Abstract
Free full text

neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas
Abstract
In the mammalian pancreas, the endocrine cell types of the islets of Langerhans, including the α-, β-, δ-, and pancreatic polypeptide cells as well as the exocrine cells, derive from foregut endodermal progenitors. Recent genetic studies have identified a network of transcription factors, including Pdx1, Isl1, Pax4, Pax6, NeuroD, Nkx2.2, and Hlxb9, regulating the development of islet cells at different stages, but the molecular mechanisms controlling the specification of pancreatic endocrine precursors remain unknown. neurogenin3 (ngn3) is a member of a family of basic helix–loop–helix transcription factors that is involved in the determination of neural precursor cells in the neuroectoderm. ngn3 is expressed in discrete regions of the nervous system and in scattered cells in the embryonic pancreas. We show herein that ngn3-positive cells coexpress neither insulin nor glucagon, suggesting that ngn3 marks early precursors of pancreatic endocrine cells. Mice lacking ngn3 function fail to generate any pancreatic endocrine cells and die postnatally from diabetes. Expression of Isl1, Pax4, Pax6, and NeuroD is lost, and endocrine precursors are lacking in the mutant pancreatic epithelium. Thus, ngn3 is required for the specification of a common precursor for the four pancreatic endocrine cell types.
The pancreatic islets of Langerhans are composed of four different cell types, α-, β-, δ-, and pancreatic polypeptide (PP) cells, which produce the hormones glucagon, insulin, somatostatin, and PP, respectively. During embryogenesis, the pancreas develops by fusion of a dorsal and a ventral epithelial bud generated by evagination of the foregut. In the mouse, the dorsal bud appears at embryonic day 9.5 (E9.5) concomitantly with the first differentiated glucagon-producing α-cells (reviewed in ref. 1). The following day (E10.5), insulin-producing cells are detected. Cells that produce somatostatin and PP are first found at E15.5 and at birth, respectively. Recently, the generation of mice deficient for a number of pancreatic transcription factors, including the homeodomain proteins Pdx1 (2–4), Hlxb9 (5, 6), and Nkx2.2 (7), the LIM-homeodomain protein Isl1 (8), the paired and homeodomain proteins Pax4 (9) and Pax6 (10, 11), and the basic helix–loop–helix (bHLH) protein NeuroD (12), has provided important insights into the control of islet cell development. Although all these genes are involved either in pancreas morphogenesis or in pancreatic endocrine cell proliferation, survival, or differentiation, their mutation does not seem to affect the early steps of endocrine cell-type specification. For example, despite the fact that in Pdx1-deficient mice pancreas development is blocked at the bud stage, insulin- and glucagon-positive cells still differentiate (3, 4). Similarly, inactivation of Pax4 results in the absence of mature β-cells, but insulin-producing cells are present in the pancreatic epithelium at E10.5 (9). Indeed, although it is thought that the endocrine cells of the pancreas arise from a common progenitor (13), little is known about the molecular mechanisms that control endocrine lineage determination from pluripotent pancreatic progenitors.
Proteins of the bHLH family are important transcriptional regulators of cell fate determination in a number of cell types in both invertebrate and vertebrate species. For example, the proneural bHLH genes achaete-scute and atonal are required for the determination of neural precursors in Drosophila (reviewed in ref. 14). Similarly, in mouse, neurogenin1 (ngn1) and neurogenin2 (ngn2), two bHLH genes related to atonal, are essential for the determination of sensory lineages in the peripheral nervous system (15–17). We and others have recently cloned a third member of the neurogenin family called neurogenin3 (18–20) that is expressed in restricted domains of the developing central nervous system as well as in the embryonic pancreas (18, 20).
In this study, we show that ngn3 is expressed in pancreatic endocrine precursors. To gain insight into the function of ngn3, we have inactivated the ngn3 gene by homologous recombination in embryonic stem (ES) cells. Mice homozygous for the ngn3 null mutation develop diabetes and die 1–3 days after birth. No islet of Langerhans can be found in ngn3-deficient mice, and the four islet cell types as well as endocrine precursor cells are lacking at all stages. Taken together, these results indicate that ngn3 is required for endocrine fate determination in the developing mouse pancreas.
Materials and Methods
Targeted Disruption of the ngn3 Gene.
