmerman GA, 2004) and, potentially, other modes of control. Diverse mechanisms of this sort are now emerging as important regulators of cell- and locale-selective mRNA translation and protein expression.90,91 Thus the findings to date indicate that platelets have a rich and varied repertoire of posttranscriptional pathways that are only now being revealed.
Acknowledgments
The authors thank the many postdoctoral fellows, students, and collaborators who contributed to work cited here. We are also indebted to our technical staff for providing invaluable support. Sharren Brewer and Linn Steele prepared the manuscript and Diana Lim drafted the figures.
Sources of Funding
This work was supported by the National Institutes of Health and the American Heart Association.
Footnotes
Disclosures
None.
This article is part of a multi-part CME-certified activity titled Translational Therapeutics at the Platelet Vascular Interface. In order to achieve all of the activity’s learning objectives, please read all of the components of the activity listed in the Table of Contents and follow the “Instructions for Participation and Obtaining CME Credit” outlined prior to the Introduction.
References
Full text links
Read article at publisher's site: https://doi.org/10.1161/atvbaha.107.160218
Read article for free, from open access legal sources, via Unpaywall:
https://www.ahajournals.org/doi/pdf/10.1161/ATVBAHA.107.160218
Free to read at intl-atvb.ahajournals.org
http://intl-atvb.ahajournals.org/cgi/content/abstract/28/3/s17
Free after 12 months at intl-atvb.ahajournals.org
http://intl-atvb.ahajournals.org/cgi/content/full/28/3/s17
Free after 12 months at intl-atvb.ahajournals.org
http://intl-atvb.ahajournals.org/cgi/reprint/28/3/s17.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Bacteria-derived extracellular vesicles: endogenous roles, therapeutic potentials and their biomimetics for the treatment and prevention of sepsis.
Front Immunol, 15:1296061, 13 Feb 2024
Cited by: 5 articles | PMID: 38420121 | PMCID: PMC10899385
Review Free full text in Europe PMC
The Perspectives of Platelet Proteomics in Health and Disease.
Biomedicines, 12(3):585, 06 Mar 2024
Cited by: 0 articles | PMID: 38540198 | PMCID: PMC10968484
Review Free full text in Europe PMC
Integrating Phenotypic and Chemoproteomic Approaches to Identify Covalent Targets of Dietary Electrophiles in Platelets.
ACS Cent Sci, 10(2):344-357, 29 Jan 2024
Cited by: 1 article | PMID: 38435523 | PMCID: PMC10906253
Temperature Dependence of Platelet Metabolism.
Metabolites, 14(2):91, 26 Jan 2024
Cited by: 2 articles | PMID: 38392983 | PMCID: PMC10890334
Biochemical characterization of spleen tyrosine kinase (SYK) isoforms in platelets.
Platelets, 34(1):2249549, 01 Dec 2023
Cited by: 2 articles | PMID: 37661351
Go to all (121) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
[The synthesis of proteins in unnucleated blood platelets].
Postepy Hig Med Dosw (Online), 67:672-679, 23 Jul 2013
Cited by: 7 articles | PMID: 24018431
Review
Nucleotide-binding oligomerization domain 2 receptor is expressed in platelets and enhances platelet activation and thrombosis.
Circulation, 131(13):1160-1170, 17 Feb 2015
Cited by: 40 articles | PMID: 25825396 | PMCID: PMC4382913
mTOR-dependent synthesis of Bcl-3 controls the retraction of fibrin clots by activated human platelets.
Blood, 109(5):1975-1983, 16 Nov 2006
Cited by: 95 articles | PMID: 17110454 | PMCID: PMC1801071
NLRP3 regulates platelet integrin αIIbβ3 outside-in signaling, hemostasis and arterial thrombosis.
Haematologica, 103(9):1568-1576, 24 May 2018
Cited by: 54 articles | PMID: 29794149 | PMCID: PMC6119128
Funding
Funders who supported this work.
NHLBI NIH HHS (8)
Grant ID: R01 HL066277-07
Grant ID: R01 HL092746
Grant ID: R01 HL091754
Grant ID: R01 HL091754-01
Grant ID: R37 HL044525-19
Grant ID: R01 HL066277
Grant ID: R01 HL092746-01
Grant ID: R37 HL044525