A PGK-neor cassette was flanked by 5′ and 3′ arms consisting of a 8.5-kilobase (kb) XbaI–XhoI fragment and a 3-kb KpnI–KpnI genomic fragment from the ngn3 locus (G.G. and F.G., unpublished data), respectively. The mutation introduced by this targeting vector is a deletion of 628 bp corresponding to 468 bp of amino acid coding sequence after amino acid 59, comprising the bHLH domain and 160 bp of 3′ untranslated sequence. The vector was electroporated in R1 ES cells (21), and the G418 selection was performed as described (17). Homologous recombination events were detected by Southern blotting of ES cell genomic DNA by using a 0.4-kb 3′ external probe generated from an ngn3 genomic clone by PCR by using primers 5′-GGTACCTCCTGATGCCAATG-3′ and 5′-AGGGAATAGGTAGGCAGTTT-3′. Independently targeted ES cell clones (n = 21) were obtained from the 87 clones analyzed. Two of them were used to generate chimeras by standard procedures. Germ-line transmission was obtained by crossing the chimeras with CD1 outbred females, and the mutant phenotype was analyzed on a mixed 129/Sv × CD1 genetic background. Genotyping was performed by PCR of genomic DNA obtained from embryonic yolk sac and postnatal tails by using the primers ngn3 5′ (5′-TCTCGCCTCTTCTGGCTTTC-3′) and ngn3 3′ (5′-CGGCAGATTTGAATGAGGGC-3′) to detect the wild-type allele and neo 5′ (5′-GCAGCGCATCGCCTTCTATC-3′) and ngn3 3′ to detect the mutant allele.
RNA in Situ Hybridization, Immunohistochemistry, and Terminal Deoxynucleotidyltransferase-Mediated UTP End Labeling (TUNEL).
Whole-mount and section RNA in situ hybridizations were performed as described (19, 22). The following cRNA probes were used: ngn3 (19), ngn3 5′ (transcribed from a 1.3-kb BamHI–XhoI genomic fragment; G.G. and F.G., unpublished data), NeuroD (17), Pax4 (kindly provided by P. Gruss, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany; ref. 9), p48 (kindly provided by P. Wellauer, Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland; ref. 23), and Prox-1 (transcribed from a 3-kb cDNA fragment kindly provided by C. Fode, IGBMC; F.G., unpublished data). Immunohistochemistry was performed as described (19). The following antibodies were used: rabbit anti-Pdx1 at 1:250 (kindly provided by C. Wright, Vanderbilt University School of Medicine, Nashville, TN; ref. 4), rabbit anti-Isl1 at 1:100 (kindly provided by H. Hedlund, University of Umea, Sweden; ref. 24), mouse anti-insulin at 1:1,000 (Sigma), mouse anti-glucagon at 1:2,000 (Sigma), rabbit anti-somatostatin at 1:200 (Dako), rabbit anti-PP at 1:600 (Dako), and mouse anti-Pax6 at 1:500 (Developmental Studies Hybridoma Bank, Iowa City; developed by A. Kawakami, National Children Hospital, Tokyo). The following secondary antibodies were used: Cy3 anti-rabbit and Cy3 anti-mouse (Jackson ImmunoResearch) as well as peroxidase anti-rabbit and anti-mouse (Vector Laboratories). The TUNEL experiment was performed as described (19).
Electron Microscopy.
Pancreatic tissues were fixed in 2.5% (vol/vol) glutaraldehyde and processed as described (25).
Results and Discussion
Cells expressing ngn3 at high levels are observed in the pancreas as early as E9.5 (Fig. (Fig.11a), when the first glucagon-expressing cells differentiate. Their number increases until E15.5 and decreases thereafter, such that no ngn3 labeled cells are found at birth or in adult pancreas (Fig. (Fig.11 b–f and data not shown). ngn3-expressing cells are either isolated or grouped in small clusters in the pancreatic epithelium at E10.5 (Fig. (Fig.11 b and e). Subsequently, ngn3+ cells are found within or adjacent to pancreatic ducts (Fig. (Fig.11 c and d), which are thought to produce endocrine precursors (26). The hypothesis that ngn3+ cells could represent islet cell precursors is further reinforced by the fact that they apparently do not coexpress glucagon or insulin (Fig. (Fig.11 e and f). Finally, no ngn3 transcripts were found in the exocrine tissue (Fig. (Fig.11 c and f), and no other neurogenin gene was found to be expressed in the embryonic pancreas.
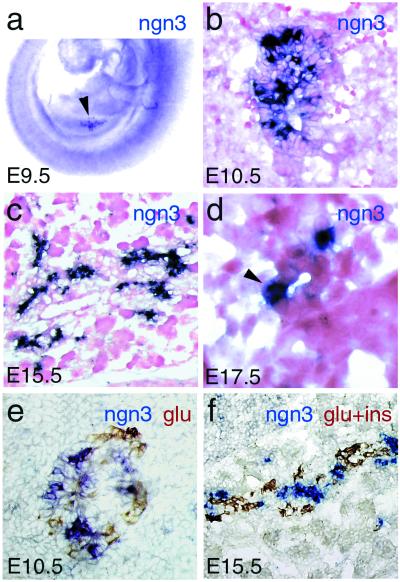
Expression of ngn3 during pancreas development. (a) Expression in the pancreatic bud (arrowhead) shown by whole-mount RNA in situ hybridization at E9.5. (b–f) Cryosections through the pancreas hybridized with an ngn3 riboprobe (dark blue) and counterstained with Safranin (b–d) or double-stained with anti-glucagon and anti-insulin antibodies (brown; e and f). ngn3 transcripts are found in cells either scattered or clustered in the pancreatic epithelium at E10.5 (b and e). A large number of ngn3 + cells are found associated with pancreatic ducts at E15.5 (c). (d) A high magnification of a transverse section through a duct at E17.5 containing an ngn3-expressing cell (arrowhead). ngn3+ cells are distinct from insulin- and glucagon-producing cells, suggesting that ngn3 is transiently expressed in endocrine precursor cells (e and f). However, we cannot completely rule out that a few cells strongly stained for ngn3 also produce glucagon or insulin. No ngn3 transcripts are detected in the exocrine tissue.
To investigate ngn3 function, the gene was deleted by homologous recombination in ES cells (Fig. (Fig.22 a–c). Animals heterozygous for a null allele of ngn3 are viable and fertile. Homozygote mutant animals are morphologically indistinguishable from their littermates and feed normally at birth, but 2 days later, they are reduced in weight and dehydrated (Fig. (Fig.22d). All of the mutants obtained died 2–3 days after birth. This phenotype is reminiscent of that of diabetic mice with mutations in NeuroD/BETA2 (12), Pdx1 (2, 4), Nkx2.2 (7), and Pax4 (9). Indeed, abnormally high glucose levels were found in the urine (from 10 to more than 20 g/liter, n = 40) and blood (334 ± 76 mg/dl, n = 15) of newborn ngn3 mutants, whereas heterozygotes and wild-type littermates had no detectable glucose in the urine and normal levels in the blood (76 ± 12 mg/dl, n = 14; 64 ± 9 mg/dl, n = 11, respectively). Inactivation of ngn3 thus results in diabetes. Although no gross morphological defects could be observed in ngn3 mutant pancreas at birth (Fig. (Fig.22e), histological analysis revealed that islets of Langerhans, which are easily observed in the pancreas of wild-type mice (Fig. (Fig.33a), are missing in mutant pancreas (Fig. (Fig.33a′).
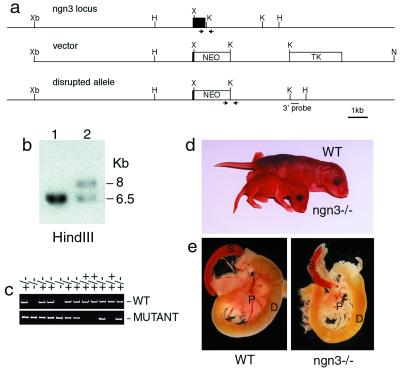
Targeted disruption of the ngn3 gene. (a) Structure of wild-type and targeted ngn3 locus. The ngn3 coding sequence (filled box) was deleted from amino acid 60 to the stop codon at amino acid 214, including the bHLH domain, and was replaced by a PGK-neo selection cassette (NEO). Restriction enzymes: H, HindIII; K, KpnI; N, NotI; X, XhoI; Xb, XbaI. (b) Genotype analysis by Southern blotting of two ES cell lines, one wild-type (lane 1) and one with a targeted ngn3 allele (lane 2), hybridized with a 0.4-kb 3′ external genomic fragment. (c) Genotyping by PCR of a litter from a heterozygous intercross. The two sets of primers used to identify the wild-type (WT) and mutant alleles (generating a 0.2- and a 0.7-kb PCR product, respectively) are indicated by arrows in a. (d) This image of two 3-day-old littermates, one wild-type (upper) and one ngn3-deficient (lower), shows the reduced size of the mutant animal. (e) Pancreas (p), spleen (s), and duodenum (d) of postnatal day 2 wild-type and ngn3 mutant mice. The mutant pancreas does not have any obvious morphological abnormality.
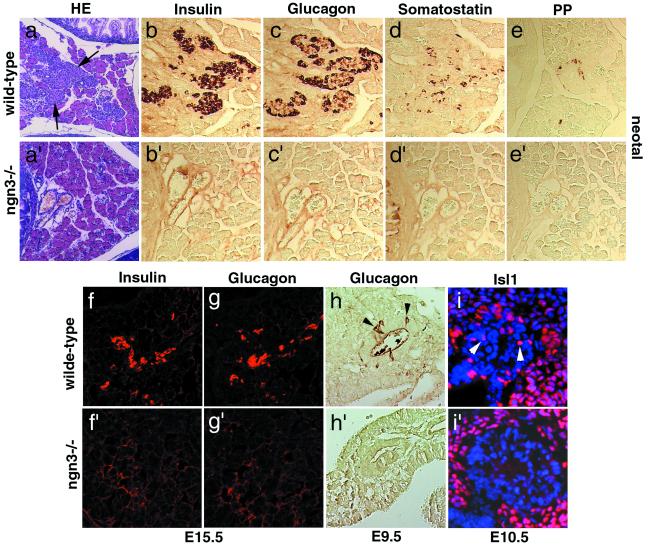
ngn3 is required for the differentiation of the four pancreatic endocrine cell lineages. Islets of Langerhans (arrows in a) and cells expressing insulin (b and f), glucagon (c, g, arrowheads in h), somatostatin (d), PP (e), and Isl1 (arrowheads in i) are observed in wild-type pancreas (a–h) but are completely missing in ngn3 mutant pancreas at birth (compare a–e with a′–e′) and also missing at early stages of differentiation: E15.5 (f, f′, g, and g′), E10.5 (i and i′), and E9.5 (h and h′). (a and a′) Hematoxylin-eosin (HE) staining.
To determine whether endocrine cells are present in the mutant pancreas at birth, even if not properly organized in islets, an immunocytochemical analysis was performed with antibodies against the four major pancreatic hormones. Cells expressing glucagon (α-cells), insulin (β-cells), somatostatin (δ-cells), and PP (PP cells) are found in wild-type pancreas (Fig. (Fig.33 b–e), whereas no hormone-containing cell could be detected in mutant mice (Fig. (Fig.33 b′–e′). ngn3 could be required for the survival of endocrine cells after they have differentiated or for the generation or differentiation of endocrine precursors. To distinguish between these possibilities, we analyzed mutant pancreas at embryonic stages (Fig. (Fig.33 f–i and f′–i′). In wild-type pancreas, we could detect glucagon as early as E9.5 (Fig. (Fig.33h) and both insulin and glucagon at E15.5 (Fig. (Fig.33 f and g), whereas neither glucagon-expressing nor insulin-expressing cells could be observed at these stages in mutant pancreas (Fig. (Fig.33f′–h′). Isl1 is a homeobox gene expressed in differentiating pancreatic endocrine cells that have left the cell cycle (8). The Isl1 protein is found in the pancreatic epithelium and in the mesenchyme surrounding the dorsal pancreatic bud at E10.5 in wild-type embryos (Fig. (Fig.33i). In contrast, Isl1 is absent from the mutant pancreatic epithelium, whereas it is maintained in the surrounding mesenchyme where ngn3 is not expressed (Fig. (Fig.33i′). Together, these results indicate that ngn3 is required for the development of endocrine pancreatic precursors at a stage that precedes their differentiation, in agreement with the observation that ngn3 is expressed only in cells that have not started to produce hormones (Fig. (Fig.11 e and f).
A number of regulatory genes expressed in endocrine cells of the embryonic pancreas have been identified recently, including the bHLH gene NeuroD/BETA2 (12), the paired and homeodomain genes Pax4 (9) and Pax6 (10, 11), and the homeodomain genes Pdx1 (3, 4, 27), Nkx6.1 (28), and Nkx2.2 (7). We studied the expression of these markers to determine whether, despite the lack of endocrine differentiation, endocrine precursors were present in ngn3 mutant pancreas. Pdx1, Nkx2.2, and Nkx6.1 are initially expressed throughout the pancreatic epithelium and are only later restricted to endocrine lineages (Fig. (Fig.44a; refs. 4, 7, 27, and 28). These genes were still expressed in ngn3 mutant pancreas at E10.5 (Fig. (Fig.44a′ and not shown); however, because of their broad early expression, it was not possible to conclude the fate of endocrine precursors. In contrast, expression of Pax4, Pax6, and NeuroD is restricted to endocrine cells in the pancreas from early stages onwards (Fig. (Fig.44 b–d; refs. 9–12). Expression of these genes is missing in ngn3-deficient mice (Fig. (Fig.44 b′–d′). In contrast, the specific expression of the bHLH gene p48 and of the homeobox gene Prox-1 in exocrine cell precursors (23, 29) is not affected in ngn3 mutants (Figs. (Figs.44 e and e′ and 5 c and c′), indicating that the exocrine cell lineage is correctly specified in the absence of ngn3.
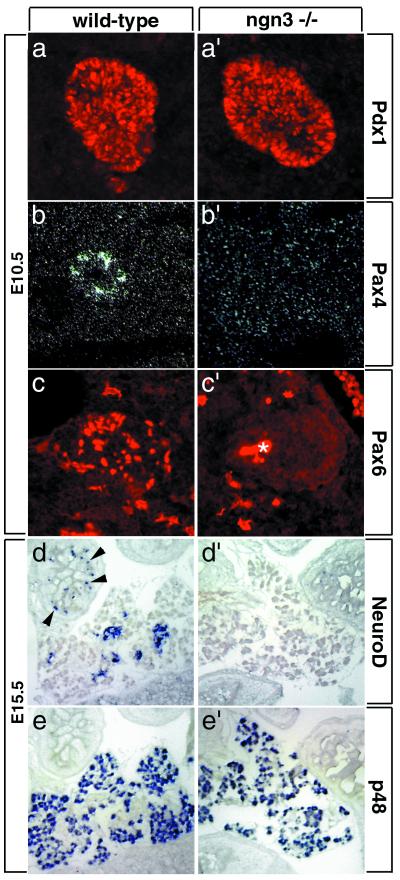
Lack of expression of several essential regulators of pancreatic development in ngn3 mutant mice. In wild-type pancreas, Pdx1 is initially expressed in endodermal progenitors in the pancreatic bud (a), whereas Pax4 (b), Pax6 (c), and NeuroD (d) are expressed in differentiating endocrine cells, and p48 (e) is expressed in the differentiating exocrine tissue. In ngn3-deficient mice, Pdx1 (a′) remains expressed in pancreatic progenitors, and p48 remains expressed in exocrine cells (e′), whereas expression of Pax4 (b′), Pax6 (c′), and NeuroD (d′) is completely missing, suggesting that ngn3 is essential for the specification or differentiation of endocrine precursors. An asterisk (*) marks the background in the lumen of the pancreatic bud also observed in control experiments. Expression of NeuroD in enteroendocrine cells of the gut (arrowheads in d) is also lost in ngn3 mutant mice (d′). (a–c and a′–c′) Pancreas at E10.5. (d–e and d′–e′) Pancreas at E15.5.
The loss of expression of NeuroD, Pax4, and Pax6 suggested that endocrine precursors are either misspecified or absent in ngn3 mutant pancreas. Alternatively, these genes could directly require ngn3 function for their expression. In particular, NeuroD is regulated by neurogenin genes in the nervous system (15–17), and loss of NeuroD expression in ngn3 mutant pancreas indeed suggests that an ngn-NeuroD cascade also operates during endocrine cell differentiation. Thus, to examine further the possibility that endocrine precursors are present in the mutant pancreas, even if not expressing appropriate markers, a 5′ ngn3 cRNA probe recognizing both wild-type and mutant transcripts (Fig. (Fig.55 a and a′) was used to mark these cells. No ngn3-expressing cell was found in ngn3-mutant pancreas at E10.5 (Fig. (Fig.55 b and b′), whereas, in a control experiment, the 5′ probe could detect the mutant transcript in spinal cord (Fig. (Fig.55a′). Although we cannot exclude the possibility that a few mutant cells have escaped detection, this result suggests that mutant endocrine precursors either are misspecified and have stopped expressing ngn3 or have been eliminated. To examine the latter possibility, we applied the TUNEL assay to E10.5 mutant pancreas. No increase in apoptosis was observed in mutant compared with wild-type pancreas (Fig. (Fig.55 d and d′), suggesting that the lack of endocrine precursors in the absence of ngn3 is not due to their elimination by apoptotic cell death but rather to their misspecification.
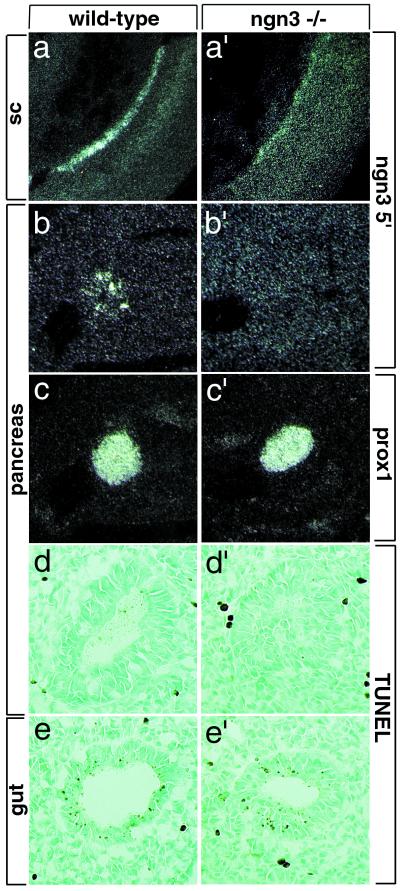
The pancreas of ngn3 mutants lacks endocrine precursors. An RNA probe recognizing both wild-type (a) and mutant (a′) ngn3 transcripts in the neural tube labels endocrine precursors in wild-type pancreas at E10.5 (b) but no cells in ngn3 mutant pancreas at the same stage (b′). Prox1 (29) transcripts mark the pancreatic epithelium in adjacent sections of the same embryos (c and c′). The absence of ngn3+ cells in mutant pancreas is unlikely to be due to their death, because no increase in apoptosis was detected in E10.5 mutant (d′) versus wild-type (d) pancreas. (d and d′) Gut cells undergoing apoptosis in the same sections shown in e and e′ are used as control for the TUNEL reaction. sc, spinal cord.
Although ngn3 is not expressed in the pancreatic exocrine tissue (Fig. (Fig.11 c and f), we observed an exocrine phenotype similar to that seen in NeuroD mutant pancreas (12), characterized by abnormal cellular polarity, with nuclei having random positions and an abundant accumulation of zymogen granules in acinar cells (Fig. (Fig.66 a and b). In the NeuroD/BETA2-deficient mice, the accumulation of secretory granules was ascribed to the loss of enteroendocrine cells producing secretin and cholecystokinin, two hormones responsible for the secretion of zymogen granules (12). It is thus likely that the defects in exocrine cells of ngn3-deficient mice are a consequence of the loss of NeuroD expression in enteroendocrine cells (see Fig. Fig.44d′).
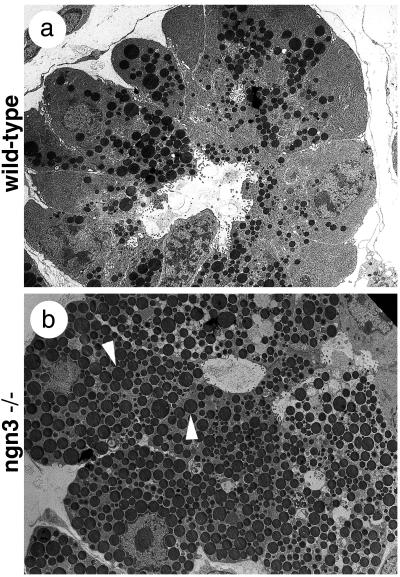
Pancreatic exocrine cells are affected by the loss of ngn3. Ultrastructural analysis of exocrine pancreas at postnatal day 1 shows an accumulation of secretory granules (arrowheads) in acinar cells in ngn3 mutant animals (b) and not in wild-type littermates (a).
In a recently reported ngn3 gain-of-function study (30), it was determined that overexpression of ngn3 in pancreatic progenitors under the Ipf1/Pdx1 promoter resulted in their premature differentiation into endocrine cells at the expense of pancreatic exocrine development (30). Conversely, we show herein that deletion of ngn3 results in the loss of all endocrine cell lineages as shown by a lack of pancreatic hormones and lack of expression of the early markers, Pax4, Pax6, NeuroD, and Isl1. The existence of a common genetic mechanism, i.e., a requirement for ngn3 function, in the generation of the four pancreatic endocrine cell types, suggests that they originate from a common endocrine precursor. The loss-of-function phenotype of ngn3 (this article), together with its gain-of-function phenotype (30), support a model in which ngn3 acts as a genetic switch that specifies an endocrine cell fate in pluripotent pancreatic progenitors, in a manner similar to the specification of a neural fate in neuroectodermal cells by neurogenin-related proneural genes (31). Another bHLH gene, p48, could specify the alternative exocrine fate (32). A second group of regulatory genes including NeuroD, Nkx2.2, Pax4, Pax6, and Isl1 would function downstream of ngn3 to regulate the proliferation, differentiation, and survival of precursors of the four endocrine cell lineages.
Acknowledgments
We thank Colette Hindelang for electron microscopy; Thomas Ding, Valérie Meyer, and Lan Chen for help with histology; Carol Fode for comments on the manuscript; Jean Luc Vonesh and Antoine Collet for help with microscopy and artwork; and Jacqueline Lee, Maike Sander, Ole Madsen, Jan Jensen, Palle Serup, Antonio Gonzalez, and Fred Lüdher for helpful discussions. This work was supported by a grant from the Association pour la Recherche sur le Cancer and funds from l'Institut National de la Recherche Scientifique, le Centre National de la Recherche Scientifique, and l'Hôpital Universitaire de Strasbourg.
Abbreviations
| PP | pancreatic polypeptide |
| En | embryonic day n |
| bHLH | basic helix–loop–helix |
| ES | embryonic stem |
| kb | kilobase |
| TUNEL | terminal deoxynucleotidyltransferase-mediated UTP end labeling |
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.97.4.1607
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC26482
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Generation and application of novel hES cell reporter lines for the differentiation and maturation of hPS cell-derived islet-like clusters.
Sci Rep, 14(1):19863, 27 Aug 2024
Cited by: 0 articles | PMID: 39191834 | PMCID: PMC11350089
Hippo pathway-mediated YAP1/TAZ inhibition is essential for proper pancreatic endocrine specification and differentiation.
Elife, 13:e84532, 25 Jul 2024
Cited by: 1 article | PMID: 39051998 | PMCID: PMC11272159
Comprehensive alpha, beta, and delta cell transcriptomics reveal an association of cellular aging with MHC class I upregulation.
Mol Metab, 87:101990, 14 Jul 2024
Cited by: 0 articles | PMID: 39009220 | PMCID: PMC11327396
The SWI/SNF chromatin remodelling complex regulates pancreatic endocrine cell expansion and differentiation in mice in vivo.
Diabetologia, 67(10):2275-2288, 03 Jul 2024
Cited by: 0 articles | PMID: 38958700
Targeting β-Cell Plasticity: A Promising Approach for Diabetes Treatment.
Curr Issues Mol Biol, 46(7):7621-7667, 18 Jul 2024
Cited by: 1 article | PMID: 39057094 | PMCID: PMC11275945
Review Free full text in Europe PMC
Go to all (935) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas.
EMBO J, 25(6):1344-1352, 02 Mar 2006
Cited by: 128 articles | PMID: 16511571 | PMCID: PMC1422151
Independent development of pancreatic alpha- and beta-cells from neurogenin3-expressing precursors: a role for the notch pathway in repression of premature differentiation.
Diabetes, 49(2):163-176, 01 Feb 2000
Cited by: 270 articles | PMID: 10868931
Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice.
Nat Genet, 23(1):71-75, 01 Sep 1999
Cited by: 206 articles | PMID: 10471502
Neurogenin3: a master regulator of pancreatic islet differentiation and regeneration.
Islets, 1(3):177-184, 01 Nov 2009
Cited by: 76 articles | PMID: 21099270
Review





